Abstract
The endless quest for the ‘fountain of youth’ has led to the discovery of a family of molecules known as sirtuins in humans, or silent mating type information regulator 2 (Sir2) in yeast, which are associated with longevity in yeast, nematodes, drosophila and rodents. Although sirtuins have yet to be proven to delay aging and promote longevity in humans, they promise ‘healthy aging’, an ideal of modern society. This review emphasizes the role of various sirtuins in maintaining glucose homeostasis, the therapeutic potential of sirtuin modulators in the prevention and treatment of diabetes, and the emerging associations of SIRT genetic polymorphisms with human longevity.
Introduction
One of the major health crises that we face today is an escalating trend of obesity and associated disorders such as type 2 diabetes mellitus (T2DM) and cardiovascular diseases (CVD). The incidence of T2DM is rising at an alarming rate, affecting more than 170 million people worldwide, and is expected to affect in excess of 300 million by 2025 [1]. Studies on the centenarian population suggest that various lifestyle factors including a lower calorie diet are major contributors to ‘healthy longevity’. Calorie restriction (CR) without malnutrition has been demonstrated to increase lifespan in yeast, nematodes, drosophila and rodents by reducing age-associated diseases such as cancer, neurological disorders, CVD and diabetes [2]. The regulatory gene that mimics CR response is the silencing information regulator 2 (Sir2) gene, a nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase enzyme that regulates important biological and metabolic pathways in eubacteria, archaea and eukaryotes [3–7]. A growing list of reviews elegantly describe the evolution, structure and metabolic regulatory functions of sirtuins which are associated with aging, cancer, neurodegenerative diseases, metabolic disorders and longevity [8–16]. The focus of this review is to compile the accumulating evidence for the role of sirtuins as prospective therapeutic targets for T2DM, and the effect of SIRT genetic polymorphism upon human health and longevity.
Mammalian sirtuins
Sirtuins are class III histone deacetylases (HDACs) that were initially associated with transcriptional repression, specifically through deacetylation of lysine 16 residues of histone H4. It is now apparent that non-histone proteins such as the nuclear factor-κB (NFκB), forkhead box type O transcription factors (FOXO), HIV Tat and the peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α), enzymes such as acetyl coenzyme A (CoA) synthetase 2 (AceCS2), and structural proteins such as α-tubulin, are also deacetylated by sirtuins.
There are seven mammalian sirtuins (SIRT1 to SIRT7) which differ in their intracellular location, substrate specificities, and biological functions (reviewed in [10,12,14,17]). The most widely characterized mammalian homolog of Sir2, SIRT1, is a NAD-dependent deacetylase which is located in the nucleus. SIRT1 acts as a nutrient-sensitive growth suppressor [18] and is a principal modulator of pathways downstream of CR that produces beneficial effects on glucose homeostasis and insulin sensitivity and is implicated in regulating lifespan. Regardless of their nuclear location, neither SIRT6 and SIRT7 co-localizes with SIRT1, but SIRT6 is associated with heterochromatin and SIRT7 is concentrated in the nucleolus [19–21]. In contrast to SIRT1, SIRT6 possess auto-ADP-ribosyltransferase activity and is implicated in DNA repair and modulation of telomeric chromatin [19,22,23]. While the substrates of SIRT6 and SIRT7 are still elusive, the biological role of SIRT7 is emerging. Overexpression of SIRT7 in HEK293T, U2OS, and NIH-3T3 cells significantly increased RNA polymerase I (Pol I)-mediated transcription, whereas knockdown of SIRT7 or inhibition of its activity resulted in reduced association of Pol I with rDNA with a concomitant reduction in Pol I transcription [24]. Knockdown of SIRT7 in mice results in heart hypertrophy and inflammatory cardiomyopathy with increases in basal apoptosis and reduced resistance to genotoxic and oxidative stress [25].
SIRT2 is localized in the nucleus as well as in the cytoplasm, and is involved in cell cycle and mitosis regulation [26–28]. SIRT3, SIRT4 and SIRT5 are mitochondrial proteins, with SIRT3 being associated with protein deacetylation and adaptive thermogenesis. SIRT4 which exhibits mono-ADP-ribosyltransferase activity, is located in mitochondria of several tissues including pancreatic β-cells and is implicated in insulin release and gluconeogenesis [29–31]. Although the function of SIRT5 remains unknown, emerging data demonstrate its possible role in translocating SIRT3 to the nucleus [32]. In general sirtuins can function either as NAD-dependent deacetylases or ribosyltransferases and play an important role in energy homeostasis, metabolic adaptations, aging and longevity
Sirtuins and insulin secretion
Both insulin resistance and β-cell insufficiency, with eventual decreased insulin secretion are major features within the pathophysiology of T2DM. Under normal conditions, postprandial increase in circulating glucose signals insulin release from pancreatic β-cells that stimulate glucose uptake and utilization by peripheral tissues. In contrast, the initial stages of T2DM are associated with resistance to insulin action in the peripheral tissues and hyperinsulinemia. With the progression of disease, high glucose and insulin concentrations induce β-cell degeneration as a result of mitochondrial toxicity [1]. Role of sirtuins in pancreatic homeostasis was initially evident when circulating insulin levels were low in SIRT1−/− mice in response to glucose challenge [33]. Alternatively, overexpression of β-cell-specific SIRT1 demonstrated an improved glucose tolerance and an increase in glucose-stimulated insulin secretion [34].
Within the pancreas, SIRT1 is expressed in β-cells of the islets of Langerhans, as well as glucagon-producing α-cells [33,34]. Increases in circulating glucose result in elevated ATP/ADP ratios in β-cells, the closing of K+ channels, the activation of voltage-gated Ca2+ channels, and Ca2+ influx, ultimately leading to insulin release. SIRT1 sensitizes β-cells by repressing uncoupling protein 2 (UCP2), a mitochondrial inner membrane protein that dissociates mitochondrial respiration and ATP synthesis [33]. UCP2-knock out mice demonstrate an increased insulin secretion and higher ATP levels in pancreatic β-cells [5], while SIRT1−/− mice exhibit low levels of circulating insulin and increased UCP2 levels in β-cells [33]. It was further demonstrated that SIRT1 inhibits UCP2 through binding to its promoter region, thereby leading to the coordinated coupling of mitochondrial respiration to ATP synthesis [33]. It is possible that effects of SIRT1 on regulation of insulin secretion may not be confined to only UCP2 repression, but may also involve other transcription factors such as p53, Ku70, NF-κB, FOXO1, PGC-1α, PPARγ and p300, all of which are important regulators of gene expression within pancreatic β-cells. Chronic exposure to high circulating glucose levels plays an important role in β-cell degeneration in diabetic patients due to oxidative stress. Kitamura et al [35] demonstrated that SIRT1-associated protection of glucose-induced cytotoxicity in pancreatic β-cells was due to nuclear translocation of FOXO1 and sustained FOXO1-mediated activation of MafA and NeuroD transcription factors, involved in Insulin 2 gene expression and apoptosis prevention.
SIRT1 is also highly expressed in pancreatic α-cells which are involved in the fasting response by increasing hepatic gluconeogenesis [34]. In contrast to SIRT1, SIRT4 appears to negatively regulate both glucose stimulated- and amino acid-stimulated insulin secretion (GSIS and AASIS), respectively, in β-cells [29–31]. Depletion of SIRT4 from insulin-producing INS-1E cells significantly increased glucose-stimulated insulin secretion [31]. SIRT4 is a mitochondrial ADP-ribosyltransferase that represses pancreatic glutamate dehydrogenase (GDH) during adequate calorie intake, and prevents glutamine-stimulated insulin release [29]. Interestingly, GDH activity is increased during CR due to alleviation of SIRT4-associated repression, resulting in the activation of AASIS in β-cells [29]. Mass-spectroscopy studies indicated that SIRT4 interacts with insulin degrading enzyme (IDE) and the ADP/ATP carrier proteins, ANT2, ANT3, and negatively regulated insulin secretion [31]. The contradictory roles of these two sirtuins, SIRT1 and SIRT4, within the same tissue is intriguing, and are possibly elicited to regulate insulin secretion under different dietary conditions, from acute starvation to chronic food limitation.
Sirtuins and the regulation of liver glucose homeostasis
The liver is the primary organ regulating glucose homeostasis, and does so by differentially regulating glycogenolysis, gluconeogenesis, glycolysis and fatty acid oxidation within both the starved and fed state. SIRT1 was initially identified as a regulator of hepatic gluconeogenesis through its regulation of FOXO1 [36]. In fasting mice, hepatic SIRT1 deacetylates PGC-1α which promotes its association with hepatic nuclear factor 4 (HNF-4) to induce gluconeogenesis and inhibit the glycolytic pathway [37], ensuring that glucose-dependent tissues such as brain and red blood cells (RBCs) have an ample supply of glucose during starvation or fasting. Upregulation of PGC-1α within the liver of fasted mice was associated with increased levels of NAD+ and pyruvate, further implying that the catalytic activity of SIRT1 is under metabolic control [37]. In contrast, hepatic PGC-1α activity is also upregulated in diabetic mice, increasing gluconeogenesis and contributing to hyperglycemia. The importance of hepatic gluconeogenesis is emphasized by the fact that anti-diabetic drugs such as metformin regulate blood glucose levels by inhibiting gluconeogenesis [38]. Similar to its action in the pancreas, CR ameliorates SIRT4 repression of GDH and increases gluconeogenesis in the liver [29].
In insulin-stimulated liver cells the effects of SIRT1 upon the hepatic insulin signaling cascade are opposite to those on PGC-1α. Zhang et al [39] demonstrated that inhibition of SIRT1 in liver cells impaired insulin signaling due to reduced Akt activation and the inhibition of insulin receptor substrate 2 (IRS-2) phosphorylation. These studies suggest that insulin-induced phosphorylation of the insulin receptor (IR) is insufficient to phosphorylate IRS2. Instead, insulin-induced increase in SIRT1 is necessary to deacetylate the lysine residues of IRS-2 before its tyrosine residues can be phosphorylated [39]. Based on the above evidence, both SIRT1 and SIRT4 are attractive targets in the modulation of gluconeogenesis, the major contributor of elevated glucose levels during diabetes.
Sirtuins and metabolic adaptations in peripheral tissues
T2DM is associated with impaired glucose uptake and utilization in peripheral tissues such as muscle and adipose tissue, resulting in increased blood glucose and secondary hyperinsulinemia. Studies by Sun et al [40] indicate that resveratrol-associated activation of SIRT1 improves insulin sensitivity and ameliorates peripheral insulin resistance in part due to suppression of protein tyrosine phosphatase 1B (PTP1B), a negative regulator of insulin signaling. Most of the anti-diabetic drugs are prone to secondary failure due to secondary weight gain as a result of increased adipose differentiation. SIRT1 regulators such as CR and resveratrol demonstrate reductions in fat accumulation within adipose tissue of mice fed a high-fat-diet (HFD [41,42]). PPARγ stimulated fat synthesis and differentiation in white adipose tissue during CR or resveratrol treatment [43]. Binding of SIRT1 to PPARγ promoter ensures the docking of the nuclear receptor corepressor (NcoR) and thyroid hormone receptor, and recruitment of this complex to adipose tissue-specific fatty acid binding protein (aP2) promoter [41], thereby preventing the accumulation of TG in adipose tissue. It is further possible that anti-adipogenic effects of SIRT1 are mediated through the activation of FOXO1 since FOXO1 represses PPARγ activity [44]. SIRT1 also regulates the secretion of adiponectin in the adipocytes, an adipocytokine that is involved in sensitizing the liver and muscle to insulin.
In skeletal muscles SIRT1 has an opposing effect on myogenesis, depending upon glucose availability. In the absence of glucose, insulin induces massive upregulation of SIRT1 and FoxO3a, whilst inhibiting myocyte differentiation [45]. During fasting, metabolic control in the skeletal muscle of mice was achieved by SIRT1 through deacetylation of PCG-1α and activating mitochondrial fatty acid oxidation genes [46]. The switch from glucose to fatty acid oxidation during low nutrient supply was dependent upon SIRT1 activation in these fasting animals, and was correlated to high NAD+ levels in skeletal muscle [46]. In another study, feeding of fructose resulted in SIRT1 dependent induction of the α-myosin heavy chain (MHC) in cardiac muscle and prevented ischemia- and reperfusion-mediated cell injury in the heart [47]. It is evident from the above studies that in the muscle SIRT1 functions not only to allow metabolic adaptations to nutrient supply, but also switches energy homeostasis in muscle from the oxidation of glucose to that of fatty acids, enabling glucose utilization by neurons and RBCs during nutrient deprived states.
Uncoupling of mitochondrial respiration in skeletal muscles due to increased UCP1 expression was correlated with a reduced age-related disease burden in mice demonstrating reduced adiposity, increased temperature and metabolic rate, elevated muscle SIRT and AMP kinase, and increased serum adiponectin [48]. Long-term fasting or diabetes induces the release of large amounts of acetate from the liver due to increased ketogenesis [8]. Mitochondrial acetyl-CoA synthetase (AceCS) expressed in heart, muscle, brain and kidney, but not in the liver, converts the circulating acetate into acetyl CoA that enters the tricarboxylic cycle (TCA) for energy production. Cytoplasmic (AceCS1) and mitochondrial (AceCS2) are differentially regulated by SIRT3 [49,50]. Fasting induced AceCS2 and reduced AceCS1 in muscle, thereby diverting the acetate to TCA cycle in extrahepatic tissues. In streptozocin-induced diabetic mice SIRT3 mRNA is down-regulated [51], possibly accounting for the increased ketogenesis observed during diabetes.
SIRT1 and diabetic complications
The importance of tight glycemic control in the prevention of diabetic complications such as dyslipidemia, neuropathy and nephropathy, is widely recognized. Diabetic nephropathy (DN) is one of the leading causes of end stage renal disease and is associated with proteinurea resulting from a damaged kidney filtration system. Glomerular lesions with thickening of basement membrane, endothelial cell proliferation and apoptosis in tubular and interstitial cells are observed in DN [52]. Pathophysiological studies associate stress-activated signaling pathways, such as NF-κB, p38 mitogen activated protein kinase (MAPK) and Jun kinases, with diabetic complications [52]. Intermittent fasting in Sprague-Dawley rats demonstrated a nephroprotective effect in diabetic conditions which was associated with activation of Sir2 and a reduction in proaopoptotic p53 [52]. Resveratrol prevented the decrease of SIRT1 expression as well as an increase in p38, p53 expression and dephosphorylation of histone H3 in rat diabetic kidney [53]. Sirtuins therefore present attractive targets not only in the treatment of insulin resistance and diabetes, but also their associated complications.
Sirtuin regulation
Although several SIRT substrates have been identified and characterized, very little is known about the actual regulators of SIRT activity. Two models were initially proposed in Saccharomyces cerevisiae to explain how CR might stimulate Sir2 activity. The first model proposed a role for increased NAD+ or the NAD+:NADH ratio to increase Sir2 activity, while the second model suggested depletion of nicotinamide (NAM), a feedback inhibitor of Sir2 metabolism per se [54,55]. In yeast, bacteria, worms and flies, NAD is synthesized de novo in four steps from tryptophan, and is recycled from nicotinamide via the NAD+ salvage pathway [56], while in mammals it is a two step process [57,58]. In yeast, the first step is catalyzed by Pnc1 while in mammals NAM phosphoribosyltransferase (Nampt, also known as PBEF to visfatin) is the rate-limiting enzyme [58,59]. Studies indicate that yeast Sir2 activity is enhanced by Pnc1, a stress- and calorie-responsive gene catalyzing the first rate-limiting step in NAD+ biosynthesis [59–61]. SIRT1 activity is increased due to Nampt over expression [62] and was demonstrated to protect cells from apoptosis due to PARP overexpression [63]. In mammals, fasting significantly increased levels of Nampt with a concomitant increase in mitochondrial NAD+. Genotoxic stress-associated cell death was inhibited by increased levels of Nampt and mitochondrial NAD+ through mitochondrial specific sirtuins, SIRT3 and SIRT4.
Besides an increase in NAD+ biosynthetic pathway, inhibition of NAD+ hydrolysis also is an important regulator of SIRT activity. CD38 is located on the inner nuclear membrane and is involved in the synthesis of secondary messengers such as cADPR, NAADP, and ADPR, as well as in the hydrolysis of NAD+ ([64] and references within). Hepatic levels of NAD+ and nuclear SIRT1 activity were significantly increased in CD38 knockout mice, with an increase in deactylation of p53, a SIRT1 substrate, suggesting that CD38 is a major regulator of nuclear NAD+ levels and SIRT1 activity in mammals [64]. The oxidation and reduction of NAD and NADH are involved in anabolic and catabolic reactions within mitochondria. Under aerobic conditions the mitochondrial electron transport chain reoxidizes NAD from NADH. However, the mitochondrial membrane is impermeable to NAD/NADH. Thus, the malate-aspartate NADH shuttle is extremely critical to maintaining the NAD:NADH ratio within mitochondria and the cytosol. Recent studies indicate that malate-aspartate shuttle is vital to metabolic regulation and CR-induced lifespan extension in yeast [65].
SIRT1 is also regulated at the transcriptional level by various enhancers and suppressors. SIRT1 transcription is suppressed by p53 feedback regulation in unstimulated rodent cells which was relieved by the association of Foxo3a [66]. Tumor suppressor HIC1 and E2F1 also transcriptionally represses SIRT1 activity by binding to its promoter region [67–69], suggesting that SIRT1 expression is subject to feedback autoregulation. SIRT1 is also subject to regulation by various transcriptional enhancers and protein suppressors such as active regulator of SIRT1 (AROS) and deleted in breast cancer-1 (DBC1), respectively [70–73]. Studies by Kim et al. demonstrate that AROS directly binds to SIRT1 and enhances SIRT1-mediated p53 deacetylation both in vitro and in vivo, thereby regulating p53-mediated transcriptional activity as well as p53-induced cell growth responses to DNA damage [72]. SIRT1 activity can be further regulated by direct-protein-protein interactions by a DBC1 protein which forms a stable complex with SIRT1 and reduces its activity. DBC1 directly binds to the catalytic domain of SIRT1, preventing substrate access and inhibiting genotoxic stress-induced apoptosis [72,73]. Studies by Abdelmohsen demonstrate that stability of SIRT1 mRNA was increased by binding of HuR protein to its 3′ untranslated region, with a concomitant increase in its protein expression [74,75]. Oxidative stress triggered the dissociation of the HuR SIRT1 mRNA complex due to increased phosphorylation of HuR by stress-associated increases in Chk2 cell cycle kinase activity [74,75], and promoted SIRT1 mRNA decay, thereby reducing SIRT1 expression and cell survival. It is therefore evident that different SIRT regulators may influence the downstream targets of SIRT and thus differentially modulate disease and lifespan outcomes.
Sirtuin modulators
The obvious role of SIR2 in regulating lifespan within diverse organisms and their coupling to NAD+ metabolism has stimulated a prevailing interest in the possibilities of identifying therapeutic SIRT regulators as CR mimetics that will reduce the age-associated disease burden, disease-associated morbidity and mortality, and thereby increase lifespan in humans. SIRT1 is now considered a ‘metabolic master switch’ [76] that extends lifespan in various species including mammals, by modulating pathways downstream of CR. Sirtuins regulate apoptosis, DNA repair, stress resistance, metabolism, aging and endocrine signaling, suggesting that they could be targeted for therapeutic benefit. CR is an extremely daunting task and the quest is therefore directed towards identifying prospective modulators of SIRT activity.
To date, numerous chemical activators and inhibitors have been identified and their detailed mechanisms have been reviewed elsewhere [77]. Much attention has been given to resveratrol, a polyphenolic SIRT1 activator, present in red wine, that was demonstrated to mimic anti-aging effects, ameliorate insulin resistance in mice fed HFD, increase mitochondrial biogenesis and promote longevity [78,79]. Since then, various other small molecules have been identified with varying SIRT activity, but lower than that of resveratrol [80–86]. Interestingly a recent study has identified SIRT1 activators that are structurally unrelated but which are 1,000-fold more potent than resveratrol [78]. These compounds were demonstrated to improve whole body glucose tolerance, insulin sensitivity, and mitochondrial function in diet-induced and genetically obese mice [78]. Although not direct activators of CR, vitamins such as niacin (B3) are important for NAD+ biosynthesis, and are implicated in regulating SIRT1 activity [87–89]. Another interesting study demonstrated the efficacy of the yeast Kluyveromyces biopeptides in activating SIRT1 in human skin cells, leading to improved resistance to stress and senescence and its usefulness in treating aging skin in humans [90], while resveratrol containing skin care formulae have demonstrated 17-fold greater antioxidant activity than idebenone [91]. These data suggest that CR mimetics and/or SIRT activators may offer a new therapeutic approach to aging and related disorders such as T2DM [78]. However, due to contradictory roles of sirtuins in different tissues, one has to exercise caution in selecting prospective targets for sirtuin therapy. For example induction of SIRT1 in diabetics may reduce adipogenesis [41] and increase insulin release in pancreas [33,34], but will concomitantly increase gluconeogenesis in liver [92], leading to increased blood glucose.
CR in humans
CR extends lifespan and produces a metabolic profile desirable for treating diseases of ageing such as T2DM. Regardless of all the available data on CR, sirtuins and longevity in various species, it is still unclear how CR and/or sirtuins or their downstream targets influence diabetes, aging and longevity in humans. In the animal kingdom, CR is the only proven way to extend life. Now, a group of individuals are trying to pursue 30–40% CR. They refer to themselves as ‘CRONies’, for calorie restricted, on optimal nutrition. Studies on CR in humans (www.jhu.edu/jhumag/1101web/eat.html) demonstrate that 25% reduction in calorie intake alone or in combination with exercise significantly reduced 24 hour energy expenditure, and improved mitochondrial function by increasing mitochondrial mass and gene expression of peroxisome proliferator-activated receptor gamma coactivator 1α (PPARGC1α), transcription factor A mitochondrial (TFAM), endothelial nitric oxide synthase (eNOS), SIRT1, and presenilin associated rhomboid-like protein (PARL), involved in nutrient sensing and mitochondrial biogenesis [93]. Furthermore, CR significantly reduced mitochondrial oxidative stress and increased mtDNA content although it had no significant effect on the activities of key mitochondrial enzymes of the TCA cycle (citrate synthase), beta-oxidation (beta-hydroxyacyl-CoA dehydrogenase), or electron transport chain (cytochrome C oxidase II [93]). These studies suggest a role for CR in improving mitochondrial function in humans. Similarly, the frequency of mtDNA J haplogroup was higher among Italian centenarians than in younger individuals and was associated with longevity [94].
Longevity and SIRT polymorphisms
Longevity has a strong genetic component, as apparent from studies in yeast through to humans. Allelic association studies among centenarians demonstrated a strong correlation between longevity and polymorphism of four genes located on 11p15.5 chromosome region viz. the tyrosine hydroxylase (TH) gene, as well as the haplotypes defined by this marker and RFLPs of proinsulin (INS), Insulin-like growth factor 2 (IGF2) and HRAS1 genes [95]. The human SIRT3 gene is also located at the telomeric terminal on 11p15.5 chromosome. Analysis of G477T, a silent marker of SIRT3 gene among Italian male centenarians demonstrated that TT genotype increases (p= 0.0272); while the GT genotype decreases (p=0.0391) survival in the elderly [95]. Linkage-disequilibrium studies between G477T alleles and alleles of the longevity genes, TH, INS, IGF2 and HRAS1, indicated that disequilibrium was not significant in any case, thus suggesting that either SIRT3, or another gene linked to SIRT3, may play an important role in human longevity [95]. A follow up study by the same group further identified a functional variant that could account for the association observed in the above study, between the silent marker of SIRT3 and survival [96]. These functional studies indicated that the allele completely lacking enhancer activity is virtually absent in males older than 90 years. Thus reduced expression of a human sirtuin gene seems to be detrimental for longevity as in other organisms [96].
Another pivotal study investigated the association between common allelic variation in the SIRT1 gene and human longevity among German centenarians [97]. In this study, five single nucleotide polymorphisms (SNPs) of SIRT1, distributed across the entire gene, including the promoter region, were genotyped among centenarians and nonagenarians and matched with younger controls. No association was detected between any of the tested SIRT1 SNPs and longevity among this population [97]. Besides longevity, studies have investigated the relation between SIRT1 polymorphism and age-related disease such as CVD and cognitive functioning. In the prospective ‘Leiden 85-plus Study’ amongst older inhabitants of Leiden in the Netherlands, Kuningas et al. assessed five SIRT1 SNPs in 1,245 participants and demonstrated that the rs3758391 T-allele carriers had a better cognitive function [98]. In addition, Kuningas et al. further demonstrated a trend for lower CVD mortality for carriers of haplotype 2 and rs3758391 T-allele [98]. Similarly, SIRT1 protein levels and its activity were shown to be reduced in peripheral lung tissue and macrophages of smokers and patients with chronic obstructive pulmonary disease [99]. Reduction in SIRT1 activity was found to be a result of post-translational modification leading to increased acetylation of RelA/p65 subunits of NF-κB, an important modulator of inflammation [99]. Such functional studies indicate the possibility that although allelic variants of SIRT demonstrate limited correlation with longevity, SIRT1 does play an important role in age-related disease ultimately affecting lifespan in humans.
A genome wide association study for longevity-related traits among Framingham Study participants revealed a strong association between age at death and SNPs intronic to FOXO1A, a gene implicated in lifespan extension in animal models, and a known target of SIRT1 [100]. Similarly, genetic variations in other target genes of SIRT1, such as insulin-like growth factor 1 (IGFI) and its receptor (IGFR [101]) and p53 [102], have been associated with longevity in Ashkenazi Jewish centenarians and Danish populations, respectively. Overall, association studies between genetic variants of SIRT and longevity in humans have yielded limited information. Similar studies are needed within different long-lived populations. Moreover, further studies on gene-environment and gene-diet interactions are warranted to understand the genotype/phenotype relationships between SIRT polymorphisms and exposure to SIRT modulators (such as the consumption of red wine). Last, but not least, the precise role of sirtuins in disease development and prevention has yet to be elucidated.
Conclusions
Healthy aging and longevity depend upon successful and dynamic interactions among biological, psychological, and environmental factors. We are just beginning to assimilate the various findings for CR and sirtuin functions. Manipulation of genes such as sirtuins and their downstream metabolic targets will unveil novel strategies for intervention and prevention within age-associated diseases such as T2DM. Gene therapy, stem cells, and modulation of sirtuins through functional foods, nutriceuticals, cosmeceuticals and lifestyle alterations, including mild stress-induced hormesis, are examples of such strategies that may in the fullness of time increase longevity and reduce disease burden.
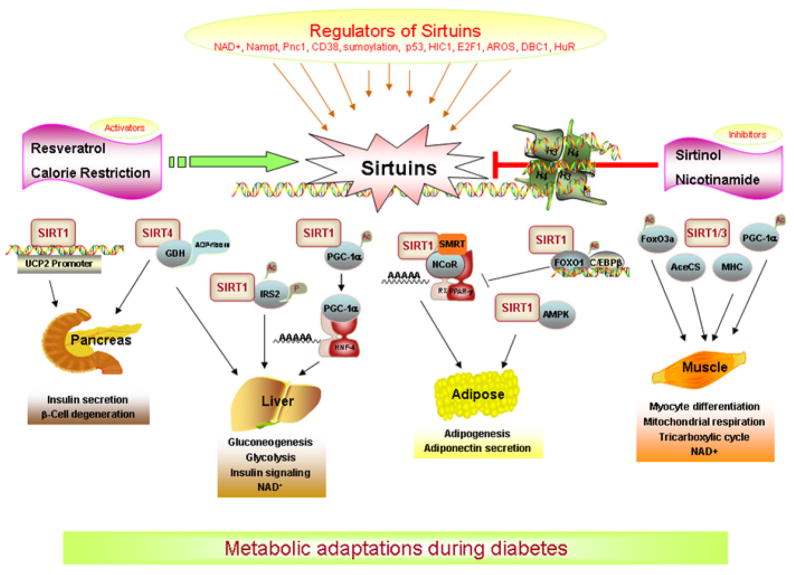
Figure 1. Role of sirtuins in diabetes.
During calorie restriction, sirtuins inhibit insulin release in the pancreas and inhibit or prevent β-cell degeneration, promote gluconeogenesis and insulin signaling, inhibit glycolysis and adipose differentiation, and prevent ketogenesis. Overall, sirtuins promote metabolic adaptations during diabetes.
Acknowledgments
This work was supported in part by U.S. Public Health Service grants from the National Institutes of Health, National Center for Complementary and Alternative Medicine (NCCAM, R21AT003719), Research Centers in Minority Institutions (RCMI, G12 RR003061), National Center for Research Resources (NCRR, P20 RR011091), National Center on Minority Health and Health Disparities (NCMHD, P20 MD000173) and the Hawaii Community Foundation (20041652).
References
- 1.Rolo AP, Palmeira CM. Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol. 2006;212:167–78. doi: 10.1016/j.taap.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 3.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–6. [PubMed] [Google Scholar]
- 4.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 5.Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb Symp Quant Biol. 2000;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- 6.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–63. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Pallas M, Verdaguer E, Tajes M, Gutierrez-Cuesta J, Camins A. Modulation of sirtuins: new targets for antiageing. Recent Patents CNS Drug Discov. 2008;3:61–9. doi: 10.2174/157488908783421492. [DOI] [PubMed] [Google Scholar]
- 10.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med. 2007;39:335–45. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 11.Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007;6:1011–8. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 13.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–21. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 15.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–68. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol Metab. 2006;17:186–91. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Anastasiou D, Krek W. SIRT1: linking adaptive cellular responses to aging-associated changes in organismal physiology. Physiology (Bethesda) 2006;21:404–10. doi: 10.1152/physiol.00031.2006. [DOI] [PubMed] [Google Scholar]
- 18.Narala SR, Allsopp RC, Wells TB, Zhang G, Prasad P, Coussens MJ, Rossi DJ, Weissman IL, Vaziri H. SIRT1 Acts as a Nutrient-sensitive Growth Suppressor and Its Loss Is Associated with Increased AMPK and Telomerase Activity. Mol Biol Cell. 2008;19:1210–9. doi: 10.1091/mbc.E07-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–6. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–35. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 22.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–41. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 24.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–80. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–10. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 26.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–61. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–85. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 29.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 30.Argmann C, Auwerx J. Insulin secretion: SIRT4 gets in on the act. Cell. 2006;126:837–9. doi: 10.1016/j.cell.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–92. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–9. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 33.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–17. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–63. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–95. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 37.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–6. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radziuk J, Bailey CJ, Wiernsperger NF, Yudkin JS. Metformin and its liver targets in the treatment of type 2 diabetes. Curr Drug Targets Immune Endocr Metabol Disord. 2003;3:151–69. doi: 10.2174/1568008033340298. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J. The direct involvement of SirT1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. J Biol Chem. 2007;282:34356–64. doi: 10.1074/jbc.M706644200. [DOI] [PubMed] [Google Scholar]
- 40.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–19. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–14. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 44.Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–91. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- 45.Nedachi T, Kadotani A, Ariga M, Katagiri H, Kanzaki M. Ambient glucose levels qualify the potency of insulin myogenic actions by regulating SIRT1 and FoxO3a in C2C12 myocytes. Am J Physiol Endocrinol Metab. 2008;294:E668–78. doi: 10.1152/ajpendo.00640.2007. [DOI] [PubMed] [Google Scholar]
- 46.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pillai JB, Chen M, Rajamohan SB, Samant S, Pillai VB, Gupta M, Gupta MP. Activation of SIRT1, a class III histone deacetylase, contributes to fructose feeding-mediated induction of the {alpha}-myosin heavy chain expression. Am J Physiol Heart Circ Physiol. 2008;294:H1388–97. doi: 10.1152/ajpheart.01339.2007. [DOI] [PubMed] [Google Scholar]
- 48.Gates AC, Bernal-Mizrachi C, Chinault SL, Feng C, Schneider JG, Coleman T, Malone JP, Townsend RR, Chakravarthy MV, Semenkovich CF. Respiratory uncoupling in skeletal muscle delays death and diminishes age-related disease. Cell Metab. 2007;6:497–505. doi: 10.1016/j.cmet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, Kahn CR. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A. 2004;101:16525–30. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tikoo K, Tripathi DN, Kabra DG, Sharma V, Gaikwad AB. Intermittent fasting prevents the progression of type I diabetic nephropathy in rats and changes the expression of Sir2 and p53. FEBS Lett. 2007;581:1071–8. doi: 10.1016/j.febslet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Tikoo K, Singh K, Kabra D, Sharma V, Gaikwad A. Change in histone H3 phosphorylation, MAP kinase p38, SIR 2 and p53 expression by resveratrol in preventing streptozotocin induced type I diabetic nephropathy. Free Radic Res. 2008;42:397–404. doi: 10.1080/10715760801998646. [DOI] [PubMed] [Google Scholar]
- 54.Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–4. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 55.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 56.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–90. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 57.Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25:683–90. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 58.Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–34. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 59.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–76. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 61.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 63.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–30. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 64.Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN. Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun. 2006;349:353–9. doi: 10.1016/j.bbrc.2006.08.066. [DOI] [PubMed] [Google Scholar]
- 65.Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–44. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–8. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 67.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–48. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–31. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 69.Nowak K, Killmer K, Gessner C, Lutz W. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim Biophys Acta. 2007;1769:244–52. doi: 10.1016/j.bbaexp.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Verdin E. AROuSing SIRT1: identification of a novel endogenous SIRT1 activator. Mol Cell. 2007;28:354–6. doi: 10.1016/j.molcel.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 71.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 72.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 73.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–92. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 75.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–57. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leibiger IB, Berggren PO. Sirt1: a metabolic master switch that modulates lifespan. Nat Med. 2006;12:34–6. doi: 10.1038/nm0106-34. discussion 36. [DOI] [PubMed] [Google Scholar]
- 77.Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 78.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 80.Quideau S. Plant ‘polyphenolic’ small molecules can induce a calorie restriction-mimetic life-span extension by activating sirtuins: Will ‘Polyphenols’ someday be used as chemotherapeutic drugs in western medicine? ChemBioChem. 2004;4:427–430. doi: 10.1002/cbic.200300835. [DOI] [PubMed] [Google Scholar]
- 81.de Boer VC, de Goffau MC, Arts IC, Hollman PC, Keijer J. SIRT1 stimulation by polyphenols is affected by their stability and metabolism. Mech Ageing Dev. 2006;127:618–27. doi: 10.1016/j.mad.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 82.Morris BJ. A forkhead in the road to longevity: the molecular basis of lifespan becomes clearer. J Hypertens. 2005;23:1285–309. doi: 10.1097/01.hjh.0000173509.45363.dd. [DOI] [PubMed] [Google Scholar]
- 83.Opie LH, Lecour S. The red wine hypothesis: from concepts to protective signalling molecules. Eur Heart J. 2007;28:1683–93. doi: 10.1093/eurheartj/ehm149. [DOI] [PubMed] [Google Scholar]
- 84.Grubisha O, Smith BC, Denu JM. Small molecule regulation of Sir2 protein deacetylases. Febs J. 2005;272:4607–16. doi: 10.1111/j.1742-4658.2005.04862.x. [DOI] [PubMed] [Google Scholar]
- 85.Zhou B, Wu LJ, Li LH, Tashiro S, Onodera S, Uchiumi F, Ikejima T. Silibinin protects against isoproterenol-induced rat cardiac myocyte injury through mitochondrial pathway after up-regulation of SIRT1. J Pharmacol Sci. 2006;102:387–95. doi: 10.1254/jphs.fpj06005x. [DOI] [PubMed] [Google Scholar]
- 86.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 87.Denu JM. Vitamin B3 and sirtuin function. Trends Biochem Sci. 2005;30:479–83. doi: 10.1016/j.tibs.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 88.Denu JM. Vitamins and aging: pathways to NAD+ synthesis. Cell. 2007;129:453–4. doi: 10.1016/j.cell.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 89.Sauve AA. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther. 2008;324:883–93. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- 90.Moreau M, Neveu M, Stephan S, Noblesse E, Nizard C, Sadick NS, Schnebert S, Bonte F, Dumas M, Andre P, Perrier E. Enhancing cell longevity for cosmetic application: a complementary approach. J Drugs Dermatol. 2007;6:s14–9. [PubMed] [Google Scholar]
- 91.Baxter RA. Anti-aging properties of resveratrol: review and report of a potent new antioxidant skin care formulation. J Cosmet Dermatol. 2008;7:2–7. doi: 10.1111/j.1473-2165.2008.00354.x. [DOI] [PubMed] [Google Scholar]
- 92.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 93.Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafé M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 95.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–70. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 96.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–63. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 97.Flachsbart F, Croucher PJ, Nikolaus S, Hampe J, Cordes C, Schreiber S, Nebel A. Sirtuin 1 (SIRT1) sequence variation is not associated with exceptional human longevity. Exp Gerontol. 2006;41:98–102. doi: 10.1016/j.exger.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 98.Kuningas M, Putters M, Westendorp RG, Slagboom PE, van Heemst D. SIRT1 gene, age-related diseases, and mortality: the Leiden 85-plus study. J Gerontol A Biol Sci Med Sci. 2007;62:960–5. doi: 10.1093/gerona/62.9.960. [DOI] [PubMed] [Google Scholar]
- 99.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an Anti-Inflammatory and Anti-Aging Protein, is Decreased in Lungs of Patients with COPD. Am J Respir Crit Care Med. 2008 doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lunetta KL, D’Agostino RB, Karasik D, Benjamin EJ, Guo CY, Govindaraju R, Kiel DP, Kelly-Hayes M, Massaro JM, Pencina MJ, Seshadri S, Murabito JM. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Medical Genetics. 2007;8 (suppl 1):S1–S13. doi: 10.1186/1471-2350-8-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Nat Acad Sci (USA) 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bojesen SE, Nordestgaard BG. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle. 2008;7:158–163. doi: 10.4161/cc.7.2.5249. [DOI] [PubMed] [Google Scholar]