Abstract
Objective
To assess technical and operational performance of a dried blood spot (DBS)-based HIV-1 RNA service for remote healthcare facilities in a low-income country.
Design
A method comparison and operational evaluation of DBS RNA against conventional tests for early infant diagnosis of HIV and HIV RNA quantitation under field conditions in Tanzania.
Methods
DBS were prepared and plasma was frozen at −80°C. DBS were mailed and plasma couriered to a central laboratory for testing using the Abbott m2000 system. Infant diagnosis DBS were also tested for HIV-1 DNA by ROCHE COBAS AmpliPrep/COBAS TaqMan System. Results of DBS RNA were compared with conventional tests; program performance was described.
Results
Among 176 infant diagnosis participants, using a threshold of ≥1,000 copies/mL, sensitivity and specificity of DBS versus plasma RNA were 1.00 and 0.99, and of DBS RNA versus DBS DNA were 0.97 and 1.00. Among 137 viral load monitoring participants, when plasma and DBS RNA were compared R was 0.9709; R was 0.9675 ≥5,000 copies/mL but was 0.7301 <5,000 copies/mL. The highest plasma RNA value at which DBS RNA was not detected was 2,084 copies/mL. Median (range) turnaround time from sample collection to result receipt at sites was 23 (4–69) days. The Tanzania mail service successfully transmitted all DBS and results between sites and the central laboratory.
Conclusion
Under program conditions in Tanzania, DBS provided HIV-1 RNA results comparable to conventional methods to remote healthcare facilities. DBS RNA testing is an alternative to liquid plasma for HIV-1 RNA services in remote areas.
Keywords: Tanzania, HIV, diagnosis, laboratory techniques and procedures, reverse transcriptase polymerase chain reaction, blood specimen collection
BACKGROUND
In 2007 an estimated 33 million people were living with HIV globally [1]. Access to antiretroviral therapy (ART) has increased markedly since 2004, particularly in low- and middle-income countries. In order to initiate life saving ART early, HIV care and treatment programs in resource-constrained settings are increasing efforts to diagnose HIV infection among infants using nucleic acid amplification testing (NAT) [2, 3]. Furthermore, the recognition that virologic failure among patients receiving ART is poorly predicted by clinical and immunologic monitoring [4, 5] has led to growing interest in expansion of NAT services for monitoring plasma HIV-1 RNA levels to patients receiving ART in low- and middle-income countries [3, 4].
Conventional HIV NAT has relied on liquid plasma samples. In many resource-constrained settings, NAT services are scarce and are highly centralized. Because of centralization of testing and the fact that liquid plasma samples must be assayed within 6 hours or frozen to −80°C to avoid deterioration, HIV NAT has been available only to persons able to travel to reference centers or to clinics with a robust cold chain. Dried blood spots (DBS) represent an alternative sample type to liquid plasma that are easy to prepare, robust, and that do not require a cold chain [6, 7]. HIV-1 DNA PCR of DBS (DBS DNA) compares favorably with HIV-1 DNA PCR of liquid plasma (plasma DNA) for early infant diagnosis of HIV [8]. Furthermore, HIV-1 RNA PCR of plasma samples (plasma RNA) has been shown to be a valid test for early infant diagnosis of HIV infection [9–11]. Similarly, several studies have evaluated DBS HIV-1 RNA (DBS RNA) for measurement of HIV-1 RNA concentration [12] and for early infant diagnosis [13, 14] under laboratory conditions. Taken together, these findings suggest that it might be possible to establish a DBS RNA service suited to the needs of remote health facilities that uses a single platform in centralized laboratories that provide simultaneous early infant diagnosis of HIV with baseline HIV-1 RNA measurement as well as quantitation of HIV-1 RNA levels for monitoring patients on ART.
In order to investigate the feasibility of a DBS RNA service for early infant diagnosis and for HIV-1 RNA concentration monitoring under field conditions in a resource-constrained setting, we studied the technical and operational performance of a DBS RNA program in Tanzania in partnership with two rural and remote healthcare facilities. The assessment included a method comparison of DBS RNA against conventional assays following transportation of DBS by mail, an evaluation of DBS sample stability over time, and operations research that monitored the reliability of the mail service for delivering DBS and the measured turnaround times.
METHODS
Study sites
Participants were recruited among HIV care and treatment clinic attendees and pediatric inpatients at two rural healthcare facilities, Magunga Hospital in Korogwe District and Teule Hospital in Muheza District. These hospitals are located in northeastern Tanzania approximately 300 and 350 km, respectively, from the Kilimanjaro Christian Medical Centre (KCMC) Biotechnology Laboratory in Moshi, Tanzania that served as the central laboratory. The climatic conditions in this area have been described elsewhere [15]. ART has been available free through the national care and treatment program since 2004, starting at consultant referral hospitals, and subsequently decentralized to regional and then district hospital level. DBS RNA and plasma RNA testing were performed at the central laboratory and DBS DNA testing was done at the University of Witwatersrand, Parktown, South Africa.
Participant selection
Infants <18 months old who were HIV-1 exposed or were clinically suspected to have HIV infection were eligible for enrollment in Part A. HIV-infected persons ≥18 months were eligible for enrollment in Part B. After obtaining informed consent, patients were administered a standardized questionnaire that assessed demographic, epidemiologic, clinical, and treatment information.
Sample collection, preparation and transport
Five to 10mL of blood was drawn by venipuncture and collected in EDTA tubes. To prepare DBS, 50 uL whole blood aliquots were spotted onto 903 filter paper cards and air dried for ≥4 hours on a drying rack on the laboratory bench. Up to 10 dried blood spots were prepared per subject for testing and to provide surplus spots in case of losses during mailing or testing. After drying, the cards were placed in a gas-impermeable zip locked bag with desiccant and stored in a safe location at ambient temperature. The DBS were mailed weekly from the two rural hospital sites to the central laboratory using a mail service of the Tanzania Posts Corporation. The remaining EDTA blood was separated ≤6 hours after collection by centrifugation, the plasma frozen at −80°C, and transported weekly on dry ice to the central laboratory via courier.
Sample testing and result reporting
Upon receipt at the KCMC Biotechnology Laboratory, DBS were cut from the 903 cards using a 16mm diameter card cutter. Two DBS were transferred to a 50 mL conical tube with 1.7 mL of lysis buffer (Abbott m Sample Preparation System buffer, Abbott Laboratories, Abbott Park, IL). These tubes were incubated at room temperature for 2 hours with intermittent mixing; 1.0 mL of the resulting solution was assayed for HIV-1 RNA by the Abbott m2000 system (Abbott Laboratories, Abbott Park, IL) using the HIV-1 RNA DBS quantitative protocol. A 0.6 mL aliquot of liquid plasma from each patient was tested using the Abbott m2000 system and results were compared.
DBS from part A were also sent to the University of Witwatersrand, South Africa, under ambient conditions for HIV-1 DNA testing. Each spot was transferred to a 1.8mL S-tube and 1,100 µL of COBAS® AmpliPrep/COBAS® TaqMan® Specimen Pre-Extraction Reagent (SPEX) was added. The tubes were incubated in an Eppendorf Thermomixer Comfort at 56°C and 1,000 rpm continuous shaking for 10 minutes; ≥1 mL of the resulting solution was assayed for HIV-1 DNA using the ROCHE COBAS AmpliPrep/COBAS TaqMan System (Roche Diagnostics, Indianapolis, IN)[13]. The quality of results of the Tanzania Abbott m2000 assay and the South Africa ROCHE COBAS AmpliPrep/COBAS TaqMan were assured by successful participation in the AIDS Clinical Trials Group Viral Quality Assurance program.
To assess the stability of DBS samples under field conditions, DBS from 32 patients enrolled in part B were stored for approximately 10 weeks before re-testing. The baseline HIV-1 RNA level was defined as the first DBS RNA measurement ≤40 days from sample collection. The follow-up level was defined as the second DBS RNA measurement taken 41–80 days following collection. DBS were sealed in gas-impermeable shipment bags with desiccant, placed in plastic shipping envelopes, and were grouped by date received. DBS were kept in the central laboratory which is temperature monitored and maintained between 18–25°C.
Assessment of program performance
To assess operational performance of the DBS program under field conditions in Tanzania, we monitored samples and results transported by mail, measuring the duration of transit times and the proportion of samples damaged or lost. Turnaround times were calculated using time in transit, time in the central laboratory prior to testing, time from sample collection time to shipping to the central laboratory, and time from sample testing to result shipment. Assay performance was tracked by documenting the number and underlying causes of run failures. The cost of sample transport was recorded.
Statistical analyses
The technical performance of the DBS RNA method was compared against conventional plasma RNA and against DBS DNA for early infant diagnosis. Methods were compared with linearity plot and Bland-Altman difference plot. Sensitivity, specificity, positive predictive value, negative predictive value, and accuracy were estimated along with 95% confidence intervals. Plasma RNA thresholds examined for early infant diagnosis were ≥1,000 copies/mL [16] and the American Academy of Pediatrics threshold of ≥10,000 copies/mL [11]. Plasma RNA thresholds examined for HIV-1 RNA quantitation were ≥400 copies/mL and the 2004 National Antiretroviral Treatment Guidelines of South Africa threshold of ≥5,000 copies/mL [17]. Stability was assessed comparing the differences in log HIV-1 RNA levels between baseline and follow-up DBS. Results were entered into Microsoft Access (Microsoft Corporation, Redmond, WA) database using Teleform (Verity, Inc. Sunnyvale, CA). Analysis was done using Microsoft Excel (Microsoft Corporation, Redmond, WA), EP Evaluator (David G. Rhoads Associates, Kennett Square, PA), and JMP software (SAS worldwide, Cary, NC).
Research ethics
This study was approved by the KCMC Research Ethics Committee, the Tanzania National Institutes for Medical Research National Research Ethics Coordinating Committee, and an Institutional Review Board of Duke University Medical Center.
RESULTS
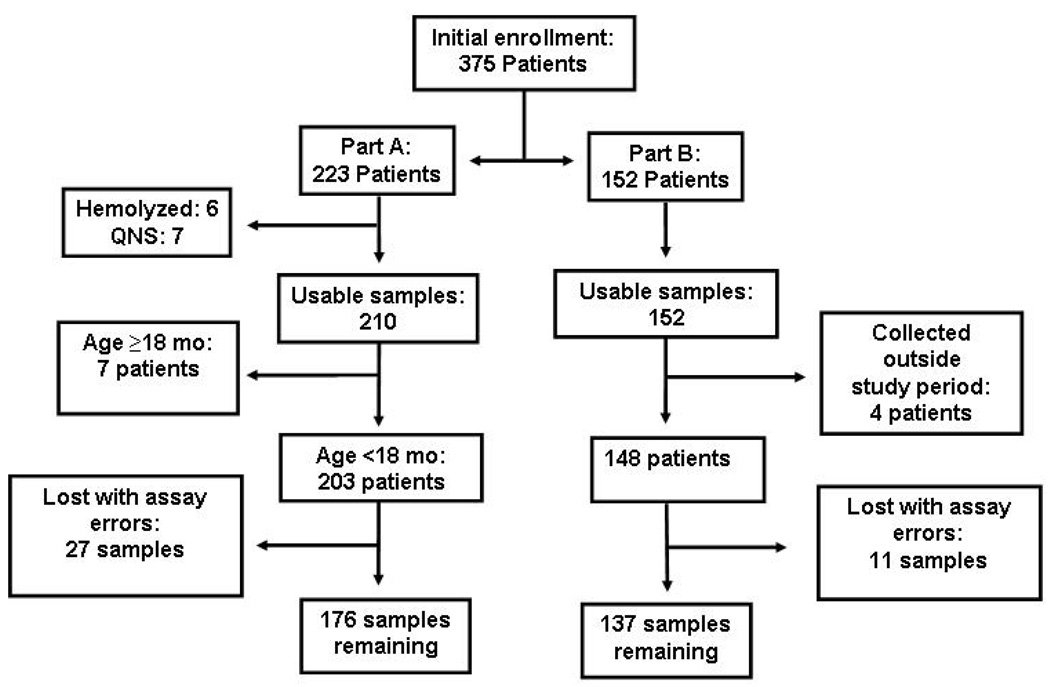
Of 375 participants enrolled from 29 October 2008 through 6 March 2009, 313 (83.5%) had samples available for analysis. Of potential participants approached, there were no refusals. The remaining samples were unavailable owing to hemolysis, insufficient volume, collection outside the study period, failure to meet enrollment criteria, or were lost owing to assay errors (Figure 1). Of the 313 subjects, 176 (56%) were enrolled into part A. The Magunga Hospital site enrolled 133 (42%) of all participants.
Figure 1.
Flow diagram, field study of a dried blood spot nucleic amplification method for infant HIV diagnosis and viral load determination in Tanzania, 2008-9
Part A: early infant diagnosis; Part B: viral load monitoring; QNS= Quantity not sufficient; Lost with assay errors= instrument flags, usually associated with inadequate volume detection, resulting in sample loss.
Part A: Early infant diagnosis
Among Part A participants, the median (range) age was 6 months (1 day to 17 months) and 82 (46%) were female; 101 (58%) were enrolled because they were HIV-exposed, 5 (3%) were enrolled because of clinical suspicion for HIV, and 70 (40%) met both criteria.
In the early infant diagnosis group, 39 (22%) had detectable plasma RNA compared with 35 (20%) by DBS RNA. At the threshold of ≥1,000 copies/mL, 34 (19%) infants were classified as infected by plasma RNA compared with 35 (20%) by DBS RNA. At the threshold of ≥10,000 copies/mL, 31 (18%) infants were classified as infected by plasma RNA compared with 31 (18%) for DBS RNA. In one participant aged 17 months with a plasma RNA level of 53 copies/mL no HIV RNA was detected from DBS while DBS HIV DNA was detected. Excluding this sample, there was complete concordance between DBS RNA and DBS DNA at the DBS RNA threshold of ≥1,000 copies/mL. Of 36 samples positive by DBS DNA, 4 (11.1%) had DBS RNA levels 1,000–10,000 copies/mL from patients aged 0, 1, 7 and 15 months. There were also 3 patients aged 10, 13, and 14 months, negative by DBS DNA, with DBS RNA not detected and plasma RNA levels of 70, <40, and 237 copies/mL, respectively. None was on ART.
Comparing plasma RNA with DBS RNA at the threshold of ≥1,000 copies/mL estimated sensitivity and specificity (95% confidence interval) were 1.00 (0.90, 1.00) and 0.99 (0.96, 1.00), and were 1.00 (0.89, 1.00) and 1.00 (0.97, 1.00) at the threshold of ≥10,000 copies/mL. Comparing DBS RNA with DBS DNA at the threshold of ≥1,000 copies/mL, estimated sensitivity and specificity (95% CI) were 0.97 (0.86, 1.00) and 1.00 (0.97, 1.00), and were 0.86 (0.71, 0.94) and 1.00 (0.97, 1.00) at the threshold of ≥10,000 copies/mL (Table 1).
Table 1.
Classification of infant HIV infection comparing DBS HIV-1 RNA results with HIV-1 plasma RNA results at the threshold of ≥1,000 copies/mL (panel a) and ≥10,000 copies/mL (panel b) and comparing DBS HIV-1 RNA results with DBS HIV-1 DNA results at the threshold of ≥1,000 copies/mL (panel c) and ≥10,000 copies/mL (panel d), Tanzania, 2008-9
| a | Plasma RNA | b | Plasma RNA | ||||||
| ≥1,000 | <1,000 | total | ≥10,000 | <10,000 | total | ||||
| ≥1,000 | 34 | 1 | 35 | ≥10,000 | 31 | 0 | 31 | ||
| DBS RNA | <1,000 | 0 | 141 | 141 | DBS RNA | <10,000 | 0 | 145 | 145 |
| total | 34 | 142 | 176 | total | 31 | 145 | 176 | ||
| SN 1.00 (0.90, 1.00), SP 0.99 (0.96, 1.00) PPV 0.97, NPV 1.00 |
SN 1.00 (0.89, 1.00), SP 1.00 (0.97, 1.00) PPV 1.00, NPV, 1.00 |
||||||||
| c | DBS DNA | d | DBS DNA | ||||||
| Positive | Negative | total | Positive | Negative | total | ||||
| ≥1,000 | 35 | 0 | 35 | ≥10,000 | 31 | 0 | 31 | ||
| DBS RNA | <1,000 | 1 | 140 | 141 | DBS RNA | <10,000 | 5 | 140 | 145 |
| total | 36 | 140 | 176 | total | 36 | 140 | 176 | ||
| SN 0.97 (0.86, 1.00), SP 1.00 (0.97, 1.00) PPV 1.00, NPV 0.99 |
SN 0.86 (0.71, 0.94), SP 1.00 (0.97, 1.00) PPV 1.00, NPV 0.97 |
||||||||
SN = sensitivity
SP = specificity
PPV = positive predictive value
NPV = negative predictive value
95% confidence intervals are shown in brackets
Part B: Viral load monitoring
Among Part B participants the median (range) age was 34 years (21 months to 77 years) and 108 (79%) were female. Of the 137 patients, 110 (80%) had CD4-positive T-lymphocyte counts (CD4 count) performed and the median (range) CD4 count was 253 (6–2,586) cells/mm3. All 137 patients provided ART information and 73 (53%) reported receiving it. All patients were taking fixed-dose combination stavudine, lamivudine, and nevirapine.
At the threshold of ≥400 copies/mL, 82 (60%) participants were classified as having virologic failure by plasma RNA compared with 88 (64%) by DBS RNA. At the threshold of ≥5,000 copies/mL, 74 (54%) participants were classified as having virologic failure by plasma RNA compared with 76 (55%) by DBS RNA. Compared with plasma RNA, estimated sensitivity and specificity (95% CI) for classifying patients with virologic failure of DBS RNA at the threshold of ≥400 copies/mL was 0.99 (0.93, 1.00) and 0.87 (0.76, 0.94), respectively, and at the threshold of ≥5,000 copies/mL was 1.00 (0.95, 1.00) and 0.97 (0.89, 0.99), respectively (Table 2).
Table 2.
Classification of virologic failure comparing plasma HIV-1 RNA results with DBS HIV-1 RNA results at the threshold of ≥400 copies/mL (panel a) and ≥5,000 copies/mL (panel b), Tanzania 2008-9
| a | plasma | b | plasma | ||||||
| ≥400 | <400 | total | ≥5,000 | <5,000 | total | ||||
| ≥400 | 81 | 7 | 88 | ≥5,000 | 74 | 2 | 76 | ||
| DBS RNA | <400 | 1 | 48 | 49 | DBS RNA | <5,000 | 0 | 61 | 61 |
| total | 82 | 55 | 137 | total | 74 | 63 | 137 | ||
| SN 0.99 (0.93, 1.00), SP 0.87 (0.76, 0.94) PPV 0.92, NPV 0.98 |
SN 1.00 (0.95, 1.00), SP 0.97 (0.89, 0.99) PPV 0.97, NPV 1.00 |
||||||||
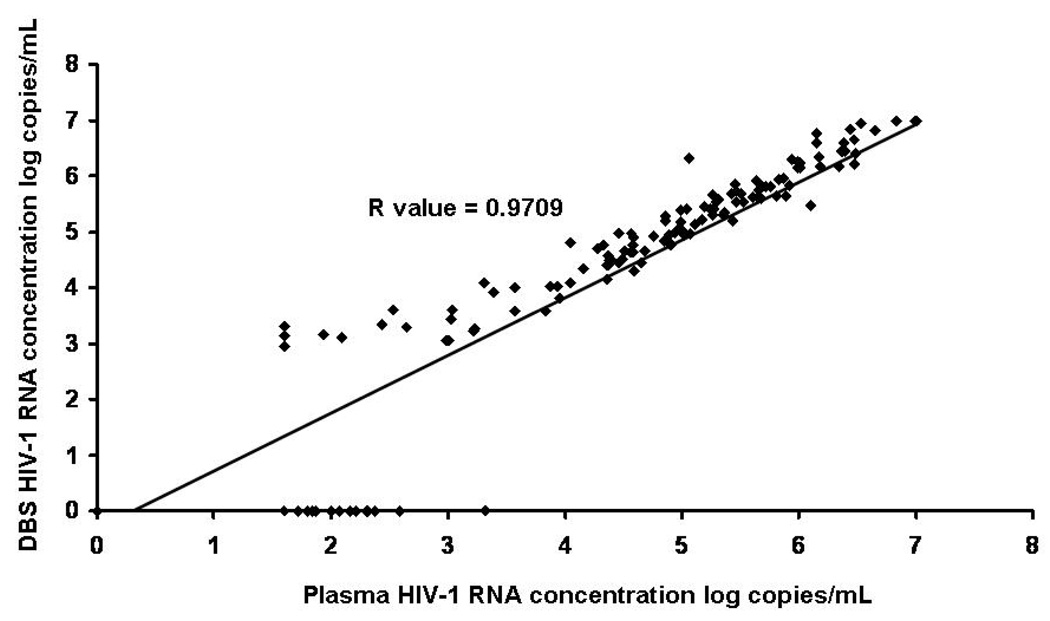
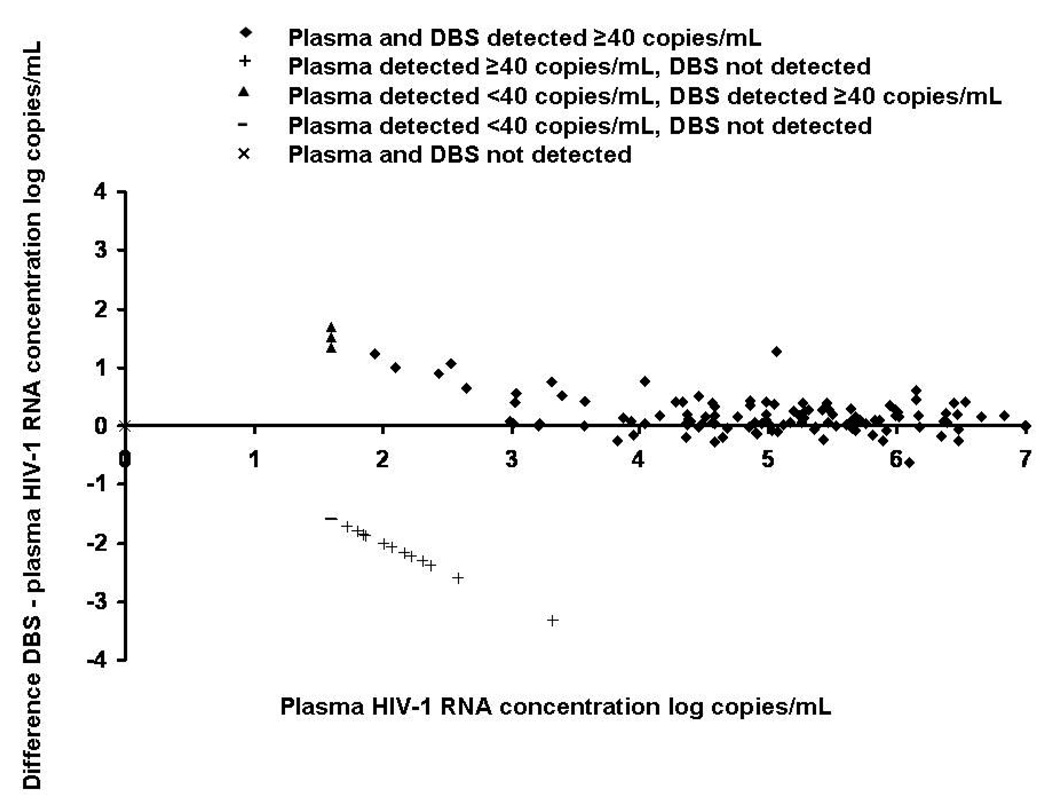
The relationship between DBS RNA and plasma RNA levels is shown in Figure 2. Using data from both parts A and B of the study, the R value produced was 0.9709; R was 0.9675 ≥5,000 copies/mL but was 0.7301 <5,000 copies/mL. Twenty-five (8.0%) of samples had HIV-1 RNA detected in plasma but not on DBS; 24 of these had plasma HIV-1 RNA concentrations <400 copies/mL. The highest plasma HIV-1 RNA level at which DBS RNA was not detected was 2,084 copies/mL. A Bland-Altman difference plot comparing Abbott m2000 system plasma and DBS RNA showed a range of difference between DBS RNA and plasma RNA levels for detectable results of 1.60 to −0.62 log copies/mL and 97% of difference values were within 2 logs of 0 (Figure 3).
Figure 2.
Linearity plot of plasma versus DBS log HIV-1 RNA concentration, Tanzania, 2008-9
Figure 3.
Bland Altman difference plot comparing DBS with plasma HIV-1 RNA concentration, Tanzania 2008-9
Stability testing
Thirty-two samples were re-tested to assess stability of DBS RNA results during approximately 10 weeks of storage. Baseline DBS RNA testing was done at a median (range) of 21 (9–37) days after sample collection, whereas follow up testing was done at 68 (42–76) days following sample collection. There was no statistically significant change in log RNA levels between the baseline and follow up time points.
Program performance
All 28 packages containing DBS, 14 (50%) from each study site, were received at the central laboratory. During the course of the study 29 packages were sent to the sites, 7 with supplies, 2 with case report forms and consent forms needing clarification, and 20 with plasma RNA results. All documents and samples reached their destination and none was damaged.
The interval from sample collection to shipping from the sites to the KCMC Biotechnology was 0–7 days. Of 20 packages sampled, the median (range) transit time from sites to the KCMC Biotechnology Laboratory was 1.5 (1–2) days. The interval from receipt of a patient care sample to completion of HIV RNA testing at the KCMC Biotechnology Laboratory was median (range) 13 (1–51) days. When this was calculated excluding a vacation period during which plasma collection was suspended and the laboratory was operating with reduce staffing levels, the median (range) interval from sample receipt to completion of testing was 7 (1–21) days. The interval from testing to shipping of results to sites was 0–7 days and the median (range) transit time of results from the KCMC Biotechnology Laboratory was estimated to be 1.5 (1–2) days. The median total turnaround time from sample collection to receipt of the result at the site was 23 days overall and was 17 days excluding the vacation period, ranging from 4 to 69 days or from 4 to 39 days excluding the vacation period. The interval from sample receipt to completion of testing in the KCMC Biotechnology Laboratory contributed the most to total turnaround time.
Factors contributing to delays in the KCMC Biotechnology Laboratory or loss of samples included three plasma run failures; two runs failed during the sample preparation, one because of a problem with the automated liquid handler, and one owing to a power outage. The third run failed at the amplification and detection stage. DBS RNA samples were lost owning to six runs with errors; three with problems during sample preparation resulting in loss of the whole run; two with out of range controls; and one with inadequate sample volume detected in 50% of samples because of technician error.
At the time of the study, the weekly cost of mailing dried blood spots from healthcare facilities to the central laboratory was 6 United States Dollars (USD), whereas the weekly ground transport of frozen plasma samples on dry ice cost USD 515.
DISCUSSION
We demonstrate under field conditions in Tanzania that an HIV-1 RNA dried blood spot program performs well against liquid plasma HIV-1 RNA for viral load monitoring and against quantitative liquid plasma HIV-1 RNA and qualitative dried blood spot DNA for early infant diagnosis. Performance was best for samples from patients with plasma RNA levels above 5,000 copies/mL. DBS RNA levels appear to be stable after approximately 10 weeks of storage compared with a baseline sample tested at a median of 21 days following collection. Furthermore, we show that a DBS program performs well using existing staff at remote health care facilities, a robust central laboratory, and that using the local mail service for DBS transport is reliable and is less expensive than ground transportation of frozen plasma on dry ice.
For early infant diagnosis, our study showed agreement between DBS RNA and both plasma RNA and DBS DNA using an RNA threshold of ≥1,000 copies/mL. Although we were able to test only one sample rather than the two recommended by the American Academy of Pediatrics (AAP), several participant samples positive by DBS DNA would have been classified as HIV-uninfected using the AAP threshold of ≥10,000 copies/mL [11]. Since untreated HIV-infected infants usually have high HIV RNA levels, it is unclear why four patients positive by DBS DNA had low HIV RNA levels, between 1,000 and 10,000 copies/mL. Low plasma HIV RNA levels have been seen in other studies [16]. Possible explanations include that these patients are able to control HIV RNA levels [18, 19], they had early acute HIV infection with low HIV RNA levels prior to viral load ramp up [20] either following breastfeeding [21] or peripartum transmission, or that HIV-1 subtype variation resulted in underquantitation. Additionally, cross-contamination of DBS samples at sites could cause false positive results. The training provided to staff for this study would make cross-contamination unlikely but difficult to rule out. Other groups have shown that most infants with initial low levels of plasma RNA followed over time subsequently are confirmed to be HIV-infected [14, 22], but a proportion are found to be HIV-uninfected [16, 22–24].
Our findings on the use of DBS RNA for early infant diagnosis are consistent with those of others. A study in Thailand yielded sensitivity of 97–100% and specificity of 100% using DBS RNA with the ROCHE Amplicor, NucliSens, and an in-house assay [7]. A South African study found sensitivity and specificity of 99.7% and100% using the Cobas AmpliPrep/Cobas TaqMan HIV-1 Qual test [13].
Compared with plasma RNA, DBS RNA was highly sensitive and specific for diagnosis of virologic failure at a threshold of ≥5,000 copies/mL and retained high sensitivity but lower specificity for virologic failure at a threshold of ≥400 copies/mL. Across a range of HIV-1 RNA levels, there was excellent agreement ≥5,000 copies/mL and fair agreement <5,000 copies/mL between plasma and DBS RNA (Figure 2 and 3). HIV-1 RNA levels from DBS tended to be higher than from plasma <5,000 copies/mL and we identified 25 samples with HIV RNA not detected on DBS but with low levels of plasma RNA detected. Entrapment of RNA in filter paper and amplification of proviral DNA may contribute to these findings. Our study was consistent with others showing disagreement and the frequency of lack of detection of DBS RNA increasing below 4,000–6,000 copies/mL [6, 25].
Our data suggests that DBS are stable over an 80 day period under laboratory conditions. DBS RNA samples for early infant diagnosis using a qualitative assay have been shown to remain 99.2% sensitive and 100% specific on retesting 4 years later [26]. Using a quantitative HIV-1 RNA assay, DBS RNA concentration was shown stable 9 months when stored at a range of temperatures [7] and in another study stable over 1 year if stored at room temperature or −70°C [6]. Thus DBS RNA samples appear to be stable over a duration that exceeds the period of clinical value of the result.
We demonstrate that a DBS RNA program performs well under field conditions in Tanzania. The local mail service rapidly and reliably transported DBS samples to the central laboratory and plasma RNA results to the clinical sites. The program achieved a median turnaround time from sample collection to receipt of the result at the remote site of 23 days. Since many follow-up visits in HIV Care and Treatment programs in Tanzania are scheduled monthly, most results were available to the clinician at the next follow-up visit. Some results took longer, leading to inconvenience for clinicians and for patients. The main contributor to total turnaround time was the interval from sample receipt to testing at the central laboratory. Laboratory turnaround time could be shortened by improved maintenance of back-up electricity infrastructure, increased technical support, and greater experience of laboratory staff with the instrument and with DBS. Total turnaround time could be further reduced by using electronic or telephone transmission of results. Despite the total turnaround time, all RNA results were deemed to be clinically useful for patient management.
A DBS RNA service with centralized testing could reach a very large population requiring or receiving ART. In 2007 it was estimated that 136,000 people were receiving ART in Tanzania [27]. The World Health Organization (WHO) recommends that, where available, HIV-1 RNA testing be offered to patients receiving ART. With annual testing, given that the assay evaluated in this study can process 93 patient samples per run, and assuming that an instrument at a central laboratory completes one run per day 365 days a year then 26,242 DBS samples could be processed per year. If two runs were done per day then 52,484 DBS RNA tests could be done per year. These test volumes would cover 39% of Tanzanians receiving ART in 2007.
Our study had a number of limitations. The remote healthcare facilities in this study had active research programs and capacities which may not be found in all rural and remote areas. The sites were served by a reliable and rapid mail service which may not be available everywhere. Our stability testing data are limited by a small sample size and would need to be verified with large numbers of samples. A number of samples were lost with assay errors and could not be included in the final analysis, possibly resulting in bias. Finally, because the study used patient samples the range of HIV-1 RNA levels may not have thorough evaluated the entire dynamic range of the assay.
We demonstrate that a DBS HIV RNA program serving rural and remote healthcare settings is feasible in Tanzania both in terms of the technical quality of the results and the operation of the program. We suggest DBS are a viable alternative to a plasma sample type within RNA programs and DBS RNA services could be scaled up to a national level. Having established the feasibility of such a program technically and operationally, we suggest that detailed cost-effectiveness analyses be conducted to allow Ministries of Health to determine costs and benefits of such a program. Larger studies that evaluate DBS program performance at non-research sites and that incorporate the impact of HIV-1 RNA results on patient management are needed.
ACKNOWLEDGEMENTS
This study was supported by Abbott Laboratories. We thank the clinicians and patients who participated in the program and Daan Potgieter, Sven Thamm, and Robert Dintruff for technical support. We also thank the Department of Molecular Medicine and Haematology, University of Witwatersrand, South Africa, for assaying DNA DBS. Hawa Chubwa, Naftali Vadwiga, Monica Mhilu, Gerald Kulwa, Omary Abdul, Samwel Gesase, Alvan Butichi, Upendo Mvungi, Emma Msuya, and Isaac Afwamba assisted with enrolling patients, preparing DBS, or testing at the KCMC Biotechnology Laboratory. Investigator support was obtained by a Stead/Hubert Yeargan Center for Global Health Scholarship (SML), the Fogarty International Center (D43 PA-03-018, JAB, JAC), the Duke Clinical Trials Unit and Clinical Research Sites (U01 AI06984-01, WS, JAB, JAC), the Duke University Center for AIDS Research (JAB), and International Studies on AIDS Associated Coinfections award (ISAAC) (U01 AI-03-036 ABM, JAB, JAC).
JAC, WS, and JAB conceived the study, and JAC and JAB obtained funding. All authors contributed to the study design, implementation and manuscript writing, SML, AIM, and SE contributed to the daily follow-up of study participants. ABM, CCC, BA, and DJS coordinated laboratory aspects of the study. SML and JAC did statistical analyses and prepared the first draft of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented in Part: 3rd International Workshop on HIV Treatment, Pathogenesis and Prevention Research in Resource-Poor Settings, Lusaka, Zambia, 26–29 May 2009 and the 5th International AIDS Society meeting, Cape Town, South Africa, 19–22 July 2009.
Financial Disclosure: This study was funded by Abbott Laboratories (Abbott Park, IL). None of the authors was employed by Abbott. SML and JAC have received speaking honoraria from Abbott Laboratories and JAB has received speaking honoraria and has served as a consultant for Abbott.
REFERENCES
- 1.UNAIDS. HIV and AIDS Estimates and Data, 2007 and 2001. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS (UNAIDS); 2008 Report on the Global AIDS Epidemic. 2008
- 2.WHO. Geneva: WHO; Priority Interventions, HIV/AIDS preventions, treatment and care in the health section. 2009 Edited by Department HA.
- 3.Calmy A, Ford N, Hirschel B, et al. HIV viral load monitoring in resource-limited regions: optional or necessary? Clin Infect Dis. 2007;44:128–134. doi: 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- 4.Bagchi S, Kempf MC, Westfall AO, Maherya A, Willig J, Saag MS. Can routine clinical markers be used longitudinally to monitor antiretroviral therapy success in resource-limited settings? Clin Infect Dis. 2007;44:135–138. doi: 10.1086/510072. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds SJ, Nakigozi G, Newell K, et al. Failure of immunologic criteria to appropriately identify antiretroviral treatment failure in Uganda. AIDS. 2009;23:697–700. doi: 10.1097/QAD.0b013e3283262a78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brambilla D, Jennings C, Aldrovandi G, et al. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: measurement, precision, and RNA stability. J Clin Microbiol. 2003;41:1888–1893. doi: 10.1128/JCM.41.5.1888-1893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leelawiwat W, Young NL, Chaowanachan T, et al. Dried blood spots for the diagnosis and quantitation of HIV-1: stability studies and evaluation of sensitivity and specificity for the diagnosis of infant HIV-1 infection in Thailand. J Virol Methods. 2009;155:109–117. doi: 10.1016/j.jviromet.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Sherman GG, Stevens G, Jones SA, Horsfield P, Stevens WS. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J Acquir Immune Defic Syndr. 2005;38:615–617. doi: 10.1097/01.qai.0000143604.71857.5d. [DOI] [PubMed] [Google Scholar]
- 9.Delamare C, Burgard M, Mayaux MJ, et al. The French Pediatric HIV Infection Study Group. HIV-1 RNA detection in plasma for the diagnosis of infection in neonates. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:121–125. doi: 10.1097/00042560-199706010-00004. [DOI] [PubMed] [Google Scholar]
- 10.Lambert JS, Harris DR, Stiehm ER, et al. Performance characteristics of HIV-1 culture and HIV-1 DNA and RNA amplification assays for early diagnosis of perinatal HIV-1 infection. J Acquir Immune Defic Syndr. 2003;34:512–519. doi: 10.1097/00126334-200312150-00011. [DOI] [PubMed] [Google Scholar]
- 11.Read JS. Diagnosis of HIV-1 infection in children younger than 18 months in the United States. Pediatrics. 2007;120:e1547–e1562. doi: 10.1542/peds.2007-2951. [DOI] [PubMed] [Google Scholar]
- 12.Marconi A, Balestrieri M, Comastri G, et al. Evaluation of the Abbott Real-Time HIV-1 quantitative assay with dried blood spot specimens. Clin Microbiol Infect. 2009;15:93–97. doi: 10.1111/j.1469-0691.2008.02116.x. [DOI] [PubMed] [Google Scholar]
- 13.Stevens W, Erasmus L, Moloi M, Taleng T, Sarang S. Performance of a novel human immunodeficiency virus (HIV) type 1 total nucleic acid-based real-time PCR assay using whole blood and dried blood spots for diagnosis of HIV in infants. J Clin Microbiol. 2008;46:3941–3945. doi: 10.1128/JCM.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young NL, Shaffer N, Chaowanachan T, et al. Early diagnosis of HIV-1-infected infants in Thailand using RNA and DNA PCR assays sensitive to non-B subtypes. J Acquir Immune Defic Syndr. 2000;24:401–407. doi: 10.1097/00126334-200008150-00001. [DOI] [PubMed] [Google Scholar]
- 15.Drakeley CJ, Carneiro I, Reyburn H, et al. Altitude-dependent and - independent variations in Plasmodium falciparum prevalence in northeastern Tanzania. J Infect Dis. 2005;191:1589–1598. doi: 10.1086/429669. [DOI] [PubMed] [Google Scholar]
- 16.Rouet F, Montcho C, Rouzioux C, et al. Early diagnosis of paediatric HIV-1 infection among African breast-fed children using a quantitative plasma HIV RNA assay. AIDS. 2001;15:1849–1856. doi: 10.1097/00002030-200109280-00015. [DOI] [PubMed] [Google Scholar]
- 17.National Department of Health. National Antiretroviral Treatment Guidelines. 1st ed. Johannesburg: Minuteman Press; 2004. Edited by Health Do. [Google Scholar]
- 18.Saksena NK, Rodes B, Wang B, Soriano V. Elite HIV controllers: myth or reality? AIDS Rev. 2007;9:195–207. [PubMed] [Google Scholar]
- 19.Suzuki Y, Yamamoto N. HIV infection and long-term non progressor. Nippon Rinsho. 1997;55:1563–1571. [PubMed] [Google Scholar]
- 20.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 21.Kreiss J. Breastfeeding and vertical transmission of HIV-1. Acta Paediatr Suppl. 1997;421:113–117. doi: 10.1111/j.1651-2227.1997.tb18332.x. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham CK, Charbonneau TT, Song K, et al. Comparison of human immunodeficiency virus 1 DNA polymerase chain reaction and qualitative and quantitative RNA polymerase chain reaction in human immunodeficiency virus 1-exposed infants. Pediatr Infect Dis J. 1999;18:30–35. doi: 10.1097/00006454-199901000-00009. [DOI] [PubMed] [Google Scholar]
- 23.Bryson YJ, Pang S, Wei LS, Dickover R, Diagne A, Chen IS. Clearance of HIV infection in a perinatally infected infant. N Engl J Med. 1995;332:833–838. doi: 10.1056/NEJM199503303321301. [DOI] [PubMed] [Google Scholar]
- 24.Nesheim S, Palumbo P, Sullivan K, et al. Quantitative RNA testing for diagnosis of HIV-infected infants. J Acquir Immune Defic Syndr. 2003;32:192–195. doi: 10.1097/00126334-200302010-00011. [DOI] [PubMed] [Google Scholar]
- 25.Garrido C, Zahonero N, Corral A, Arredondo M, Soriano V, de Mendoza C. Correlation between human immunodeficiency virus type 1 (HIV-1) RNA measurements obtained with dried blood spots and those obtained with plasma by use of Nuclisens EasyQ HIV-1 and Abbott RealTime HIV load tests. J Clin Microbiol. 2009;47:1031–1036. doi: 10.1128/JCM.02099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr RJ, Player G, Fiscus SA, Nelson JA. Qualitative human immunodeficiency virus RNA analysis of dried blood spots for diagnosis of infections in infants. J Clin Microbiol. 2009;47:220–222. doi: 10.1128/JCM.01521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UNAIDS/WHO Working Group on Global HIV/AIDS and STI Surveillance. Geneva, Switzerland: UNAIDS/WHO; Epidemiological Fact Sheet on HIV and AIDS: Core data on epidemiology and response, United Republic of Tanzania. 2008 Edited by UNAIDS/WHO.