Abstract
The ability to form biofilms is a universal attribute of bacteria. Biofilms are multicellular communities held together by a self-produced extracellular matrix. The mechanisms that different bacteria employ to form biofilms vary, frequently depending on environmental conditions and specific strain attributes. In this review, we emphasize four well-studied model systems to give an overview of how several organisms form biofilms: Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. Using these bacteria as examples, we discuss the key features of biofilms as well as mechanisms by which extracellular signals trigger biofilm formation.
Bacteria produce extracellular matrices that allow them to respond to antibiotics and other stimuli by forming aggregates that contain phenotypically distinct cell types.
Bacteria are able to grow adhered to almost every surface, forming architecturally complex communities termed biofilms. In biofilms, cells grow in multicellular aggregates that are encased in an extracellular matrix produced by the bacteria themselves (Branda et al. 2005; Hall-Stoodley and Stoodley 2009). Biofilms impact humans in many ways as they can form in natural, medical, and industrial settings. For instance, formation of biofilms on medical devices, such as catheters or implants often results in difficult-to-treat chronic infections (Hall-Stoodley et al. 2004; Donlan 2008; Hatt and Rather 2008). Moreover, infections have been associated with biofilm formation on human surfaces such as teeth, skin, and the urinary tract (Hatt and Rather 2008). However, biofilms on human surfaces are not always detrimental. For example, dental plaque biofilms comprise dozens of species and the community composition frequently determines the presence or absence of disease. In dental plaque, there is a progression of colonization and the presence of beneficial species antagonizes colonization by detrimental organisms (Kreth et al. 2008). But biofilms form everywhere. For example, biofilms form on the hulls of ships and inside pipes where they cause severe problems (de Carvalho 2007). On the other hand, in many natural settings, biofilm formation often allows mutualistic symbioses. For instance, Actinobacteria often grow on ants, allowing the ants to maintain pathogen-free fungal gardens (Currie 2001; Danhorn and Fuqua 2007). Given the vast potential benefits and detriments that biofilms can confer, it is essential that we understand how bacteria thrive in these communities.
There are numerous benefits that a bacterial community might obtain from the formation of biofilms. Biofilms confer resistance to many antimicrobials, protection from protozoan grazing, and protection against host defenses (Mah and O’Toole 2001; Matz and Kjelleberg 2005; Anderson and O’Toole 2008). One possible reason for the increased resistance to environmental stresses observed in biofilm cells appears to be the increase in the portion of persister cells within the biofilm (Lewis 2005). Despite being genetically identical to the rest of the population, persister cells are resistant to many antibiotics and are nondividing. Persister cells have been proposed to be protected from the action of antibiotics because they express toxin–antitoxin systems where the target of the antibiotics is blocked by the toxin modules (Lewis 2005). In addition to an increase in persisters, the presence of an extracellular matrix protects constituent cells from external aggressions. Extracellular matrices also act as a diffusion barrier to small molecules (Anderson and O’Toole 2008; Hall-Stoodley and Stoodley 2009). Related to this, in biofilms the diffusion of nutrients, vitamins, or cofactors is slower resulting in a bacterial community in which some of cells are metabolically inactive. Furthermore, the rate of bacterial growth is influenced by the fact that cells within a biofilm are confined to a limited space (Stewart and Franklin 2008). This condition is similar to the stationary phase created in laboratory conditions. Hence, biofilm formation in a way represents the natural stationary phase of bacterial growth. During stationary phase, bacteria profoundly change their physiology by increasing production of secondary metabolites such as antibiotics, pigments, and other small-molecules (Martin and Liras 1989). These secondary metabolites also function as signaling molecules to initiate the process of biofilm formation or to inhibit biofilm formation by other organisms that inhabit the same habitat (Lopez and Kolter 2009). In this article, we review the metabolic processes that characterize biofilm formation for a handful of well-studied bacterial organisms: Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, and Bacillus subtilis. In addition, we address the function of secondary metabolites and their role as signaling molecules during biofilm formation.
STRUCTURAL COMPONENTS OF BIOFILMS
The molecular mechanisms that regulate biofilm formation vary greatly among different species, and even vary between different strains of the same species. However, some features are recognized as general attributes of biofilm formation (Monds and O’Toole 2009). For instance, all biofilms contain an extracellular matrix that holds cells together. This matrix is often composed of a polysaccharide biopolymer along with other components such as proteins or DNA (Branda et al. 2005). The nature of the matrix exopolysaccharide greatly varies depending on growth conditions, medium, and substrates.
P. aeruginosa is a gram-negative pathogen that makes biofilms by producing three distinct exopolysaccharides: alginate, PEL, and PSL. The importance and contribution of each exopolysaccharide to the matrix varies depending on the strain studied (Ryder et al. 2007; Tart and Wozniak 2008). For example, alginate is a produced by mucoid strains of P. aeruginosa that are often isolated from lungs of cystic fibrosis patients. The pel gene cluster, encoding a glucose-rich polymer termed PEL, is found in most of the strains analyzed to date, yet its expression strongly varies among strains (Branda et al. 2005). The reference strain PA14 used in many laboratories harbors a partial deletion of the psl locus, which prevents the PSL mannose-rich polysaccharide from being made (Friedman and Kolter 2004).
The soil-dwelling Gram-positive bacterium B. subtilis is also studied as a model organism for biofilm formation. Different B. subtilis strains are able to secrete two distinct polymers: the polysaccharide EPS and poly-δ-glutamate (PGA). Both of these molecules have been described to participate in the process of biofilm formation (Stanley and Lazazzera 2005; Branda et al. 2006). Yet, they contribute differently depending on the strain and conditions studied. For example, in colony biofilms the undomesticated strain NCIB3610 requires exopolysaccharide EPS for biofilm formation (Fig. 1). However, no colony biofilm defect is observed in a mutant strain lacking the ability to produce PGA (Branda et al. 2006). Instead, cells that overproduced PGA formed structureless, mucoid colonies. Another undomesticated strain of B. subtilis, RO-FF-1 naturally produces PGA and forms mucoid colonies. PGA production is important for surface-adhered biofilm formation in both RO-FF-1 and the laboratory strain JH642 (Stanley and Lazazzera 2005). In contrast, the strain NCIB3610 is unable to form robust surface-adhered biofilms (Branda et al. 2006).
Figure 1.
Colony morphology of B. subtilis strain 3610 wild type and matrix mutant (eps). Top view of cells after 3 d of growth on 1.5% agar MSgg medium. Bar is 5 mm.
Another bacterial model used to study biofilm formation is the Gram-positive pathogen S. aureus. Most strains of S. aureus use a polymer of N-acetyl glucosamine (PNAG) also referred to as polysaccharide intercellular adhesin (PIA), to form biofilms (O’Gara 2007). The ica operon encodes the machinery that synthesizes this polymer, yet not all strains carry this operon. Even in some of those strains that carry the ica operon, deletion of the operon does not impair their ability to make biofilm via an ica-independent pathway (O’Gara 2007; Otto 2008). This alternative mechanism relies on the ability of S. aureus to express a variety of adhesin proteins that allow cells to attach and colonize a large number of different surfaces (Lasa and Penades 2006).
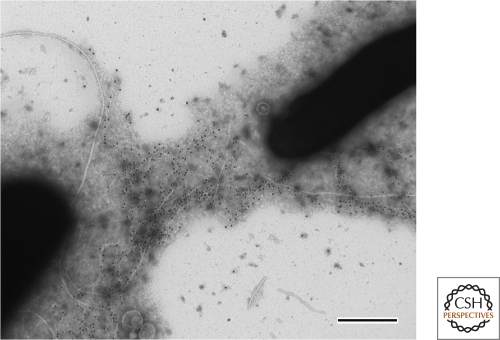
As alluded to above, the extracellular matrix of biofilms also harbors adhesive proteins. For instance, S. aureus matrix harbors biofilm-associated proteins (termed Bap) that are required for biofilm formation (Lasa and Penades 2006; Latasa et al. 2006). These proteins are found anchored to the cell wall of S. aureus and serve to hold cells together within the biofilm, probably by interacting with other proteins on the surface of neighboring cells. In certain strains, the expression of Bap proteins eliminates the requirement for exopolysaccharides for biofilm formation (Cucarella et al. 2004). Unlike the multitude of Bap proteins found in S. aureus biofilms, B. subtilis expresses a single major protein associated with the extracellular matrix termed TasA. Mutants deficient in TasA fail to form biofilms despite the fact that they can still produce exopolysaccharide (Branda et al. 2006). TasA has recently been shown to form extracellular filaments that have amyloid-like properties and is thought to play a structural or architectural role in the extracellular matrix (Fig. 2) (Romero et al. 2010). TasA is not the first amyloid-like protein to be implicated in biofilm formation. In E. coli, the curli protein also forms amyloid filaments and is critical for biofilm formation (Chapman et al. 2002; Barnhart and Chapman 2006). Other proteinaceous structures important for biofilm formation are pili and fimbriae. These cell appendages are used to adhere cells to each other or to different surfaces. E. coli produces Type I fimbriae that are required for adherence to mannose-containing receptors. These fimbriae are important for biofilm formation on plastic surfaces as well as on host cells during urinary tract infections (Pratt and Kolter 1998; Wright et al. 2007). P. aeruginosa also has many surface proteins that contribute to biofilm formation. For example, mutants unable to produce type IV pili or the CupA fimbriae were defective in surface-adhered biofilms (O’Toole and Kolter 1998; Vallet et al. 2001; D’Argenio et al. 2002). In addition, there are other matrix-associated lectin-binding proteins that recognize and bind carbohydrate moieties. These facilitate cell–matrix or cell–cell interactions within the biofilm. P. aeruginosa, has two lectin-binding proteins involved in biofilm formation (LecA and LecB) (Tielker et al. 2005; Diggle et al. 2006).
Figure 2.
Electron micrograph of B. subtilis strain 3610 immunogold labeled with anti-TasA antibody (black dots). Bar is 0.5 µm. Image courtesy of Diego Romero.
In addition to the exopolysaccharides and proteins, extracellular DNA (eDNA) also provides structural integrity to the biofilm. Biofilm matrix in P. aeruginosa contains significant amounts of eDNA. The addition of DNase to cultures inhibits biofilm formation and dissolves mature biofilms (Whitchurch et al. 2002). S. aureus biofilms also have eDNA in the matrix and it functions to provide stability to the biofilms. It is thought that this eDNA is caused by cell lysis and subsequent release of genomic DNA (Rice et al. 2007).
The participation of multiple types of molecules such as polysaccharides, DNA, and proteins in the formation of extracellular matrix makes it impossible to present a single summary of biofilm matrix. Similarly, although it is tempting to suggest matrix is a biofilm feature that could be targeted for biofilm control, the variation between matrices of different strains and species makes it impossible to find a unifying attribute. Furthermore, as described earlier, depending on the conditions, different components of the matrix become more or less important for the integrity of the biofilm.
CELL HETEROGENEITY WITHIN BIOFILMS
Although single-species biofilms can arise from a single cell and should therefore be clonal populations, they are often composed of phenotypically distinct subpopulations. Within biofilms specialized cell types often arise because of differences in gene expression but not in gene composition (Fux et al. 2005; An and Parsek 2007; Spormann 2008; Stewart and Franklin 2008). Cell differentiation in bacterial communities depends on the extracellular conditions to which cells are exposed. The formation of gradients of nutrients, oxygen or electron acceptors throughout the biofilm creates microenvironments to which cells respond by altering their gene expression (Spormann 2008; Stewart and Franklin 2008). For instance, in P. aeruginosa oxygen only penetrates the outer regions of the biofilm. When the enzymatic activity of the oxygen-dependent enzyme alkaline phosphatase was measured in cross sections of biofilms the activity of the enzyme was correlated with the cells located in areas more exposed to the oxygen (Xu et al. 1998). Similarly, S. aureus biofilms displayed an aerobic zone closer to the surface of the biofilm. When the metabolically active zones of the biofilm were identified by localizing the areas where DNA and protein synthesis occured, two distinct strata were observed. One correlated with the area exposed to oxygen and a second was at the base of the colony at the agar surface closer to the nutrients. This indicates that in S. aureus about two-thirds of the biofilm was metabolically inactive (Rani et al. 2007).
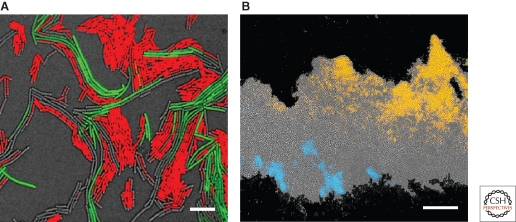
The physiological state of cells within a biofilm can also be monitored by analyzing cell-type specific gene expression for each defined subpopulation of cells. This technique can be applied only when each cell differentiation pathway is well understood at the molecular level, as is the case for the model organism B. subtilis. This organism sporulates during starvation, forming metabolically inactive spores resistant to many environmental stresses (Piggot and Hilbert 2004). Using lacZ or GFP transcriptional fusions to sporulation-specific genes, sporulating cells were observed to preferentially localize in the aerial structures that form on colony biofilms (Branda et al. 2001; Veening et al. 2006). The localization of spores to the apical region of the aerial structures was confirmed at the single-cell level by thin sectioning frozen colony biofilms of cells that had cell-type specific promoters fused to fluorescent proteins. This technique was applied to the localization of other subpopulation of cells such as matrix producers or motile cells, cells that express flagella that allow these cells to swim. Sporulation, matrix production, and motility were shown to occur in distinct subpopulations within the biofilm (Fig. 3) (Vlamakis et al. 2008). Furthermore, the percentage and localization of each cell type was dynamic. Early stages of biofilm formation show an abundance of motile cells whereas as the biofilm matured, many of the motile cells differentiated into matrix producing cells. At later time points the subpopulation of sporulating cells arose primarily from the matrix producing subpopulation (Vlamakis et al. 2008). Strains harboring fluorescent reporters have also been used to detect the presence of surfactin-producing cells, the signaling molecule that triggers the differentiation of matrix producers (Lopez et al. 2009c). As we discuss later, this unidirectional signaling represents a remarkable event in the field of bacterial cell differentiation.
Figure 3.
Heterogeneity in B. subtilis biofilms. (A) Top view of cells at the onset of colony development. Overlay of fluorescence images with DIC (gray), motile (red), and matrix-producing (green) cells. Bar 5 µm. (B) Thin-sectioned three-day-old biofilm. Agar is at the bottom and the center of the colony is on the right. Overlay of fluorescence images with DIC (gray), motile (blue), and sporulating (orange) cells. Bar 50 µm.
SIGNALING IN BIOFILM FORMATION
Because the formation of a biofilm can be considered a mechanism to protect the bacterial community from external insults, it seems reasonable that specific extracellular cues regulate activation of the metabolic pathways that lead to biofilm formation. These external cues come from diverse sources. Signals can be produced and secreted by the bacterial community itself, in which case the molecules are termed autoinducers. Autoinducers accumulate extracellularly and the concentration of autoinducer can be correlated with population density. At high concentrations, autoinducers trigger signal transduction cascades that lead to multicellular responses in the bacterial population. This mechanism of cell–cell communication in bacteria (termed quorum sensing) controls a large number of developmental processes included those related to biofilm formation (Camilli and Bassler 2006).
QUORUM-SENSING MOLECULES THAT INDUCE BIOFILM FORMATION
In P. aeruginosa, along with many other Gram-negative organisms, quorum-sensing systems respond to a class of autoinducer termed acyl homoserine lactones (AHLs). P. aeruginosa possesses two AHL quorum-sensing systems: las and rhl. Each system has its own AHL synthase (LasI and RhlI) and its own transcriptional regulator (LasR and RhlR). The AHL signals produced by the synthases are N-(3-oxododecanoyl)-HSL and N-butyryl-HSL, respectively (de Kievit 2009). Work by many groups has found that depending on the strain and experimental conditions, the importance of these two systems in biofilm formation varies (Hentzer et al. 2004; de Kievit 2009). In strain PAO1 both Las and Rhl systems were important for extracellular DNA release, which plays a role in biofilm matrix and structure (Allesen-Holm et al. 2006). In the strain PA14 the Las system is essential for biofilm architecture probably through control of the production of the PEL exopolysaccharide (Sauer et al. 2002; Sakuragi and Kolter 2007; Yang et al. 2007).
In Gram-positive organisms, the autoinducers are often peptides and these are detected outside the cell. To be detected extracellularly, autoinducer molecules are generally sensed by membrane-associated sensor kinases, which activate cognate response regulators by phosphorylation. That, in turn, activates the expression of the target genes (Novick and Geisinger 2008). In S. aureus, the autoinducer molecule is a peptide (AIP) derived from the product of the agrD gene. This peptide is processed to yield a cyclic peptide containing a thiolactone ring. AIP is secreted and detected by AgrC, which activates the regulator AgrA. AgrA positively regulates the transcription of genes including those that code for several extracellular proteases involved in the dispersal of the biofilm (Balaban and Novick 1995; Ji et al. 1995; Yarwood et al. 2004; O’Gara 2007). Thus, in the case of S. aureus, quorum-sensing negatively regulates biofilm formation (Boles and Horswill 2008). Biofilm formation in S. aureus involves several sequential stages such as initial attachment, cell-to-cell adhesion, maturation, and final detachment (Otto 2004). Adhesion to a surface is the initial step to transition from planktonic cells to biofilm formation in S. aureus. This step is favored only when the agr quorum-sensing systems is inhibited. Once cells successfully attach to a surface, bacteria accumulate forming a complex architecture, which involves the production of exopolysaccharide (known as PNAG or PIA). Eventually small clusters of cells detach from the mature biofilm; a step that is important for the dispersal of the community (Yarwood et al. 2004; Yao and Strauch 2005). Mutants lacking the agr gene form thicker biofilms than wild type. This is not attributed to cell growth or death, but rather to the inability of cells to detach from the mature biofilm (Vuong et al. 2000).
For B. subtilis, the production and secretion of the quorum-sensing molecule surfactin is important for biofilm formation (Lopez et al. 2009a). Aside from its surfactant properties, surfactin causes potassium leakage from the cytoplasm. Potassium leakage is sensed by a membrane-associated sensor kinase, KinC, to specifically trigger the expression of the genes involved in extracellular matrix production (Lopez et al. 2009a). Because of the nature of the stimulus, various small-molecules were identified that induce matrix production via KinC. These differ largely in their molecular structure. What is conserved is their ability to induce potassium leakage by making ion-selective pores in the membrane of B. subtilis. Among these molecules are the macrolide polyenes nystatin and amphotericin as well as the peptide antibiotics gramidicin and valinomycin, all of which are produced by soil-dwelling bacteria. This particular mechanism of quorum-sensing that recognizes the mode of action of the signaling molecule rather than its structure permits B. subtilis to sense and respond to a diverse number of signals. These signals are not only to self-produced molecules but also natural products that are secreted by other soil-dwelling organisms (Lopez et al. 2009a).
Despite the fact that surfactin is a secreted molecule that should be able to interact with all of the cells within the population, not all of the cells respond to the presence of the molecule. This heterogeneity in response can be explained by the mechanism of gene activation (Lopez et al. 2009a). Once surfactin activates the sensor kinase KinC, that leads to activation of the kinase’s cognate regulator, Spo0A. Spo0A is a transcriptional regulator whose activity depends on the level of phosphorylated protein within the cell. Matrix gene expression is triggered only when Spo0A∼P accumulates above certain levels (Fujita et al. 2005). This mechanism restricts the expression of matrix-related genes to the subpopulation of cells that accumulates the required level of Spo0A∼P, triggering a cascade of events resulting in a bimodal population where some cells produce matrix and others do not (Chai et al. 2008).
Interestingly, the subpopulation of surfactin producers is different from the subpopulation of cells (matrix producers) that respond to surfactin (Lopez et al. 2009c). The srfAA-AD operon is responsible for surfactin production and is directly controlled by another regulator termed ComA. Activation of ComA by phosphorylation (ComA∼P) is driven by another quorum-sensing system that senses the presence of the extracellular pheromone ComX via the membrane-associated sensor kinase ComP (Magnuson et al. 1994; Nakano 1991). This system, in which a primary signal controls the production of secondary system, might serve as a timing mechanism to regulate the activation of diverse metabolic pathways sequentially during the course of development.
Surfactin acts as a uni-directional signal in this particular quorum-sensing system, in which one subpopulation of cells produces the molecule, whereas another population of cells responds to it and produces matrix (Lopez et al. 2009c). This mechanism adds more complexity to the concept of “quorum sensing” or, as we have referred previously “autoinduction,” where all cells are physiologically similar thus, able to produce the signal and respond to it (Fuqua et al. 1994; Camilli and Bassler 2006). In the typical quorum-sensing scenario, signaling would be referred as autocrine. The uni-directional signaling recently described in B. subtilis is the first example of paracrine signaling in bacteria where the subpopulation producing the signal does not respond to it (Lopez et al. 2009c).
MOLECULES THAT INDUCE BIOFILM FORMATION INDEPENDENT OF QUORUM SENSING
In addition to quorum-sensing molecules, a diversity of other signals trigger biofilm formation. These include secondary metabolites such as antibiotics, pigments, and siderophores. At subinhibitory concentrations many antibiotics function not to kill cells, but rather as signals that trigger changes in gene expression (Yim et al. 2007). Subinhibitory concentrations of the antibiotic imipenem induced expression of the polysaccharide alginate in P. aeruginosa biofilms (Bagge et al. 2004). Hence, imipenem-exposed biofilms were thicker and covered more of the substratum than nontreated biofilms. Similarly, subinhibitory concentrations of the aminoglycoside antibiotic tobramycin induced biofilm formation in P. aeruginosa and E. coli (Hoffman et al. 2005). The mechanism of action of this signaling process is not understood.
In addition to sensing the presence of antibiotics produced by other organisms, P. aeruginosa responds to small-molecules that it produces. For example, the redox-active pigments termed phenazines have been described to have antibiotic activity or function as virulence factors in eukaryotic hosts (Price-Whelan et al. 2006). Within biofilms, the phenazine pyocyanin functions in extracellular electron transfer to generate energy for growth. Having a small, diffusible molecule to shuttle electrons in a biofilm where the diffusion solubility may be limited is beneficial for the community (Hernandez and Newman 2001). Phenazines in P. aeruginosa also function as signaling molecules in biofilm formation, as a mutant unable to produce phenazines produced dramatically more wrinkled colony morphology than a wild-type strain (Dietrich et al. 2008). This difference was because of the induction of SoxR-regulated genes in response to phenazines; both phenazine overproducing strains and soxR mutant strains formed flat, featureless colonies.
In the case of S. aureus, the activation of the quorum-sensing system inhibits biofilm formation. Thus, small molecules that inhibit quorum sensing also favor biofilm formation. This was recently described for the furanones, which are natural products derived from marine algae (de Nys et al. 2006). These small molecules are able to inhibit the quorum-sensing systems of many Gram-negative bacteria (Wu et al. 2004). Additionally, the molecules were tested for inhibition of the quorum-sensing system in Staphylococci. At high concentrations, furanones had bactericidal effect on S. epidermidis and S. aureus Interestingly, S. aureus treated with subinhibitory concentrations of the marine furanones resulted in an inhibition of the quorum-sensing system coupled with an increase in the ability of S. aureus to make biofilms (Kuehl et al. 2009). Similar results were previously observed with subinhibitory concentrations of other well-known antimicrobials such as tetracycline or quinupristin-dalfopristin. Cultures of Staphylococcus epidermidis treated with subinhibitory concentrations of these molecules enhance the expression of genes responsible for exopolysaccharide production. Also, a weaker induction was observed when treated with sub-inhibitory concentrations of the antibiotic erythromycin. The mechanism underlying this effect is not well understood yet (Rachid et al. 2000).
Other small-molecules induce biofilm formation in B. subtilis independent of quorum sensing. Specific molecules with antibiotic properties trigger differentiation of cells into the subpopulation of matrix producers. This occurs though the Spo0A genetic pathway that triggers differentiation into matrix producers. In addition to regulating matrix gene expression, Spo0A∼P also triggers a second differentiation pathway called cannibalism (Gonzalez-Pastor et al. 2003; Ellermeier et al. 2006; Claverys and Havarstein 2007). Cells expressing cannibalism genes produce, and are resistant to, two toxins: Skf and Sdp. As described earlier, different gene sets are regulated by different levels of Spo0A phosphate, thus only the cells in the population that have achieved high enough levels of Spo0A∼P are able to express the cannibalism toxins and resistance machinery (Fujita et al. 2005). This leaves a sensitive portion of the population that has not achieved high enough levels of Spo0A∼P. It is this sensitive portion of the population that lyses once some cells express skf and sdp genes. Dead siblings serve as food for the community to overcome nutritional limitation, and this delays the onset of sporulation. This could benefit the community because spore development is energy intensive and, once committed, cells may not exit this state for prolonged periods. Thus, by sacrificing a portion of the population, B. subtilis can delay the entry into sporulation for as long as possible (Gonzalez-Pastor et al. 2003; Ellermeier et al. 2006).
Both matrix production and cannibalism are triggered by the same genetic cascade, and these two traits are indeed expressed concomitantly in the same subpopulation of cells (Lopez et al. 2009b). Consistent with this, the signaling molecule surfactin, which is responsible for differentiation of the subpopulation of matrix producers, also triggers cannibal toxin production. When the cannibalism toxins are secreted to the extracellular space, only the subpopulation of matrix producers are favored to grow, because it is the only subpopulation that expresses the immunity machinery to the action of the cannibalism toxins. This gives that subpopulation the advantage in that they can use the nutrients released by their killed siblings causing the matrix/cannibal cells to increase relative to other cell types. With the increase in the relative number of matrix-producing cells, these communities are able to produce more extracellular matrix and form stronger biofilms. This behavior constitutes a mechanism to eliminate cell types that might no longer be required for the development of the community to promote the growth of other subpopulations such as matrix producers (Lopez et al. 2009b).
Other antimicrobial peptides could mimic the effect of the cannibalism toxins. This is because of the ability of the cannibalism resistance machinery to work nonspecifically for several similar molecules (Butcher and Helmann 2006). One such molecule is nisin, a peptide antibitiotic putatively similar to the Skf toxin. The presence of nisin therefore promotes the subpopulation of matrix producers in B. subtilis communities much like the cannibalism toxins (Fig. 4) (Lopez et al. 2009b). This ability to sense small-molecules produced by diverse soil microorganisms suggests a broad mechanism that B. subtilis uses to respond to surrounding bacterial communities by altering its development.
Figure 4.
Effect of the antimicrobial nisin on B. subtilis biofilm morphology. Cells closer to the disk containing nisin are more wrinkled due to the presence of more matrix-producing cells. Bar is 3 mm. (Reprinted, with permission, from Lopez et al 2009b [© Wiley].)
SUMMARY
As presented, the majority of microbes are able to develop multicellular biofilm communities. These communities are composed of subpopulations of different cell types that provide additional benefits to the organisms. There are numerous differences among the mechanisms that induce biofilm formation in different species. Even considering only four of the best-known bacterial models (E. coli, P. aeruginosa, S. aureus, and B. subtilis), the discrepancies between the processes involved in the formation of biofilms among these examples are broad. A vast array of exopolysaccharides, secreted proteins and cell-surface adhesins contribute to the structural integrity of biofilms. Small molecules such as homoserine lactones, antibiotics, and other secondary metabolites play a critical role in the development and maintenance of biofilm communities.
Footnotes
Editors: Lucy Shapiro and Richard Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- An D, Parsek MR 2007. The promise and peril of transcriptional profiling in biofilm communities. Curr Opin Microbiol 10:292–296 [DOI] [PubMed] [Google Scholar]
- Anderson GG, O’Toole GA 2008. Innate and induced resistance mechanisms of bacterial biofilms. in Bacterial Biofilms (ed. Romeo T.), pp. 85–105 Springer, Heidelberg; [DOI] [PubMed] [Google Scholar]
- Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Hoiby N 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and β-lactamase and alginate production. Antimicrob Agents Chemother 48:1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N, Novick RP 1995. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc Natl Acad Sci 92:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238 [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci 98:11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R 2005. Biofilms: The matrix revisited. Trends Microbiol 13:20–26 [DOI] [PubMed] [Google Scholar]
- Butcher BG, Helmann JD 2006. Identification of Bacillus subtilis σ-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol 60:765–782 [DOI] [PubMed] [Google Scholar]
- Camilli A, Bassler BL 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67:254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS 2007. Cannibalism and fratricide: Mechanisms and raisons d’etre. Nat Rev Microbiol 5:219–229 [DOI] [PubMed] [Google Scholar]
- Cucarella C, Tormo MA, Ubeda C, Trotonda MP, Monzon M, Peris C, Amorena B, Lasa I, Penades JR 2004. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun 72:2177–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie CR 2001. A community of ants, fungi, and bacteria: A multilateral approach to studying symbiosis. Annu Rev Microbiol 55:357–380 [DOI] [PubMed] [Google Scholar]
- D’Argenio DA, Calfee MW, Rainey PB, Pesci EC 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol 184:6481–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn T, Fuqua C 2007. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422 [DOI] [PubMed] [Google Scholar]
- de Carvalho CC 2007. Biofilms: Recent developments on an old battle. Recent Pat Biotechnol 1:49–57 [DOI] [PubMed] [Google Scholar]
- de Kievit TR 2009. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol 11:279–288 [DOI] [PubMed] [Google Scholar]
- de Nys R, Givskov M, Kumar N, Kjelleberg S, Steinberg PD 2006. Furanones. Prog Mol Subcell Biol 42:55–86 [DOI] [PubMed] [Google Scholar]
- Dietrich LE, Teal TK, Price-Whelan A, Newman DK 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K 2006. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol 8:1095–1104 [DOI] [PubMed] [Google Scholar]
- Donlan RM 2008. Biofilms on central venous catheters: Is eradication possible? Curr Top Microbiol Immunol 322:133–161 [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R 2006. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124:549–559 [DOI] [PubMed] [Google Scholar]
- Friedman L, Kolter R 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186:4457–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP 1994. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P 2005. Survival strategies of infectious biofilms. Trends Microbiol 13:34–40 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R 2003. Cannibalism by sporulating bacteria. Science 301:510–513 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Stoodley P 2009. Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P 2004. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108 [DOI] [PubMed] [Google Scholar]
- Hatt JK, Rather PN 2008. Role of bacterial biofilms in urinary tract infections. in Bactarial Biofilms (ed. Romeo T.), 163–192 Springer, Heidelberg; [DOI] [PubMed] [Google Scholar]
- Hentzer M, Givskov M, Eberl L 2004. Quorum sensing in biofilms: Gossip in slime city. in Microbial Biofilms (ed. O’Toole M.G.a.G.), pp. 118–140 ASM Press, Washington DC [Google Scholar]
- Hernandez ME, Newman DK 2001. Extracellular electron transfer. Cell Mol Life Sci 58:1562–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175 [DOI] [PubMed] [Google Scholar]
- Ji G, Beavis RC, Novick RP 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci 92:12055–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl R, Al-Bataineh S, Gordon O, Luginbuehl R, Otto M, Textor M, Landmann R 2009. Furanone enhances biofilm of staphylococci at subinhibitory concentrations by luxS repression. Antimicrob Agents Chemother 53:4157–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I, Penades JR 2006. Bap: A family of surface proteins involved in biofilm formation. Res Microbiol 157:99–107 [DOI] [PubMed] [Google Scholar]
- Latasa C, Solano C, Penades JR, Lasa I 2006. Biofilm-associated proteins. C R Biol 329:849–857 [DOI] [PubMed] [Google Scholar]
- Lewis K 2005. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc) 70:267–274 [DOI] [PubMed] [Google Scholar]
- Lopez D, Kolter R 2009. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu F, Losick R, Kolter R 2009a. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci 106:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R 2009b. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol 74:609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Losick R, Kolter R 2009c. Paracrine signaling in a bacterium. Genes Dev 23:1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R, Solomon J, Grossman AD 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77:207–216 [DOI] [PubMed] [Google Scholar]
- Mah TF, O’Toole GA 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39 [DOI] [PubMed] [Google Scholar]
- Martin MF, Liras P 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol 43:173–206 [DOI] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S 2005. Off the hook–how bacteria survive protozoan grazing. Trends Microbiol 13:302–307 [DOI] [PubMed] [Google Scholar]
- Monds RD, O’Toole GA 2009. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol 17:73–87 [DOI] [PubMed] [Google Scholar]
- Nakano MM, Xia LA, Zuber P 1991. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol 173:5487–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564 [DOI] [PubMed] [Google Scholar]
- O’Gara JP 2007. ica and beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 270:179–188 [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304 [DOI] [PubMed] [Google Scholar]
- Otto M 2004. Quorum-sensing control in Staphylococci – a target for antimicrobial drug therapy? FEMS Microbiol Lett 241:135–141 [DOI] [PubMed] [Google Scholar]
- Otto M 2008. Staphylococcal biofilms. in Bacterial Biofilms (ed. Romeo T.), pp. 207–228 Springer, Heidelberg [Google Scholar]
- Piggot PJ, Hilbert DW 2004. Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586 [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R 1998. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293 [DOI] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LE, Newman DK 2006. Rethinking ‘secondary’ metabolism: Physiological roles for phenazine antibiotics. Nat Chem Biol 2:71–78 [DOI] [PubMed] [Google Scholar]
- Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother 44:3357–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS 2007. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol 189:4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci 104:8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R 2010. Amyloid fibers provide structural integrity to Bacillus subtilis BIOFILMS. Proc Natl Acad Sci 107:2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C, Byrd M, Wozniak DJ 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuragi Y, Kolter R 2007. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol 189:5383–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spormann AM 2008. Physiology of microbes in biofilms. in Bacterial Biofilms (ed. Romeo T.), pp. 17–36 Springer, Heidelberg; [DOI] [PubMed] [Google Scholar]
- Stanley NR, Lazazzera BA 2005. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-dl-glutamic acid production and biofilm formation. Mol Microbiol 57:1143–1158 [DOI] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210 [DOI] [PubMed] [Google Scholar]
- Tart AH, Wozniak DJ 2008. Shifting paradigms in Pseudomonas aeruginosa biofilm research. in Bacterial Biofilms (ed. Romeo T.), pp. 193–206 Springer, Heidelberg; [DOI] [PubMed] [Google Scholar]
- Tielker D, Hacker S, Loris R, Strathmann M, Wingender J, Wilhelm S, Rosenau F, Jaeger KE 2005. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151:1313–1323 [DOI] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: Identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci 98:6911–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Kuipers OP, Brul S, Hellingwerf KJ, Kort R 2006. Effects of phosphorelay perturbations on architecture, sporulation, and spore resistance in biofilms of Bacillus subtilis. J Bacteriol 188:3099–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Saenz HL, Gotz F, Otto M 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182:1688–1693 [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ 2007. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol 9:2230–2241 [DOI] [PubMed] [Google Scholar]
- Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, Hoiby N 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J Antimicrob Chemother 53:1054–1061 [DOI] [PubMed] [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol 64:4035–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153:1318–1328 [DOI] [PubMed] [Google Scholar]
- Yao F, Strauch MA 2005. Independent and interchangeable multimerization domains of the AbrB, Abh, and SpoVT global regulatory proteins. J Bacteriol 187:6354–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, Bartels DJ, Volper EM, Greenberg EP 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J 2007. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci 362:1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]