Abstract
The origin of the p53 superfamily predates animal evolution and first appears in unicellular Flagellates. Invertebrate p53 superfamily members appear to have a p63-like domain structure, which seems to be evolutionarily ancient. The radiation into p53, p63, and p73 proteins is a vertebrate invention. In invertebrate models amenable to genetic analysis p53 superfamily members mainly act in apoptosis regulation in response to genotoxic agents and do not have overt developmental functions. We summarize the literature on cnidarian and mollusc p53 superfamily members and focus on the function and regulation of Drosophila melanogaster and Caenorhabditis elegans p53 superfamily members in triggering apoptosis. Furthermore, we examine the emerging evidence showing that invertebrate p53 superfamily proteins also have functions unrelated to apoptosis, such as DNA repair, cell cycle checkpoint responses, compensatory proliferation, aging, autophagy, and innate immunity.
Invertebrate p53 has an ancient structure more reminiscent of vertebrate p63. It regulates apoptosis in response to genotoxic agents and also functions in DNA repair, compensatory proliferation, and innate immunity.
The vertebrate p53 family of proteins consists of three members, p53, p63, and p73. p53 has received considerable attention because of the fact that it is mutated in approximately 50% of all human cancers and plays an important role in protecting cells against DNA damage and cellular stressors. p63 and p73 on the other hand, seem to be less involved in tumorigenesis but play important roles in epithelial development and neurogenesis, respectively. p53 related sequences also exist in invertebrate species. We review the functional data on invertebrate p53 superfamily proteins, largely focusing on the model organisms, Caenorhabditis elegans and Drosophila melanogaster. Invertebrate p53 superfamily members act in apoptosis regulation in response to genotoxic agents and the deletion of invertebrate p53 superfamily proteins does not lead to overall developmental defects. Nevertheless, there is emerging evidence that invertebrate p53-like proteins also have functions unrelated to apoptosis.
There has been a debate whether invertebrate p53 superfamily proteins are phylogenetically more related to vertebrate p53 or p63. Taking advantage of recent genome sequencing projects, we analyze the phylogenetic relationships of the p53 superfamily from vertebrates and invertebrates. Consistent with previous reports, our phylogenetic analysis supports the conclusion that a p63-like domain structure is evolutionarily more ancient. It thus appears that a protein with a p63-like domain structure originally evolved, possibly to mediate apoptosis of damaged cells. In vertebrates, this earlier role of p53-like proteins is largely performed by p53. However, it appears that p63 has maintained the evolutionary ancient role of apoptosis in the female germline (Suh et al. 2006)
PHYLOGENETIC RELATIONSHIP OF INVERTEBRATE P53 FAMILY PROTEINS
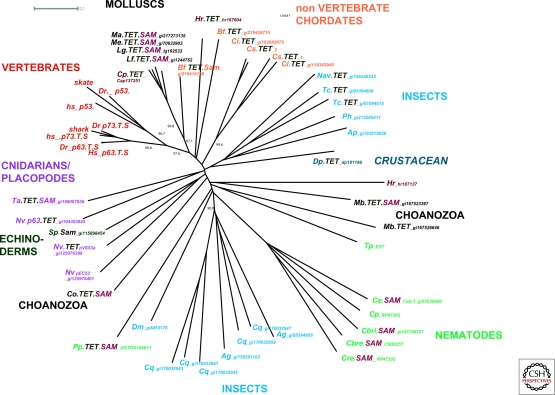
A host of recent genome sequencing projects allows for a comprehensive analysis of the evolutionary origins and phylogenetic relationships of the p53 superfamily, which includes all the sequences in vertebrates and invertebrates that are related to the vertebrate p53 protein (Fig 1). The most ancient of the p53 superfamily proteins can be found in Choanozoans, single celled organisms thought to have preceded animal evolution. Monosiga brevicollis (Mb) encodes two p53 superfamily members whereas Capsaspora owczarzaki contains one (Nedelcu and Tan 2007; King et al. 2008; Broad Institute, ongoing sequencing project). Choanozoa, together with animals, fungi, and Microsporidia, are part of the Opisthokonts, which is one of the eight major groups of eukaryotes (for review see Baldauf 2003). Using sensitive profile searches we could not find any p53 superfamily member within fungi or in any other group besides Opisthokonts. Within Choanozoans Capsaspora owczarzaki appears to be the more ancient species, preceding Choanoflagellates (Monosigia) and animals (Shalchian-Tabrizi et al. 2008).We could not confirm a previously reported Entamoeba histolytica p53 homolog (Mendoza et al. 2003), which would have indicated conservation of p53 extending to the group of the Discicristates. Thus, as family members of most of the eight major groups of eukaryotes, including plants, are sequenced it appears likely that the p53 superfamily members are only encoded in Opisthokonts. Within the Opisthokonts a p53 superfamily protein appears to have first arisen in Choanozoa but not in Fungi (data not shown). We expect that further p53 family members will be identified in the Broad Institute sequencing project focusing on species at the boundary between animals and fungi.
Figure 1.
Phylogenetic tree of the p53 superfamily. Sequences corresponding to the conserved DNA binding domain (Supplementary File 1) was used to generate an Unrooted Phylogenetic Tree using the Splits Tree 4 program (Huson and Bryant 2006). Full sequences and annotations can be found in Supplementary File 2. Abbreviations used are: Ag, Anopheles gambiae; Ap, Apisum; Bf, Branchiostoma floridae, (Amphioxus); Cbr, Caenorhabditis brenneri; Cbri, Caenorhabditis briggsae; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Co, Capsaspora owczarzaki, Cp, Capitella sp; Cq, Culex quinquefasciatus; Cre, Caenorhabditis remanei; Cs, Ciona savignyi; Dp, Daphina pulex; Dm, Drosophila melanogaster; Dr, Danio rerio (Zebrafish); Hr, Helobdella robusta; Hs, Homo sapiens; Lf, Loligo forbesi; Lg Lottia gigantea; Ma, Mya arenaria; Mb, Monosiga brevicollis; Me Mytilus edulis; Nav, Nasonia vitripennes; Nv, Nematostella vectensis; Ph, Pediculus humanus; Pp Pristionchus pacificus; shark (Elephant shark); Little skate (Leucoraja erinacea); Strongylocentrotus purpuratus; Ta, Trichoplax sp; Tc, Tribolium castaneum; Tp, Trichonella sp.
Within animals p53 superfamily sequences are encoded in almost all sequenced genomes and one or more p53 superfamily sequences are apparent in basal animals with radial symmetry, which include the cnidaria (corals, sea anemones, and jellyfish) and placozoa. The starlet sea anemone Nematostella vectensis (Nv) encodes for three p53 superfamily proteins. We also found three p53-like sequences in Hydra that appear to be incorrectly assembled so we have not included them in our phylogenetic analysis (not shown). The placozoa Trichoplax adherens (Ta) encodes for one p53-like protein. Trichoplax is representative of a basal eumetazoan lineage (all animal clades except sponges) that diverged before the separation of cnidarians and bilaterians (Srivastava et al. 2008).
At least one p53 family member is encoded in insects, nematodes, and the echinoderm, Strongylocentrotus purpuratus (Sp). The analysis of genomic data of multiple species within insects and nematodes shows that, at least within these groups, the p53 superfamily proteins appear to have rapidly evolved.
Within the superphylum Lophotrochozoa (annelids, leeches, and molluscs) most p53 superfamily members cluster together as expected from the phylogenetic relationship of these species. We found two p53 superfamily members in the leech, Helobdella robusta (Hr) and one p53 superfamily member each encoded in the annelid worm, Capitella (Cp) and in various molluscs including the cephalopod, Loligo forbesi (Lf), the bivalves, Mya arenaria (Ma) and Mytilus edulis (Me), and the gastropod, (snails and slugs) Lottia gigantea (Lg).
Analyzing the phylogenetic relationships of p53 superfamily members clearly supports previous literature that argue that all vertebrate p53, p63, and p73 proteins form a monophyletic group distinct from invertebrate p53 superfamily members (Fig. 1) (Nedelcu and Tan 2007; Pintus et al. 2007; Fernandes and Atchley 2008). This radiation of p53, p63, and p73 proteins seems to have occurred only in the vertebrate lineage as sequenced non vertebrate chordates such as the cephalochordate Florida lancelet, Brachistoma floridae (Bf) and the two urochordates, Ciona intestinalis (Cs, sea squirt) and Ciona savignyi (Cs) only contain two p53 superfamily members that don't cluster with the vertebrate proteins. It is likely that the radiation into three distinct vertebrate proteins occurred early in vertebrate evolution as we found evidence for distinct p53- and p63-like sequences in shark and ray EST databases. These genomes represent cartilaginous fish, which are considered to be the most basal group of vertebrates.
Based on sequence similarity, Drosophila (Dm), C. elegans (Ce), and Nematostella vectensis (Nv) p53 superfamily genes appear closer to the vertebrate p63 family than to the p53 family (Derry et al. 2001; Schumacher et al. 2001; Suh et al. 2006). This conclusion is further supported by the presence of a carboxy-terminal SAM domain in many invertebrate p53 superfamily proteins (Fig. 1). This protein interaction domain is found in all vertebrate p63-like proteins but is absent in p53-like proteins. Initially this domain was missed in the identification of the C. elegans p53 superfamily gene cep-1 (C. elegans p53-like) but became clear in structural studies on CEP-1 (Ou et al. 2007) and can now be detected in most nematode p53 superfamily genes using improved search algorithms. A SAM domain is also found in the carboxyl terminus of one of the two Monosiga brevicollis (Mb) superfamily genes and in the Capsaspora owczarzaki (Co) p53 superfamily gene. Gene predictions for the second Monosiga p53 superfamily gene are likely to be incomplete and might miss such a domain. Thus, given the occurrence of p53 superfamily genes with an amino-terminal tetramerization domain, a central DNA binding domain, as well as a carboxy-terminal SAM domain in Choanozoa and in early invertebrate lineages, it is likely that a p63-like protein structure is evolutionarily ancient and has been preserved in mollusc p53 superfamily sequences and in vertebrate p63. The absence of a SAM domain in other invertebrate species could be because of its loss in some lineages, such as in insects where p53 superfamily genes from all species we analysed did not contain a SAM domain, or not being identified because of incomplete gene predictions or significant divergence.
THE FUNCTION OF P53 SUPERFAMILY MEMBERS IN EARLY ANIMALS
Although we and others have been able to identify p53 superfamily proteins in the genomes of many animals and the choanoa (Fig. 1) (Nedelcu and Tan 2007; Pintus et al. 2007; Fernandes and Atchley 2008) data on the molecular characterization or function of most of these proteins are lacking. Apart from studies in the commonly used model organisms D. melanogaster and C. elegans, published data on p53-superfamily proteins is limited to the starlet sea anemone Nematostella vectensis and to various molluscs.
Within the Nematostella vectensis genome there are three p53 superfamily proteins (Nedelcu and Tan 2007; Pankow and Bamberger 2007). One of the proteins, Nvp63, appears to be required for apoptosis in early gametes following UV radiation as siRNA knockdown in adult N. vectensis polyps resulted in fewer apoptotic germ cells following treatment (Pankow and Bamberger 2007). Recombinant Nvp63 can bind to a consensus p53 element in vitro and Nvp63 transfection can induce a p53 transcriptional reporter in a UV dose dependent manner. Similar to human p63, removal of the last 34 amino acids in the carboxyl terminus of Nvp63 resulted in increased transcriptional activity showing that Nvp63 also contains a transactivation inhibitory domain (TID). Together these data show that Nvp63 acts to eliminate damaged germ cells. Currently there are no data published on the roles of NvpVS53 or NvpEC53. Originally NvpEC53 was reported to be more related to p53 superfamily proteins from Ecdysozoa, a group that includes insects and nematodes (Pankow and Bamberger 2007) but this conclusion is not supported by our phylogenetic analysis (Fig. 1).
p53 SUPERFAMILY ISOFORMS AND FUNCTION IN MOLLUSCS
p53 superfamily coding sequences have been published for six mollusc species, two in the soft-shell clam Mya arenaria (Kelley et al. 2001), five each for the mussels Mytilus edulis and Mytilus trossulus, including the only known deltaN isoforms in invertebrates (Muttray et al. 2005; Muttray et al. 2007), two in the Hawaiian bobtail squid Euprymna scolopes (Goodson et al. 2006), four in the surf clam Spisula solidissima (Jessen-Eller et al. 2002; Cox et al. 2003), and one in the Pacific oyster Crassostrea gigas (Farcy et al. 2008). Interestingly, whereas within some species a number of different proteins have been identified, it appears that all isoforms may arise from alternate splicing as regions that overlap between different proteins from the same species are 100% identical as seen in Mytilus trossulus and Mytilus edulis (Muttray et al. 2007), Mya arenaria (Kelley et al. 2001), Euprymna scolopes (Goodson et al. 2006), and possibly S. solidissima (Jessen-Eller et al. 2002).
At present only correlative evidence for functional roles of mollusc p53 superfamily proteins exist. In species in which developmental expression has been studied, such as S. solidissima (Jessen-Eller et al. 2002), M. arenaria (Kelley et al. 2001), and C. gigas (Farcy et al. 2008), it has been observed that different p53 superfamily isoforms are expressed at either different developmental stages or tissues, indicating that they may play different roles in normal development. A possible role for p53 superfamily proteins in developmentally controlled apoptosis has been reported in the Hawaiian bobtail squid Euprymna scolopes. This squid enters a symbiotic interaction with the marine luminescent bacterium Vibrio fischeri to form a light organ. Once the bacteria have colonised the light organ a number of symbiont induced developmental changes occur including the apoptosis of the cilliated epithelial cells of the light organ. Given that at least three p53 superfamily proteins are more abundant in the nuclei of light organs from symbiotic animals as compared with non symbiotic animals it was postulated that the accumulation of p53 superfamily proteins may result in the apoptosis of the cilliated epithelial cells (Goodson et al. 2006).
Evidence for a role for mollusc p53 superfamily proteins in tumourigenesis arises from studies in the mussels M. trossulus (bay mussel) and Mytilus edulis (blue mussel) and the soft-shell clam M. arenaria. Animals of all three species develop a leukaemic like disease of the heamocytes. Examination of leukaemic heamocytes show differential up-regulation of p53 superfamily isoforms in diseased compared with healthy cells in M. trossulus (Muttray et al. 2008) or accumulation of p53 superfamily proteins in the cytoplasm instead of the nucleus (as happens in healthy cells) in M. arenaria (Kelley et al. 2001).
p53 SUPERFAMILY PROTEIN FUNCTION IN DROSOPHILA MELANOGASTER
The sole p53 superfamily protein in D. melanogaster (Dmp53) was initially identified by three laboratories (Brodsky et al. 2000; Jin et al. 2000; Ollmann et al. 2000). Conservation with the mammalian p53 family is restricted to the DNA binding domain. In accordance with the conservation of the DNA binding domain, Dmp53 can bind to DNA containing p53 binding sequences (Jin et al. 2000; Ollmann et al. 2000) and transcribe a p53 reporter construct in Drosophila S2 cells (Brodsky et al. 2000; Jin et al. 2000). Yeast two-hybrid experiments indicate that Dmp53 can oligomerize and analytical ultracentrifugation and NMR shows that Dmp53 forms a tetramer (Ou et al. 2007). The ability to oligomerize is important to Dmp53 function as mutation of conserved residues important for oligomerization of human p53 results in a dominant negative Dmp53 protein (Brodsky et al. 2000; Jin et al. 2000; Ollmann et al. 2000).
Experiments performed with the dominant negative Dmp53 and Dmp53 mutants indicated that mutants are largely defective in inducing apoptosis following DNA damage, whether it be because of irradiation (Brodsky et al. 2000; Ollmann et al. 2000; Lee et al. 2003; Sogame et al. 2003), telomere loss (Titen and Golic 2008), or under- or re-replication stress in mitotic cells (LaRocque et al. 2007; Mehrotra et al. 2008). Flies lacking Dmp53 are viable (Lee et al. 2003; Sogame et al. 2003) and do not have an overt developmental defect, besides a modest reduction in the apoptosis of primordial germ cells (Yamada et al. 2008) possibly because of a requirement for p53 in regulating hid expression in these cells (Maezawa et al. 2009). Surprisingly, Dmp53 null flies still undergo cell cycle arrest following irradiation. This contrasts with vertebrate p53, which is required for both cell cycle arrest and apoptosis following DNA damage, but is similar to C. elegans CEP-1, which also only seems to play a role in apoptosis induction following ionizing irradiation (but is required for both cell cycle arrest and apoptosis following UV irradiation, see later).
Apoptosis in Drosophila requires at least five activator genes, reaper, grim, hid, sickle and jafrac2, known collectively as the RHG genes (Hay and Guo 2006). These proteins act as direct negative regulators of the Drosophila inhibitor of apoptosis (DIAP) protein. DIAP inhibits apoptosis by directly binding to and inducing the degradation of caspases needed for apoptosis induction (Hay and Guo 2006). reaper is one known target of Dmp53 in apoptosis induction. In its promoter there is a 150 bp radiation responsive cis-element that contains a Dmp53 binding site (Brodsky et al. 2000). Dmp53 null flies fail to activate a reporter containing this enhancer element (Sogame et al. 2003) and also fail to up-regulate reaper mRNA following irradiation (Lee et al. 2003; Sogame et al. 2003). Although reaper appears to be the main Dmp53 target following irradiation (Peterson et al. 2002) other DIAP inhibitors such as sickle and hid are also dependent on Dmp53 (Sogame et al. 2003; Akdemir et al. 2007) implying that they may also play a role. Furthermore, other proteins are likely to contribute to Dmp53 induced apoptosis as various studies have identified a small set of genes that are IR responsive and Dmp53 dependent (Brodsky et al. 2004; Akdemir et al. 2007). Interestingly, these studies hint that Dmp53 may also play a role in DNA repair as the Ku70, Ku80, Mre11, and Rad50 DNA double strand break repair genes have been identified as likely Dmp53 targets (Brodsky et al. 2004; Akdemir et al. 2007) and see later). Transcriptional studies also highlight a possible role for Dmp53 in regulating constitutive gene expression. Approximately 880 genes were consistently altered when non treated Dmp53 null and wild type flies were compared (Akdemir et al. 2007).
Another downstream pathway which appears important for apoptosis induction by Dmp53 is the Hippo pathway. Hippo (Hpo) is a MST (Mammalian Sterile Twenty-like) kinase orthologue which is activated by phosphorylation in a Dmp53 dependent manner (Colombani et al. 2006). In the absence of a functional Hippo pathway irradiation dependent apoptosis was greatly reduced, indicating that in addition to the activities of the RHG proteins the Hippo pathway is also required for proper apoptosis induction.
Dmp53 also seems to play non apoptotic roles in certain tissues. In pupal retinas Dmp53 actually protects cells from undergoing apoptosis in response to UV irradiation, possibly through activating a DNA repair-only response (Jassim et al. 2003), which relates to the finding that Dmp53 may transcribe DNA repair genes discussed earlier. Other evidence for a role for Dmp53 in DNA repair comes from the observation, that unlike wildtype flies, which show an increase in histone H3 acetylation on K9 and K14 following UV treatment, Dmp53 null flies fail to induce K9 acetylation and have lower levels of basal K14 acetylation of histone H3 (Rebollar et al. 2006). Since histone modification can facilitate DNA repair, Dmp53, like human p53 (Allison and Milner 2003; Rubbi and Milner 2003), may affect DNA repair by regulating chromatin modification. Dmp53 also seems to regulate the cell cycle downstream of SNF4 (the AMPK homolog) in response to low ATP levels (Mandal et al. 2005). In addition, Dmp53 is required for compensatory proliferation of ‘undead’ cells (in which apoptosis is initiated but prevented by the expression of the caspase inhibitor p35). Dronc, via a non apoptotic mechanism, induces the expression of Dmp53 mRNA resulting in a positive feedback loop consisting of Dronc, hid, and reaper which results in compensatory proliferation (Wells et al. 2006). These other roles of Dmp53 may help to explain why Dmp53 null flies are more sensitive to DNA damage, they either lack the ability to properly repair the damage sustained and/or fail to recover the lost or damaged cells by compensatory proliferation.
Dmp53 also seems to be an important mediator of lifespan. Dmp53 null flies or flies expressing the dominant negative form in the muscle or fat body have shortened lifespans, whereas flies expressing the dominant negative Dmp53 in neuronal cells have longer lives (Bauer et al. 2005). Further studies have shown that expression of dominant negative Dmp53 solely in the fourteen insulin producing cells of the brain is sufficient for this observed increase in lifespan and is thought to act by inhibiting insulin signaling (Bauer et al. 2007). Because the dominant negative protein prevents the transcription of Dmp53 targets but is presumably still capable of carrying out possible nontranscriptional roles, it is unclear which aspect of Dmp53 function is important in mediating this increase in lifespan or even if it is not because of a neomorphic role of the overexpressed dominant negative mutant.
Like the mammalian p53 family, Drosophila p53 plays an important role in decisions as to whether to live or die and therefore must be kept under strict control. So what are the mechanisms by which Dmp53 is held in check or activated when necessary? Chk2 is necessary for the apoptotic response in Drosophila (Peters et al. 2002; Brodsky et al. 2004). Following DNA damage Dmp53 is phosphorylated in a Chk2 dependent manner and this seems to regulate the transcriptional activity of Dmp53 (Brodsky et al. 2004). Human p53 is phosphorylated by ATM, Chk1, and Chk2 on serines 15 and 37. Mutation of serines in the homologous region of Dmp53 showed that serine 4 appears to be important in the up-regulation of Dmp53 activity by Chk2 (Peters et al. 2002). However, whether Chk2 phosphorylates Dmp53 directly or through an intermediatory kinase is unknown. Interestingly, unlike mammalian p53, Chk1 does not appear to have a role in regulating Dmp53 (Peters et al. 2002; Brodsky et al. 2004).
Other posttranslation modifications of Dmp53 include sumoylation of lysines 26 and 302. Mutation of both lysines prevents sumoylation of Dmp53 and results in reduced transcriptional activity in cells with reporter constructs containing a p53 binding site or the cis-regulatory element from the reaper locus. Sumoylation appears to be important for the transcriptional activation of Dmp53 and apoptosis induction as the expression of nonsumoylatable Dmp53 in contrast to wild-type Dmp53 could not rescue the apoptosis induction defect of Dmp53 null flies (Mauri et al. 2008). Although these findings show that importance of Dmp53 sumoylation, it is unclear what regulates the sumoylation of Dmp53.
Dmp53 activity is also controlled by direct binding of regulatory proteins. For example, DLP is the homolog of Daxx, a known transcriptional and cell signaling regulator and regulator of mammalian p53 (Gostissa et al. 2004; Zhao et al. 2004). DLP binds to the carboxyl terminus of Dmp53 and can act to repress Dmp53 mediated responses (Bodai et al. 2007), however, it is not clear whether it does so by regulating Dmp53 transcription or through some other mechanism.
The identification of Drosophila p53 led to both expected and unexpected results. As expected it is required for apoptosis following DNA damage and other cellular stressors but surprisingly does not play a role in cell cycle arrest. It is also becoming increasingly clear that Dmp53 plays important non apoptotic roles including DNA repair, compensatory proliferation, and longevity.
p53 SUPERFAMILY PROTEIN FUNCTION IN C. ELEGANS
CEP-1 is the sole p53 superfamily member in C. elegans. Conservation was initially only evident in the DNA binding domain (Derry et al. 2001; Schumacher et al. 2001), however, subsequent biochemical and bioinformatic analysis has identified an oligomerization and a SAM domain (Ou et al. 2007). Interestingly, unlike mammalian p53 family members or Drosophila p53, CEP-1 only forms a dimer in solution. It is possible that a tetramer may form on target promoters, however, the presence of only a half p53 consensus binding site at the egl-1 promoter (see later) argues that CEP-1 may indeed function as a dimer (Ou et al. 2007). Despite these differences in oligomerization, CEP-1 can induce transcription from a reporter containing p53 binding sites (Schumacher et al. 2001) and can bind the p53 target site in the p21 promoter (Huyen et al. 2004), indicating that it is functionally similar to p53. Despite high binding specificities toward human p53 consensus elements, structural data indicate that CEP-1 may have several important differences in its three dimensional structure compared with human and mouse p53 proteins, in particular in some of the residues that contact the DNA bases and in the L1, L2, and L3 loops that are involved in DNA and Zn binding and at this stage it is difficult to reconcile the strong binding similarities of the two proteins with their structures (Huyen et al. 2004).
Loss of CEP-1 function, achieved through RNAi knockdown, mutation, or the expression of a dominant negative protein lacking the DNA binding domain, results in viable worms without any overt developmental defects, including no defects in somatic developmental apoptosis. cep-1 loss of function only affects DNA damage induced apoptosis occurring in female germ cells after treatment with genotoxic agents such as ionizing irradiation, UV light, or ENU treatment (Derry et al. 2001; Schumacher et al. 2001). In adult worms, the germline is the only tissue that proliferates and responds to DNA damage through arrest of mitotic cells or apoptosis of pachytene stage meiotic cells. Unlike human p53, worms lacking CEP-1 still undergo normal cell cycle arrest in the mitotic region following ionising irradiation and overexpression of CEP-1 does not induce cell cycle arrest at any stage of development. Interestingly, CEP-1 is required for both apoptosis and cell cycle arrest in the germline following UV irradiation (Derry et al. 2007; Stergiou et al. 2007), indicating that different stimuli result in different CEP-1 dependent outcomes.
Like other p53 superfamily members CEP-1 acts as a transcription factor. Two well established transcriptional targets of CEP-1 are the BH3 only protein encoding genes egl-1 (Hofmann et al. 2002) and ced-13 (Schumacher et al. 2005b). These BH3 only protein induce apoptosis in worms by binding to the Bcl-2 homolog, CED-9, which normally holds the Apaf-1 homolog CED-4 inactive. Binding of EGL-1 to CED-9 releases CED-4, which can then activate the caspase CED-3. The transcription of both egl-1 and ced-13 is up-regulated following DNA damage in a cep-1 dependent manner and both promoters contain p53 consensus binding sites (Schumacher et al. 2005b; Ou et al. 2007) although it has not yet been directly tested whether CEP-1 binds at these sites. These data suggest that CEP-1 is part of a signaling cascade that triggers apoptosis through the transcriptional induction of BH3 domain only proteins, analogous to the p53 dependent transcriptional induction of mammalian PUMA and Noxa. Another possible target of CEP-1 that may specifically mediate UV induced cell cycle arrest is phg-1 (Derry et al. 2007). However, this finding still has to be confirmed by analysis using a deletion mutant. In addition, fasn-1, which encodes a fatty acid synthase gene, has been identified as a potential cep-1 target through an in silico approach looking for genes with conserved p53 response elements between humans and worms (D'Erchia et al. 2006). Whether these or other genes are bona fide transcriptional targets of CEP-1 remains controversial at this stage. Microarray experiments have attempted to identify further CEP-1 transcriptional targets. Derry and colleagues using a single cep-1 allele found that the transcription of a large number of genes is regulated by cep-1 during normal development and following UV irradiation (Derry et al. 2007). However, none of the DNA damage independent cep-1 targets could be confirmed when more sensitive Affymetrix arrays and two independent cep-1 deletion alleles were used (Greiss et al. 2008b). In this study the only genes whose transcriptional induction was affected by both cep-1 null mutants were egl-1 and ced-13 and another, apparently nematode specific, gene (Greiss et al. 2008b). Given the large number of p53 family transcriptional targets in vertebrates it would be surprising if CEP-1 were only to be required for the transcriptional induction of the BH3 only genes egl-1 and ced-13.
We know very little about CEP-1 functions unrelated to DNA damage responses. Nevertheless, the expression pattern of CEP-1 suggests that such functions may exist. Apart from the germline, CEP-1 is also expressed in late stage oocytes, embryos, and pharynx cells. It is possible that CEP-1 impinges on cell cycle checkpoints during embryogenesis, as its loss suppressed the slowed embryonic cell divisions occurring in clk-2 DNA damage checkpoint mutants (Moser et al. 2009). It also appears that cep-1 plays a role in mediating innate immunity as cep-1 was required for the increased pathogen resistance of mutants with compromised nucleolar function (Fuhrman et al. 2009).
CEP-1 may also play a role in aging. cep-1 mutants or cep-1 RNAi treated worms have a slight increase in lifespan that is dependent on the daf-16 Foxo transcription factor (Arum and Johnson 2007; Ventura et al. 2009). This phenotype is, however, relatively weak compared with strong worm longevity mutants and such weak aging phenotypes are associated with many C. elegans mutants. Nevertheless, the life span extension phenotype associated with cep-1 mutants appears to involve autophagy as it is dependent on beclin (Tavernarakis et al. 2008) and loss of cep-1 results in up-regulation of LGG-1, the worm homolog of LC3 (Tasdemir et al. 2008). However, in contradiction to the earlier studies, cep-1 (RNAi) suppresses the increase in lifespan due to partial loss of function of the nonsense mediated decay protein SMG-1 (Masse et al. 2008). Finally, a well controlled study firmly established that CEP-1 mediates lifespan in response to mitochondrial stress. When worms suffer mild mitochondrial stress because of partial loss of mitochondrial protein function their lifespan is prolonged. This increase in lifespan requires functional cep-1, which possibly activates compensatory mechanisms. Paradoxically, under more severe mitochondrial stress conditions cep-1 contributes to a shortened lifespan (Ventura et al. 2009). In contrast to the fly system we do not know if CEP-1 mediates longevity by interacting with the insulin pathway and if the role of CEP-1 life span regulation is cell autonomous or requires its expression in select tissues or cells.
We know very little about how CEP-1 is activated but phosphorylation is likely to be important as CEP-1 becomes phosphorylated in response to DNA damage (Quevedo et al. 2007). In contrast to the situation observed in flies, it is unlikely that C. elegans CHK-2 kinase is needed for CEP-1 activation in response to ionizing irradiation but it might have a role in response to UV irradiation (Stergiou et al. 2007). Nevertheless, as expected from other systems there is ample evidence that CEP-1 acts downstream of upstream checkpoint kinases such as the worm ATR and CHK-1 orthologues (Hofmann et al. 2002; Garcia-Muse and Boulton 2007; Gao et al. 2008; Greiss et al. 2008b).
Besides CEP-1, several other genes have been implicated in DNA damage induced apoptosis and are likely to act alongside CEP-1. RNAi of the C. elegans homolog of the mammalian MAML Notch transcriptional coactivator lag-3 resulted in a decrease in apoptosis because of decreased egl-1 and ced-13 induction following irradiation, indicating that LAG-3 may act as a CEP-1 coactivator similar to the function of MAML in p53 activation (Zhao et al. 2007). These studies, however, still have to be confirmed by the analysis of a lag-3 deletion allele.
Mutations in several other genes have been shown to specifically result in reduced apoptosis induction in response to ionizing irradiation reminiscent of cep-1 mutants. Unexpectedly, egl-1 and ced-13 are still normally induced in these mutants, suggesting that the corresponding genes impinge on apoptosis induction in one or more genetic pathways that act downstream or in parallel of the cep-1 dependent transcriptional induction of egl-1. The first such factors described were components of the C. elegans retinoblastoma complex (Schertel and Conradt 2007), which also affects basal germ cell apoptosis. The worm homolog of the human SIRT1 histone deacetylase, which has been implicated in regulating transcriptional repression and aging also affects IR induced apoptosis (Greiss et al. 2008a). Other such factors are the ING-3 transcription factor (Luo et al. 2009) and components of the ceramide synthesis pathway have been shown to be required for DNA damage induced apoptosis (Deng et al. 2008). At the moment these factors cannot be placed into a simple genetic or biochemical pathway and future studies will be required to mechanistically link these genes to apoptosis regulation.
Both worm and flies do not encode an obvious MDM2 homolog. Nevertheless, several negative regulators of cep-1 dependent apoptosis have also been identified through the generation of mutant worms that show increased apoptosis following DNA damage because of deregulation of CEP-1. The ABL-1 kinase antagonizes cep-1 induced apoptosis following irradiation but its mode of action is unknown (Deng et al. 2004). Likewise, it is unknown how AKT-1 antagonizes cep-1 induced apoptosis but in its absence there are higher levels of CEP-1 protein both in its unmodified and phosphorylated forms (Quevedo et al. 2007). Loss of components of the SCFFSN-1 ubiquitin ligase and of the neddylation pathway also result in increased CEP-1 protein levels and phosphorylation and increased egl-1 transcription (Gao et al. 2008). However, it appears that SCFFSN-1 may not regulate CEP-1 directly as no physical interaction between FSN-1 and CEP-1 could be detected by co-immunoprecipitation. Interestingly, FBXO45, the human ortholog of FSN-1, specifically binds to p73 and ubiquitinylates it, thus triggering its proteasome-dependent degradation (Peschiaroli et al. 2009) FSN-1/FBOX45 mediated ubiquitinylation might therefore be a conserved mechanism regulating p73 and CEP-1.
Two negative regulators of CEP-1 were shown to directly interact with CEP-1. GLD-1 is a mRNA binding protein that inhibits the translation of multiple target messenger RNAs involved in germ cell differentiation and maintenance and germ cell sex determination. A hypomorphic allele of gld-1 was identified that, in contrast to gld-1 null alleles, shows no overt defects in germ cell differentiation but specifically leads to the hyper-activation of CEP-1 dependent apoptosis (Schumacher et al. 2005a). The corresponding mutant protein does not recognise the cep-1 3′ UTR leading to increased CEP-1 expression, whereas other developmental targets of GLD-1 are recognized normally. In the absence of GLD-1 more CEP-1 is translated, resulting in increased apoptosis. C. elegans contains only the inhibitory form of the ASSP proteins (iASSP), which can directly bind to p53 (Bergamaschi et al. 2003). Worm iASSP bound both CEP-1 and p53 in vitro, indicating that it negatively regulates CEP-1 by direct interaction. The transcriptional activity of CEP-1 is also negatively regulated. The protein arginine methyltransferase PRMT-5 can bind to both CEP-1 and the coactivator CBP-1 (p300/CBP homolog) and methylate the latter (Yang et al. 2009). When PRMT-5 is absent cep-1 induced apoptosis and egl-1 mRNA levels are increased following DNA damage indicating that it normally acts to modulate CEP-1 transcription of egl-1.
In summary, since the discovery of worm CEP-1 tremendous progress has been made to better understand its regulation and to implicate it in various pathways. Without doubt the best characterized CEP-1 function relates to DNA damage induced germ cell apoptosis.
CONCLUDING REMARKS
The p53 superfamily of proteins seems to predate the evolutionary origin of metazoans. It is estimated that Choanozoa and metazoans shared an unicellular ancestor in the late Precambrian period, more than 600 million years ago (Peterson and Butterfield 2005). It appears that at least one p53 superfamily member is present in all invertebrate species indicating that these proteins play fundamental roles in animals. As many of these animals do not develop tumors the evolutionary pressure shaping the evolution of the p53 superfamily was unlikely to protect organisms against tumorigenesis. However, in all invertebrates in which p53 superfamily protein function has been studied, apoptosis induction in response to DNA damage has been a commonly found function, indicating that this is likely to be one of the ancient roles of the p53 superfamily, at least in metazoans. We do not know if apoptosis occurs in the Choanozoa or in its last common ancestor with us, and we could not find any obvious Bcl-2, Apaf-1 or caspase like molecules in these organisms. Nevertheless, at least in animals, one shared function of the p53 superfamily might be related to germ cell apoptosis, possibly in response to mistakes occurring through the generation and resolution of DNA double strand breaks during meiotic recombination. Germ cell apoptosis, triggered by p53 superfamily members, might have evolved early in animal evolution and is preserved in p63 dependent apoptosis of germ cells in vertebrates (Suh et al. 2006), to ensure the demise of genetically damaged germ cells.
Studies of invertebrate p53 superfamily members have also began to shed light on other roles for p53 superfamily proteins such as in DNA repair or aging. Furthermore, the greatly diverse nature of invertebrate p53 superfamily protein structures will allow a better understanding of structural features of vertebrate p53 superfamily members. Further study of p53 superfamily proteins in invertebrates will further promote our understanding of not only how these proteins and their functions have evolved in invertebrates but will also contribute to our understanding of vertebrate p53, p63, and p73 function and evolution.
ACKNOWLEDGMENTS
Research in the Gartner lab was funded by a CRUK career development award and by a CRUK project grant.
Footnotes
Editors: Arnold J. Levine and David Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM 2007. p53 directs focused genomic responses in Drosophila. Oncogene 26:5184–5193 [DOI] [PubMed] [Google Scholar]
- Allison SJ, Milner J 2003. Loss of p53 has site-specific effects on histone H3 modification, including serine 10 phosphorylation important for maintenance of ploidy. Cancer Res 63:6674–6679 [PubMed] [Google Scholar]
- Arum O, Johnson TE 2007. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J Gerontol A Biol Sci Med Sci 62:951–959 [DOI] [PubMed] [Google Scholar]
- Baldauf SL 2003. The deep roots of eukaryotes. Science 300:1703–1706 [DOI] [PubMed] [Google Scholar]
- Bauer JH, Chang C, Morris SN, Hozier S, Andersen S, Waitzman JS, Helfand SL 2007. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci 104:13355–13360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL 2005. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol 15:2063–2068 [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, O'Neil NJ, Trigiante G, Crook T, Hsieh JK, O'Connor DJ, Zhong S, Campargue I, Tomlinson ML, et al. 2003. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet 33:162–167 [DOI] [PubMed] [Google Scholar]
- Bodai L, Pardi N, Ujfaludi Z, Bereczki O, Komonyi O, Balint E, Boros IM 2007. Daxx-like protein of Drosophila interacts with Dmp53 and affects longevity and Ark mRNA level. J Biol Chem 282:36386–36393 [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101:103–113 [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Polesello C, Josue F, Tapon N 2006. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr Biol 16:1453–1458 [DOI] [PubMed] [Google Scholar]
- Cox RL, Stephens RE, Reinisch CL 2003. p63/73 homologs in surf clam: novel signaling motifs and implications for control of expression. Gene 320:49–58 [DOI] [PubMed] [Google Scholar]
- Deng X, Hofmann ER, Villanueva A, Hobert O, Capodieci P, Veach DR, Yin X, Campodonico L, Glekas A, Cordon-Cardo C, et al. 2004. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat Genet 36:906–912 [DOI] [PubMed] [Google Scholar]
- Deng X, Yin X, Allan R, Lu DD, Maurer CW, Haimovitz-Friedman A, Fuks Z, Shaham S, Kolesnick R 2008. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science 322:110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Erchia AM, Tullo A, Lefkimmiatis K, Saccone C, Sbisa E 2006. The fatty acid synthase gene is a conserved p53 family target from worm to human. Cell Cycle 5:750–758 [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH 2001. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science 294:591–595 [DOI] [PubMed] [Google Scholar]
- Derry WB, Bierings R, van Iersel M, Satkunendran T, Reinke V, Rothman JH 2007. Regulation of developmental rate and germ cell proliferation in Caenorhabditis elegans by the p53 gene network. Cell Death Differ 14:662–670 [DOI] [PubMed] [Google Scholar]
- Farcy E, Fleury C, Lelong C, Dubos MP, Voiseux C, Fievet B, Lebel JM 2008. Molecular cloning of a new member of the p53 family from the Pacific oyster Crassostrea gigas and seasonal pattern of its transcriptional expression level. Mar Environ Res 66:300–308 [DOI] [PubMed] [Google Scholar]
- Fernandes AD, Atchley WR 2008. Biochemical and functional evidence of p53 homology is inconsistent with molecular phylogenetics for distant sequences. J Mol Evol 67:51–67 [DOI] [PubMed] [Google Scholar]
- Fuhrman LE, Goel AK, Smith J, Shianna KV, Aballay A 2009. Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLoS Genet 5:e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao MX, Liao EH, Yu B, Wang Y, Zhen M, Derry WB 2008. The SCF FSN-1 ubiquitin ligase controls germline apoptosis through CEP-1/p53 in C. elegans. Cell Death Differ 15:1054–1062 [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ 2007. Meiotic recombination in Caenorhabditis elegans. Chromosome Res 15:607–621 [DOI] [PubMed] [Google Scholar]
- Goodson MS, Crookes-Goodson WJ, Kimbell JR, McFall-Ngai MJ 2006. Characterization and role of p53 family members in the symbiont-induced morphogenesis of the Euprymna scolopes light organ. Biol Bull 211:7–17 [DOI] [PubMed] [Google Scholar]
- Gostissa M, Morelli M, Mantovani F, Guida E, Piazza S, Collavin L, Brancolini C, Schneider C, Del Sal G 2004. The transcriptional repressor hDaxx potentiates p53-dependent apoptosis. J Biol Chem 279:48013–48023 [DOI] [PubMed] [Google Scholar]
- Greiss S, Hall J, Ahmed S, Gartner A 2008a. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev 22:2831–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A 2008b. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics 9:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Guo M 2006. Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol 22:623–650 [DOI] [PubMed] [Google Scholar]
- Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M, Hengartner MO 2002. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol 12:1908–1918 [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267 [DOI] [PubMed] [Google Scholar]
- Huyen Y, Jeffrey PD, Derry WB, Rothman JH, Pavletich NP, Stavridi ES, Halazonetis TD 2004. Structural differences in the DNA binding domains of human p53 and its C. elegans ortholog Cep-1. Structure 12:1237–1243 [DOI] [PubMed] [Google Scholar]
- Jassim OW, Fink JL, Cagan RL 2003. Dmp53 protects the Drosophila retina during a developmentally regulated DNA damage response. Embo J 22:5622–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen-Eller K, Kreiling JA, Begley GS, Steele ME, Walker CW, Stephens RE, Reinisch CL 2002. A new invertebrate member of the p53 gene family is developmentally expressed and responds to polychlorinated biphenyls. Environ Health Perspect 110:377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, Levine AJ 2000. Identification and characterization of a p53 homolog in Drosophila melanogaster. Proc Natl Acad Sci 97:7301–7306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley ML, Winge P, Heaney JD, Stephens RE, Farell JH, Van Beneden RJ, Reinisch CL, Lesser MP, Walker CW 2001. Expression of homologs for p53 and p73 in the softshell clam (Mya arenaria), a naturally-occurring model for human cancer. Oncogene 20:748–758 [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451:783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocque JR, Dougherty DL, Hussain SK, Sekelsky J 2007. Reducing DNA polymerase α in the absence of Drosophila ATR leads to P53-dependent apoptosis and developmental defects. Genetics 176:1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee E, Park J, Kim E, Kim J, Chung J 2003. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett 550:5–10 [DOI] [PubMed] [Google Scholar]
- Luo J, Shah S, Riabowol K, Mains PE 2009. The Caenorhabditis elegans ing-3 gene regulates ionizing radiation-induced germ-cell apoptosis in a p53-associated pathway. Genetics 181:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa T, Arita K, Shigenobu S, Kobayashi S 2009. Expression of the apoptosis inducer gene head involution defective in primordial germ cells of the Drosophila embryo requires eiger, p53, and loki function. Dev Growth Differ 51:453–461 [DOI] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu-Ansah E, Banerjee U 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell 9:843–854 [DOI] [PubMed] [Google Scholar]
- Masse I, Molin L, Mouchiroud L, Vanhems P, Palladino F, Billaud M, Solari F 2008. A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PLoS One 3:e3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri F, McNamee LM, Lunardi A, Chiacchiera F, Del Sal G, Brodsky MH, Collavin L 2008. Modification of Drosophila p53 by SUMO modulates its transactivation and pro-apoptotic functions. J Biol Chem 283:20848–20856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR 2008. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev 22:3158–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza L, Orozco E, Rodriguez MA, Garcia-Rivera G, Sanchez T, Garcia E, Gariglio P 2003. Ehp53, an Entamoeba histolytica protein, ancestor of the mammalian tumour suppressor p53. Microbiology 149:885–893 [DOI] [PubMed] [Google Scholar]
- Moser SC, von Elsner S, Bussing I, Alpi A, Schnabel R, Gartner A 2009. Functional dissection of Caenorhabditis elegans CLK-2/TEL2 cell cycle defects during embryogenesis and germline development. PLoS Genet 5:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muttray AF, Schulte PM, Baldwin SA 2008. Invertebrate p53-like mRNA isoforms are differentially expressed in mussel haemic neoplasia. Mar Environ Res 66:412–421 [DOI] [PubMed] [Google Scholar]
- Muttray AF, Cox RL, Reinisch CL, Baldwin SA 2007. Identification of DeltaN isoform and polyadenylation site choice variants in molluscan p63/p73-like homologs. Mar Biotechnol 9:217–230 [DOI] [PubMed] [Google Scholar]
- Muttray AF, Cox RL, St-Jean S, van Poppelen P, Reinisch CL, Baldwin SA 2005. Identification and phylogenetic comparison of p53 in two distinct mussel species (Mytilus). Comp Biochem Physiol C Toxicol Pharmacol 140:237–250 [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Tan C 2007. Early diversification and complex evolutionary history of the p53 tumor suppressor gene family. Dev Genes Evol 217:801–806 [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, et al. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91–101 [DOI] [PubMed] [Google Scholar]
- Ou HD, Lohr F, Vogel V, Mantele W, Dotsch V 2007. Structural evolution of C-terminal domains in the p53 family. Embo J 26:3463–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankow S, Bamberger C 2007. The p53 tumor suppressor-like protein nvp63 mediates selective germ cell death in the sea anemone Nematostella vectensis. PLoS ONE 2:e782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G 2009. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene 28:3157–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M, DeLuca C, Hirao A, Stambolic V, Potter J, Zhou L, Liepa J, Snow B, Arya S, Wong J, et al. 2002. Chk2 regulates irradiation-induced, p53-mediated apoptosis in Drosophila. Proc Natl Acad Sci 99:11305–11310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K 2002. reaper is required for neuroblast apoptosis during Drosophila development. Development 129:1467–1476 [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Butterfield NJ 2005. Origin of the Eumetazoa: Testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc Natl Acad Sci 102:9547–9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintus SS, Fomin ES, Oshurkov IS, Ivanisenko VA 2007. Phylogenetic analysis of the p53 and p63/p73 gene families. In Silico Biol 7:319–332 [PubMed] [Google Scholar]
- Quevedo C, Kaplan DR, Derry WB 2007. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr Biol 17:286–292 [DOI] [PubMed] [Google Scholar]
- Rebollar E, Valadez-Graham V, Vazquez M, Reynaud E, Zurita M 2006. Role of the p53 homolog from Drosophila melanogaster in the maintenance of histone H3 acetylation and response to UV-light irradiation. FEBS Lett 580:642–648 [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J 2003. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. Embo J 22:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C, Conradt B 2007. C. elegans orthologs of components of the RB tumor suppressor complex have distinct pro-apoptotic functions. Development 134:3691–3701 [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A 2005a. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120:357–368 [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol 11:1722–1727 [DOI] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, Conradt B, Shaham S 2005b. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ 12:153–161 [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T 2008. Multigene phylogeny of choanozoa and the origin of animals. PLoS One 3:e2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogame N, Kim M, Abrams JM 2003. Drosophila p53 preserves genomic stability by regulating cell death. Proc Natl Acad Sci 100:4696–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454:955–960 [DOI] [PubMed] [Google Scholar]
- Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO 2007. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ 14:1129–1138 [DOI] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F 2006. p63 protects the female germ line during meiotic arrest. Nature 444:624–628 [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. 2008. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10:676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G 2008. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 4:870–873 [DOI] [PubMed] [Google Scholar]
- Titen SW, Golic KG 2008. Telomere loss provokes multiple pathways to apoptosis and produces genomic instability in Drosophila melanogaster. Genetics 180:1821–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE 2009. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell 8:380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells BS, Yoshida E, Johnston LA 2006. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 16:1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Davis KD, Coffman CR 2008. Programmed cell death of primordial germ cells in Drosophila is regulated by p53 and the Outsiders monocarboxylate transporter. Development 135:207–216 [DOI] [PubMed] [Google Scholar]
- Yang M, Sun J, Sun X, Shen Q, Gao Z, Yang C 2009. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet 5:e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LY, Liu J, Sidhu GS, Niu Y, Liu Y, Wang R, Liao D 2004. Negative regulation of p53 functions by Daxx and the involvement of MDM2. J Biol Chem 279:50566–50579 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Katzman RB, Delmolino LM, Bhat I, Zhang Y, Gurumurthy CB, Germaniuk-Kurowska A, Reddi HV, Solomon A, Zeng MS, et al. 2007. The notch regulator MAML1 interacts with p53 and functions as a coactivator. J Biol Chem 282:11969–11981 [DOI] [PubMed] [Google Scholar]