Abstract
Metastasis is a critical event in oral squamous cell carcinoma (OSCC) progression. To identify proteomic biomarkers for OSCC metastasis, 3 paired OSCC cell lines (UM1/UM2, 1386Tu/1386Ln, 686Tu/686Ln) with different metastatic potential were examined. Among those 3 cell lines, UM1, 1386Ln and 686Ln exhibited a higher degree of metastatic potential than their paired cell lines UM2, 1386Tu and 686Tu, respectively, as measured using an in vitro cell invasion assay. A total of 40 differentially expressed proteins were identified using 2D-PAGE/MS proteomic approach. Selected protein candidates (superoxide dismutase 2 and heat shock protein 27) were further investigated by immunohistochemistry (IHC) method using independent OSCC patient tissue samples. The statistically significantly increases in IHC staining for manganese superoxide dismutase 2 (SOD2) were observed in lymph node metastatic disease when compared with the paired primary OSCC. Our results thus indicated that elevated SOD2 levels is associated with lymph node metastasis in OSCC and may provide predictive values for diagnosis of metastasis.
Keywords: SOD2, manganese superoxide dismutase 2, oral cancer, OSCC, metastasis, proteomic biomarker, 2D-PAGE/MS
Oral cancer, predominantly oral squamous cell carcinoma (OSCC), is one of the most devastating diseases. The American Cancer Society estimated that more than 30,000 new cases of oral cancer were diagnosed in 2006, representing approximately 3% of all malignancies in men and 2% of all malignancies in women. Worldwide oral cancer is a major cancer problem (ranked as the 6th), with an estimated 300,000 new cases diagnosed annually (1, 2). Despite the tremendous improvements in surgery, radiotherapy and chemotherapy over the last decade, the prognosis for patients with OSCC is more or less unchanged for the past 3 decades. This is because patients continue to die from metastatic diseases at regional and distant sites. Improvement in patient survival requires an increased understanding of tumor metastasis so that aggressive tumors can be detected early in the disease process and targeted therapeutic interventions can be developed.
Currently, no molecular biomarkers have been included in clinical work-up strategies for the detection of nodal metastasis. Since several genes have been reported in retrospective trials to yield prognostic information independently of the TMN classification, it is reasonable to hypothesize that molecular “fingerprints” could exist that might define sub-groups of patients with significantly more aggressive disease. The tumor cells may progress via the bloodstream or the lymphatics to colonize new areas of the body. The metastasis of the OSCCs is unique in that they metastasize mainly to regional lymph nodes through the draining lymphatics, where metastasis to distant sites is relatively uncommon. Several recent gene expression studies from this and other laboratories have suggested the existence of such “fingerprints” in the primary tumor for metastasis of OSCC (3-6). In this study, we carry out the proteomic analysis of paired OSCC cells with different metastasis potentials to identify these “fingerprints” for metastasis, and further validate our findings using clinical samples.
Materials and Methods
Cell culture and in vitro cell invasion assay
Human OSSC cell lines, UM1, UM2, MDA1386TU, 1386LN, MDA686TU and 686LN cells were maintained in DMEM containing 10% FBS, 1,000 units/mL penicillin, and 500 μg/mL streptomycin (GIBCO) in a 37°C incubator with 5% CO2.
The Cultrex 96-well membrane invasion assay kit (Basement Membrane Extract, R&D Systems) was used to determine the ability of UM1, UM2, MDA1386TU, 1386LN, MDA686TU and 686LN cells to invade into Basement Membrane Extract (BME)-coated filters according to manufacturer's instruction (www.rndsystems.com/pdf/3455-096-K.pdf). Briefly, in day 1, 50 μl of 0.5×BME coating solution was placed in each well in the cell invasion device, and cells were starved in serum-Free medium. In day 2, the cells were harvested and seeded into the upper chamber at 5×105 cells/well after incubating for 24 hours in serum-Free medium. 150 μl of medium per well was added to the bottom chamber (10% FBS or without FBS). The chambers were placed in a humidified atmosphere with 5% CO2 at 37°C for 42 hours. In day 3, medium was aspirated from the top/bottom chambers and each well was washed with 1× wash buffer. 100 μl of 1× Calcein AM was added to the bottom chamber, the cell invasion device was then assembled, and incubated at 37°C for 1 hour. Experiments were run in quadruplicate. The top chamber was removed, and the bottom plate was measured at 485 nm excitation and 520 nm emission. The data was compared to the standard curve to determine the number of cells that have invaded. A separate standard curve was used for each cell type. The percentage of cell invasion was calculated as the number of the invaded cells divided by the number of the cells at the start of the assay.
Proteomic analysis
Ten million cells were centrifuged at 2000g for 10 minutes at 4°C in a microfuge. When centrifugation was complete, the medium was removed by aspiration, leaving the cell pellet as dry as possible. The proteins were extracted with rehydration buffer consisting of 8 M urea, 4% CHAPS, 65 mM DTE, 0.5% ampholytes, and a trace of bromophenol blue; the final volume was 200 μL. The protein solution was then spun at 100,000 RPM for 30 minutes at 4°C and then 130 μl was absorbed into a 7 cm immobilized pH gradient pH 3-10 IPG strip (IPG strip, GE Bioscience) overnight. The first isoelectric focusing dimension was conducted for a total of 60 kVhr (PROTEAN IEF cell, Bio-Rad) at room temperature. Before SDS-PAGE electrophoresis, IPG strips were equilibrated with 3 ml of an equilibrium solution containing 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, a trace of bromophenol blue, and DTE (1% w/v) for 20 min, followed by a second equilibration for 20 min in the same equilibrium solution containing iodoacetamide (2.5% w/v) instead of DTE. Finally, the strips were transferred to the top of 4-12% Nu-Page gels and held in position with molten 0.5% agarose in running buffer containing 25 mM Tris, 0.192 M glycine, 0.1% SDS. Gels were run at constant voltage of 200 V for 45 minutes. The gels were fixed and stained overnight with Coomassie blue staining solution [50% (v/v) methanol, 0.2% (w/v) Coomassie brilliant blue R-250, and 10% (v/v) acetic acid], and destained by washing 3 times in destaining solution [50% (v/v) methanol and 10% (v/v) acetic acid] for 1 h each time. Protein spots were quantified using the PDQuest software (Protein Databases, Inc, Huntington Station, NY, USA). All gels were also visually inspected to check for errors in spot detection. Although spot density can vary from gel to gel, it has previously been shown that the relative spot size and density remains relatively stable within known biological samples. Significantly over-expressed spots (>2-fold) were selected for analysis with mass spectrometry. Protein spots on the gel were picked by hand. In-gel tryptic digestion and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry of tryptic peptides was performed as follows. Briefly, gel plugs were washed in 50% acetonitrile, reduced of sulfide bonds in 60 mM DTT, alkylated of free sulfhydryl groups in iodoacetamide, 50 mM ammonium bicarbonate (pH 8.0) and 5 mM EDTA, and then incubated in trypsin [in 50 mM ammonium bicarbonate (pH 8.0) solution at a concentration of 2 μg/100 μl] overnight. For MALDI-TOF, residual peptides were extracted, spotted onto a MALDI-TOF target, and analyzed by a positive-ion reflector mode with delayed extraction over the m/z range 700-4000 using a Voyager DE-PRO Mass Spectrometer (Applied Biosystems, Foster City, CA, USA) equipped with a nitrogen laser. Spectra were externally and internally calibrated. Peptide mass results were used to identify the proteins using the MASCOT Peptide Fingerprint link (http://www.matrixscience.com). Conditions were set to allow for one missed cleavage, a mass tolerance of 50 ppm, and limited to occur within the proximity of the protein's approximate molecular weight.
Western blot analysis
Cells were harvested at near confluence. After washing with cold PBS 3 times, cells were lysed in RIPA buffer of 150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 0.5% Triton X-100, and protease inhibitors (Sigma), homogenized on ice, and centrifuged at 15,000 rpm at 4°C for 15 min. The supernatant was collected and stored at −80°C until use. Protein concentration was determined using the Bradford assay (Bio-Rad). Twenty five μg of protein extraction was loaded on 12% tris-polyacrylamide gels (SDS–PAGE, Bio-Rad, USA). The proteins were then transferred to nitrocellulose membrane (Whatman, Germany). The membranes were blocked in 5% non-fat dry milk, washed in TBS with 0.05% Tween 20 (TBST), and incubated in the appropriate primary antibody solution for 2 hr at room temperature in PBST: 1:3,000 both for mouse anti-SOD2 and mouse anti-HSP27 (ABCam). Mouse anti-beta-actin (ABCam) was set up for internal control. The membranes were washed three times in PBST and incubated for 1 hour with goat anti-mouse antibodies (Bio-Rad) conjugated to horseradish peroxidase at 1:5,000 dilution, washed three times in PBST, and treated with Immun-Star HRP peroxide buffer and Luminol/Enhancer (Bio-Rad) for chemiluminescence detection of protein bands.
Immunohistochemical analysis
The archived tissue samples from 21 OSCC patients with pathologically detectable lymph node metastasis were utilized in this study. Clinical characterization of the OSSC patients was summed in Table I. This study was approved by the ethical committee of The First Affiliated Hospital, Sun Yat-Sen University and the Institutional Review Boards (IRB) at University of Illinois at Chicago, IL, USA.
Table I. Clinical characterization of the OSCC patients.
| OSCC (n=21) | ||
|---|---|---|
| Age | Average | 45.1 |
| (Range) | 22-66 | |
| Gender | Male (%) | 57.1 |
| Female (%) | 42.9 | |
| Pathological T stage | Stage 4 (%) | 19.0 |
| Stage 3 (%) | 23.8 | |
| Stage 2 (%) | 52.4 | |
| Stage 1 (%) | 4.8 | |
| Pathological N stage | Stage 2 (%) | 33.3 |
| Stage 1 (%) | 66.7 |
Tumor tissue was dehydrated in an ethanol series, cleared in xylene, and embedded in paraffin. Five-micrometer sections were prepared and mounted on poly-L-lysine-coated slides. Representative sections were stained with H&E and histologically evaluated by a pathologist. Immunohistochemical analysis was done using a commercially available kit (Invitrogen, Carlsbad, CA, USA). Sections were incubated at 60°C for 30 min and deparaffinized in xylene. Endogenous peroxidase activity was quenched by incubation in a 9:1 methanol/30% hydrogen peroxide solution for 10 min at room temperature. Sections were rehydrated in PBS (pH 7.4) for 10 min at room temperature. Sections were blocked with 10% normal serum for 10 min at room temperature followed by incubation with anti-SOD2 and anti-Hsp27 antibodies (ABCam) at a dilution of 1:200 for 16 h at room temperature. After washing thrice in PBS, the sections were incubated with secondary antibody conjugated to biotin for 10 min at room temperature. After additional washing in PBS, the sections were incubated with streptavidin-conjugated horseradish peroxidase enzyme for 10 min at room temperature. Following final washes in PBS, antigen-antibody complexes were detected by incubation with a hydrogen peroxide substrate solution containing aminoethylcarbazole chromogen reagent. Slides were rinsed in distilled water, cover-slipped using aqueous mounting medium, and allowed to dry at room temperature. The relative intensities of the completed immunohistochemical reactions were evaluated using light microscopy by 3 independent trained observers who were unaware of the clinical data. A scale of 0 to 3 was used to score relative intensity, with 0 corresponding to no detectable immunoreactivity and 1, 2, and 3 equivalent to low, moderate, and high staining, respectively. Non-parametric data was analyzed using Mann-Whitney test.
Results
Three paired OSCC cell lines (UM1/UM2, 1386Tu/1386Ln and 686Tu/686Ln) were assembled. The UM1 and UM2 are a paired OSCC with different metastatic potential that generated from a single patient with OSCC of the tongue (7). The 1386Tu/1386Ln and 686Tu/686Ln are paired cell lines generated from primary tumors and lymph node metastatic diseases from the OSCC patients. A critical component of tumor metastasis is invasion of the tumor cell through the basement membrane and into the surrounding stroma. To evaluate the metastatic potential of these paired cell lines, a transwell in vitro cell invasion assay was performed (Figure 1). Under basal conditions (serum-free medium in both lower and upper chambers of the transwell), minimum cell invasion was observed for all cells. When chemoattractant (10% FBS) was present in the lower chamber, marked cell invasions were observed for all cell lines, where the UM1, 1386Ln and 686Ln exhibited statistically significantly elevated invasions when compared to their paired cell lines, UM2, 1386Tu and 686Tu, respectively (p<0.05).
Figure 1.
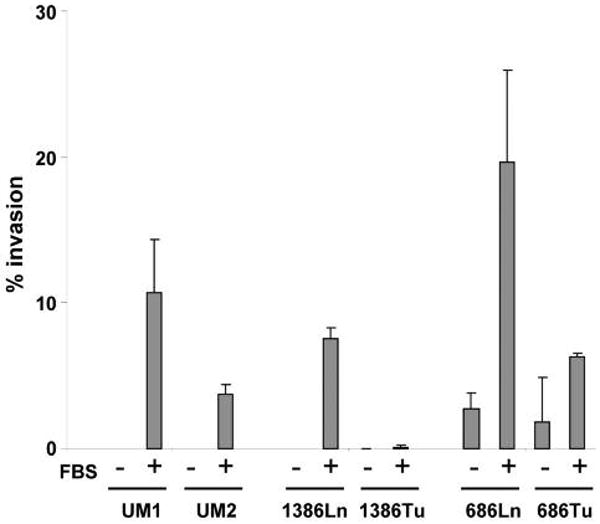
Chemoattractants induced invasion of the OSCC cells. The metastatic potentials of paired OSCC cell lines (UM1/UM2, 1386Ln/Tu, and 686Ln/Tu) were evaluated using a transwell in vitro cell invasion assay as described in the Material and Methods section. The invasions for each cell lines were measured in the presence and absence of chemoattractant (10% FBS) in the lower chamber of the transwell. Statistically significantly differences in the invasions were observed for all 3 paired cell lines in the presence of chemoattractant (p<0.05). The result represents 3 independent experiments with quadruplicates of each measurement.
Mapping proteomes, the protein complements to genomes, has being utilized extensively to discover new disease biomarkers for clinical and diagnostic applications. Here, the cellular proteomes of these paired cell lines were determined using 2D-PAGE/MS proteomic approach as described. The representative images of 2D-PAGE analysis for UM1 and UM2 cells were presented in Figure 2. The protein spots were quantified and the differentially expressed spots (>= 2-fold) were excised and in-gel tryptic digested. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry of tryptic peptides was performed by a positive-ion reflector mode with delayed extraction over the m/z range 700-4000 using a Voyager DE-PRO Mass Spectrometer (Applied Biosystems, Foster City, CA, USA) equipped with a nitrogen laser. Representative mass spectrums were shown in Figure 3.
Figure 2.
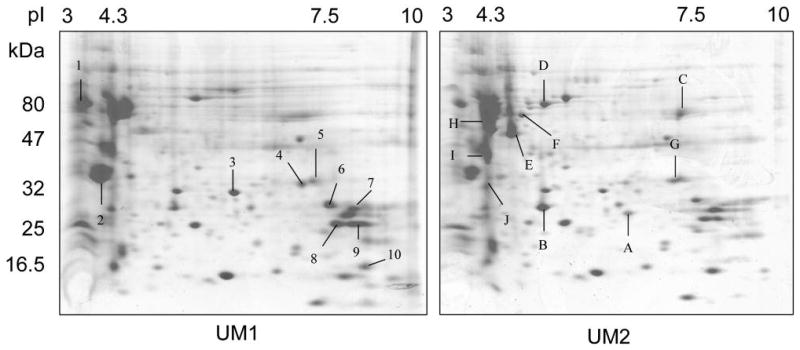
2D-PAGE based identification of differentially expressed proteins for OSCC metastasis and invasion. The proteins from OSCC cells with different metastatic potentials were separated by 2D-PAGE (1st dimension: pH gradient 3-10; 2nd dimension: 4-12% gradient SDS-PAGE). The gels were stained with Coomassie brilliant blue R-250 and quantified using the PDQuest software as described. Representative gel images for UM1 (A) and UM2 (B) were shown.
Figure 3.
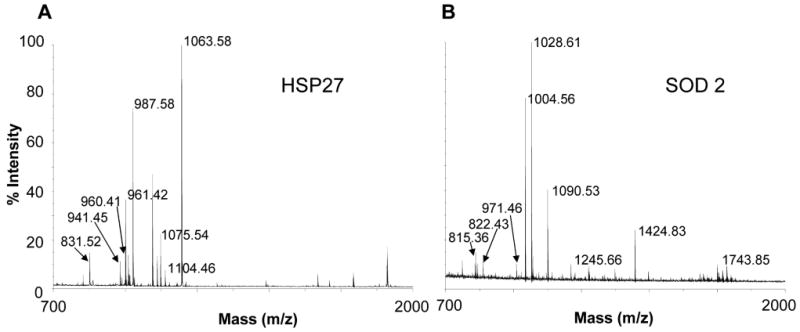
Identification of SOD2 and HSP27 as candidate biomarkers for OSCC metastasis and invasion. The MALDI-TOF mass spectrometry was used to depict the identities of the differentially expressed protein spots identified by 2D-PAGE. Peptide mass results were used to identify the proteins using the MASCOT Peptide Fingerprint. Representative Spectra were shown. A: Hsp27; B: SOD2.
A total of 40 differentially expressed proteins were identified (16 for UM1/UM2, 11 for 1386Tu/Ln, and 13 for 686Tu/Ln, respectively), as listed in Table II. Among those protein candidates, the enhanced expressions of SOD2 and Hsp27 were observed in cells with elevated metastatic potential in 2 out of 3 paired cell lines (UM1/UM2 and 1386Tu/Ln for SOD2, 1386Tu/Ln and 686Tu/Ln for Hsp27, respectively). These observed expressional differences independently were validated using Western blot analyses. As shown in Figure 4, the SOD2 expression was elevated in UM1 and 1386Ln compared to their paired cell lines (UM2 and 1386Tu, respectively). No apparent difference in SOD2 expression was observed between 686Ln and 686Tu. Similarly, the Hsp27 expression was elevated in 1386Ln and 686Ln, compared to their paired cell lines (1386Tu and 686Tu, respectively). However, the Hsp27 expression in UM2 appeared to be higher than that in UM1, which was not detected by our proteomic method.
Table II. Differentially expressed protein identified by 2D-PAGE/MS.
| Accession no. | Protein name | Mascot score | Peptides matched | pI | Mr | Spot ratio |
|---|---|---|---|---|---|---|
| UM1/UM2 | ||||||
| gi|34419633 | zinc finger protein 135 | 58 | 6 | 8.44 | 77267 | >50 |
| gi|48994190 | myomesin 1 | 78 | 11 | 6.51 | 188994 | >50 |
| gi|4557701 | keratin 17 | 66 | 13 | 4.97 | 48361 | 7.4 |
| gi|22748759 | hypothetical protein LOC125144 | 47 | 5 | 9.49 | 13986 | 3.49 |
| gi|24119203 | tropomyosin 3 isoform 2 | 104 | 16 | 4.75 | 29243 | 3.3 |
| gi|10863927 | peptidylprolyl isomerase A | 76 | 8 | 7.68 | 18229 | 2.6 |
| gi|662841 | heat shock protein 27 | 85 | 8 | 7.83 | 22427 | 2.5 |
| gi|30841309 | manganese-containing superoxide dismutase | 85 | 9 | 6.87 | 23772 | 2.08 |
| gi|4507645 | triosephosphate isomerase 1 | 161 | 16 | 6.45 | 26938 | 0.76 |
| gi|494066 | chain A, glutathione S-transferase | 66 | 6 | 5.44 | 23438 | 0.47 |
| gi|119603566 | hCG2026398, isoform CRA_a | 56 | 7 | 9.60 | 45316 | 0.16 |
| gi|59806361 | hypothetical protein LOC387104 | 74 | 13 | 5.81 | 103478 | <0.1 |
| gi|4503571 | enolase 1 | 159 | 20 | 7.01 | 47481 | <0.1 |
| gi|119617057 | keratin 8 | 204 | 23 | 5.41 | 57829 | <0.1 |
| gi|4557888 | keratin 18 | 156 | 18 | 5.34 | 48029 | <0.1 |
| gi|90111766 | keratin 19 | 208 | 22 | 5.05 | 44065 | <0.1 |
| 1386LN/TU | ||||||
| gi|860986 | protein disulfide isomerase | 100 | 15 | 6.10 | 57043 | >50 |
| gi|4504919 | keratin 8 | 80 | 5 | 5.52 | 53671 | >50 |
| gi|30841309 | manganese-containing superoxide dismutase | 106 | 9 | 6.87 | 23772 | >50 |
| gi|5689551 | KIAA1107 protein | 75 | 7 | 5.93 | 142197 | 9.2 |
| gi|913159 | neuropolypeptide h3 | 71 | 8 | 7.43 | 21027 | 8.6 |
| gi|4505621 | prostatic binding protein | 71 | 8 | 7.01 | 21158 | 8.6 |
| gi|662841 | heat shock protein 27 | 199 | 13 | 7.83 | 22427 | 2.73 |
| gi|4557888 | keratin 18 | 104 | 13 | 5.34 | 48029 | 1.9 |
| gi|5031851 | stathmin 1 | 110 | 11 | 5.76 | 17292 | 0.4 |
| gi|119579750 | synovial sarcoma, X breakpoint 9 | 49 | 7 | 9.18 | 19084 | <0.1 |
| gi|39793924 | CGI-96 protein | 48 | 6 | 8.86 | 14949 | <0.1 |
| 686LN/TU | ||||||
| gi|662841 | heat shock protein 27 | 135 | 11 | 7.83 | 22427 | >50 |
| gi|56204548 | zinc finger, CCHC domain containing 11 | 52 | 4 | 8.01 | 34525 | >50 |
| gi|44890059 | Involucrin | 258 | 21 | 4.62 | 68551 | 18.8 |
| gi|21619129 | SRBD1 protein | 66 | 7 | 9.04 | 70639 | 5.42 |
| gi|4506773 | S100 calcium-binding protein A9 | 82 | 7 | 5.71 | 13291 | 4.1 |
| gi|4507835 | uridine monophosphate synthase | 56 | 7 | 6.81 | 52645 | 3.96 |
| gi|15431310 | keratin 14 | 261 | 26 | 5.09 | 51875 | 0.51 |
| gi|4557701 | keratin 17 | 70 | 11 | 4.97 | 48361 | <0.1 |
| gi|57209813 | tubulin, beta polypeptide | 69 | 10 | 4.70 | 48135 | <0.1 |
| gi|4757900 | calreticulin precursor | 67 | 10 | 4.29 | 48283 | <0.1 |
| gi|119617035 | keratin 6E | 142 | 21 | 8.38 | 60144 | <0.1 |
| gi|119395754 | keratin 5 | 105 | 18 | 7.58 | 62568 | <0.1 |
| gi|825671 | B23 nucleophosmin | 94 | 12 | 4.71 | 31090 | <0.1 |
Figure 4.
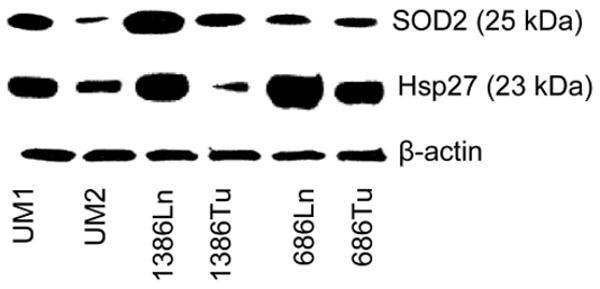
Western blot analyses of SOD2 and Hsp27 on OSCC cells. The expression levels of SOD2 and Hsp27 were determined for the paired UM1/UM2, 1386Ln/Tu, and 686Ln/Tu cell lines as described. The level of β-actin is shown as loading control. The results represent three independent experiments with similar findings.
To further validate the potential diagnostic values of SOD2 and Hsp27 for lymph node metastasis, immunohistochemistry analyses were performed on 21 cases of tongue SCC patient samples, including paired primary tumor and lymph node metastasis disease tissue samples (Figure 5). For SOD2, IHC staining was observed in the cytoplasm of epithelial layers except the superficial layers for the normal tongue tissue (Figure 5A). Positive SOD2 staining was observed in the primary SCC (Figure 5B). Diffuse cytoplasmic staining was observed in the carcinoma cell nests. The intensity of SOD2 staining varied dramatically among SCC cases, but those with higher T stages generally showed stronger SOD2 staining than those with lower T stages (data not shown). The staining intensity for SOD2 was significantly higher in lymph node metastases (Figure 5C) than those in the paired primary tumors (p<0.05). In the lymph node metastasis, SOD2 expression was higher in basaloid cells that underlying the carcinoma cells nests, when compared to the suprabasal cells.
Figure 5.
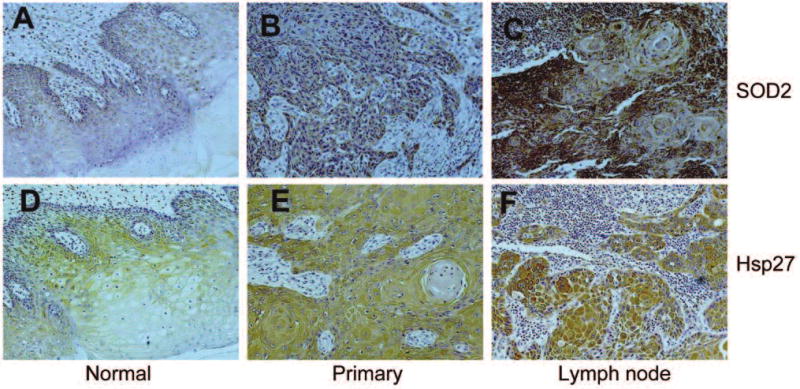
Immunohistochemistry analyses of SOD2 and Hsp27 on primary OSCC and paired lymph node metastasis diseases. Immunohistochemistry analyses for SOD2 and Hsp27 were performed on tissue samples from a panel of patients with oral tongue SCC (n=21) as described using appropriate antibodies. Representative Images (×200) were shown. A, B, C: SOD2; D, E, F: Hsp27. A, D: normal oral mucosa; B, E: primary OSCCs; C, F: lymph node metastasis diseases. A statistically significant increase in the expression of SOD2 was observed in the lymph node metastasis diseases when compared with the paired primary diseases (n<0.05). No statistically significant change was observed for the expression of Hsp27.
Hsp27 staining was observed in the cytoplasm of the spinous and granular epithelial cells, and was absent in the basal and scattered in the most superficial layers for the normal tongue tissue (Figure 5D). Diffuse positive IHC labeling for Hsp27 was shown in tongue SCC samples from both primary tumor site and lymph node metastatic diseases (Figure 5E & F). While the apparent increases in HSP27 IHC intensities were observed in lymph node metastatic diseases when compared with primary tumors, this difference was not statistically significant (p>0.05).
Discussion and Conclusion
Biomarkers are measurable and quantifiable biological parameters that can serve as indices for health-related assessments, such as disease diagnosis, environmental exposure and its effects, metabolic processes and epidemiologic studies. As proteomic tools evolve, protein biomarker discovery will become a central application of proteomics. Profiling proteins in tissues or body fluids over the course of disease progression could reveal potential biomarkers indicative of specific disease status, which may be used extensively for future medical diagnostics. For the purposes of the biomarker discovery, well-defined methods and robust technologies for both protein separation and identification are available in the form of 2D-PAGE combined with mass spectrometry. In reality, 2D-PAGE remains one of the best among all available protein separation approaches. There are two discrete separation steps in this process. Separation in the first dimension, isoelectric focusing, is based on protein's charge and separation in the second dimension SDS-polyacrylamide gel is based on protein's molecular weight. A single 2D-PAGE can resolve hundreds of proteins which, when stained, form a pattern of spots. A comparison of the intensity of individual spots yields information on the relative amounts of each protein in the sample. 2D gels, combined with MS applications, can be used to define both qualitative and quantitative changes in expression levels between biological samples, such as comparing cancer cells derived from the same patient at different stages of disease progression.
An essential characteristic of cancer is the ability to invade surrounding tissues and metastasize to regional and distant sites. Detection of local lymph node metastasis is pivotal for choosing appropriate treatment, especially for individuals diagnosed with OSCC in the oral cavity (8). In this study, we identified a set of differentially expressed proteins from paired OSCC cell lines with different metastatic potential. Selected candidates, Hsp27 and SOD2, were further investigated using independent OSCC patient tissue samples. While the Hsp27 has been shown to be upregulated in OSCC (9, 10) and may has prognostic values for OSCC patients (11, 12), no significant difference was observed at IHC level between paired primary OSCC samples and lymph node metastatic disease. On the other hand, the IHC for SOD2 was significantly different between paired primary tumor and lymph node metastatic disease. Significant difference was also observed at SOD2 mRNA level between primary OSCC samples with or without lymph node metastasis (data not shown) (13). These results suggested that SOD2 expression may be useful in order to identify cases of OSCC with a more aggressive and invasive phenotype.
Accumulating evidence has indicated that the intracellular redox state plays important roles in cellular signaling transduction and gene expression. But, the effects of redox state in malignancies are somewhat contradictory. In theory, reducing the oxidative stress may prevent DNA degeneration and therefore prevent the development of cancer. However, doing so may also offer increased growth potential to tumor cells and protect them from excess of reactive oxygen species (ROS), which would otherwise lead to apoptosis or necrosis. Adding to the complexity is superoxide dismutase 2 (SOD2), which has been considered to be one of the most important antioxidant enzymes. The major part of SOD2 is present in mitochondrial matrix, the place where oxygen is mostly consumed and where oxidative stress is most evident. The role of SODs in carcinogenesis has been widely studied but remains ambiguous. While the majority of published in vitro studies have reported a protective role of SOD2 against tumor progression in several type of cancer cell lines (14-18), including oral cancer cell lines (19), some recent studies have shown SOD2 enhancements with invasion and migration of the tumor cells (20). The in vivo studies suggest more complicated roles of SOD2 in tumorigenesis. Increased SOD2 levels have been observed from esophageal, gastric, brain astrocytic and colorectal carcinomas, and often associated with metastasis and poor prognosis (21-29). The status of SOD2 level in breast cancer is not clear, with some studies showing increased SOD2 level (30), while others showing a decrease in SOD2 (31). There seems to be a reduction in SOD2 levels also in prostatic carcinomas when compared to healthy tissue (32, 33). Our results indicated a significant increase in expression of SOD2 gene in OSCC, and this increase is associated with increased metastasis potential. These findings are in agreement with the recent observation in oral cancer (10, 34).
Several studies have shown that the SOD2-dependent production of H2O2 leads to increased expression of MMP family members (including MMP-1 and MMP-9) and that there is a strong correlation between this increase in MMP levels and enhanced metastasis (35-37). Thus, SOD2-dependent up-regulation of MMPs may, in part, contribute to increased invasion and metastatic capacity of tumors displaying elevated SOD2 levels. Interestingly, our early study shown that MMP-9 expression level can provide predictive values for lymph node metastasis and extracapsular spread (ECS) of OSCC of the tongue (3). Our recent study also demonstrated that MMP-1 is the most significantly upregulated gene in tongue OSCC (13). Also, a single nucleotide polymorphism (SNP) that creates an Ets site at the promoter region of the MMP-1 gene has been shown to be responsible for the SOD2 dependent MMP-1 expression (36). This SNP has been investigated extensively and has been shown in several populations to be associated with OSCC susceptibility and aggressiveness (38-41). While, further studies will be needed to fully understand the molecular role(s) of redox state and SOD2 in OSCC, our present study, together with previous results, clearly indicated the association of increased SOD2 level and enhanced metastasis potential of OSCC.
Acknowledgments
This work was supported in part by NIH PHS grants K22 DE014847, RO3 DE016569, RO3 CA114688 and a grant from Prevent Cancer Foundation. MDA686Tu, MDA686Ln, MDA1386Tu, MDA1386Ln cells were gifts from Dr. PG. Sacks of the New York University. We thank Ms. Katherine Long for her editorial assistance.
References
- 1.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50(1):7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Temam S, Oh M, Pungpravat N, Huang BL, Mao L, Wong DT. Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia. 2006;8(11):925–932. doi: 10.1593/neo.06430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmalbach CE, Chepeha DB, Giordano TJ, Rubin MA, Teknos TN, Bradford CR, Wolf GT, Kuick R, Misek DE, Trask DK, et al. Molecular profiling and the identification of genes associated with metastatic oral cavity/pharynx squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130(3):295–302. doi: 10.1001/archotol.130.3.295. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24(7):1244–1251. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- 6.Roepman P, Wessels LF, Kettelarij N, Kemmeren P, Miles AJ, Lijnzaad P, Tilanus MG, Koole R, Hordijk GJ, van der Vliet PC, et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37(2):182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama S, Sasaki A, Mese H, Alcalde RE, Matsumura T. Establishment of high and low metastasis cell lines derived from a human tongue squamous cell carcinoma. Invasion Metastasis. 1998;18(5-6):219–228. doi: 10.1159/000024515. [DOI] [PubMed] [Google Scholar]
- 8.Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4(6):448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 9.He QY, Chen J, Kung HF, Yuen AP, Chiu JF. Identification of tumor-associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics. 2004;4(1):271–278. doi: 10.1002/pmic.200300550. [DOI] [PubMed] [Google Scholar]
- 10.Lo WY, Tsai MH, Tsai Y, Hua CH, Tsai FJ, Huang SY, Tsai CH, Lai CC. Identification of over-expressed proteins in oral squamous cell carcinoma (OSCC) patients by clinical proteomic analysis. Clin Chim Acta. 2007;376(1-2):101–107. doi: 10.1016/j.cca.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Lo Muzio L, Leonardi R, Mariggio MA, Mignogna MD, Rubini C, Vinella A, Pannone G, Giannetti L, Serpico R, Testa NF, et al. HSP 27 as possible prognostic factor in patients with oral squamous cell carcinoma. Histol Histopathol. 2004;19(1):119–128. doi: 10.14670/HH-19.119. [DOI] [PubMed] [Google Scholar]
- 12.Mese H, Sasaki A, Nakayama S, Yoshioka N, Yoshihama Y, Kishimoto K, Matsumura T. Prognostic significance of heat shock protein 27 (HSP27) in patients with oral squamous cell carcinoma. Oncol Rep. 2002;9(2):341–344. [PubMed] [Google Scholar]
- 13.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9(1):69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan T, Oberley LW, Zhong W, St Clair DK. Manganese-containing superoxide dismutase overexpression causes phenotypic reversion in SV40-transformed human lung fibroblasts. Cancer Res. 1996;56(12):2864–2871. [PubMed] [Google Scholar]
- 15.Zhong W, Oberley LW, Oberley TD, Yan T, Domann FE, St Clair DK. Inhibition of cell growth and sensitization to oxidative damage by overexpression of manganese superoxide dismutase in rat glioma cells. Cell Growth Differ. 1996;7(9):1175–1186. [PubMed] [Google Scholar]
- 16.Church SL, Grant JW, Ridnour LA, Oberley LW, Swanson PE, Meltzer PS, Trent JM. Increased manganese superoxide dismutase expression suppresses the malignant phenotype of human melanoma cells. Proc Natl Acad Sci USA. 1993;90(7):3113–3117. doi: 10.1073/pnas.90.7.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullen JJ, Weydert C, Hinkhouse MM, Ritchie J, Domann FE, Spitz D, Oberley LW. The role of manganese superoxide dismutase in the growth of pancreatic adenocarcinoma. Cancer Res. 2003;63(6):1297–303. [PubMed] [Google Scholar]
- 18.Ough M, Lewis A, Zhang Y, Hinkhouse MM, Ritchie JM, Oberley LW, Cullen JJ. Inhibition of cell growth by overexpression of manganese superoxide dismutase (MnSOD) in human pancreatic carcinoma. Free Radic Res. 2004;38(11):1223–1233. doi: 10.1080/10715760400017376. [DOI] [PubMed] [Google Scholar]
- 19.Liu R, Oberley TD, Oberley LW. Transfection and expression of MnSOD cDNA decreases tumor malignancy of human oral squamous carcinoma SCC-25 cells. Hum Gene Ther. 1997;8(5):585–595. doi: 10.1089/hum.1997.8.5-585. [DOI] [PubMed] [Google Scholar]
- 20.Connor KM, Hempel N, Nelson KK, Dabiri G, Gamarra A, Belarmino J, Van De Water L, Mian BM, Melendez JA. Manganese superoxide dismutase enhances the invasive and migratory activity of tumor cells. Cancer Res. 2007;67(21):10260–10267. doi: 10.1158/0008-5472.CAN-07-1204. [DOI] [PubMed] [Google Scholar]
- 21.Izutani R, Asano S, Imano M, Kuroda D, Kato M, Ohyanagi H. Expression of manganese superoxide dismutase in esophageal and gastric cancers. J Gastroenterol. 1998;33(6):816–822. doi: 10.1007/s005350050181. [DOI] [PubMed] [Google Scholar]
- 22.Malafa M, Margenthaler J, Webb B, Neitzel L, Christophersen M. MnSOD expression is increased in metastatic gastric cancer. J Surg Res. 2000;88(2):130–134. doi: 10.1006/jsre.1999.5773. [DOI] [PubMed] [Google Scholar]
- 23.Kim JJ, Chae SW, Hur GC, Cho SJ, Kim MK, Choi J, Nam SY, Kim WH, Yang HK, Lee BL. Manganese superoxide dismutase expression correlates with a poor prognosis in gastric cancer. Pathobiology. 2002;70(6):353–360. doi: 10.1159/000071276. [DOI] [PubMed] [Google Scholar]
- 24.Janssen AM, Bosman CB, Sier CF, Griffioen G, Kubben FJ, Lamers CB, van Krieken JH, van de Velde CJ, Verspaget HW. Superoxide dismutases in relation to the overall survival of colorectal cancer patients. Br J Cancer. 1998;78(8):1051–1057. doi: 10.1038/bjc.1998.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toh Y, Kuninaka S, Oshiro T, Ikeda Y, Nakashima H, Baba H, Kohnoe S, Okamura T, Mori M, Sugimachi K. Overexpression of manganese superoxide dismutase mRNA may correlate with aggressiveness in gastric and colorectal adenocarcinomas. Int J Oncol. 2000;17(1):107–12. doi: 10.3892/ijo.17.1.107. [DOI] [PubMed] [Google Scholar]
- 26.Haapasalo H, Kylaniemi M, Paunul N, Kinnula VL, Soini Y. Expression of antioxidant enzymes in astrocytic brain tumors. Brain Pathol. 2003;13(2):155–164. doi: 10.1111/j.1750-3639.2003.tb00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korenaga D, Yasuda M, Honda M, Nozoe T, Inutsuka S. MnSOD expression within tumor cells is closely related to mode of invasion in human gastric cancer. Oncol Rep. 2003;10(1):27–30. [PubMed] [Google Scholar]
- 28.Nozoe T, Honda M, Inutsuka S, Yasuda M, Korenaga D. Significance of immunohistochemical expression of manganese superoxide dismutase as a marker of malignant potential in colorectal carcinoma. Oncol Rep. 2003;10(1):39–43. [PubMed] [Google Scholar]
- 29.Qi Y, Chiu JF, Wang L, Kwong DL, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5(11):2960–2971. doi: 10.1002/pmic.200401175. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi MS, Bianchi NO, Bolzan AD. Superoxide dismutase activity and superoxide dismutase-1 gene methylation in normal and tumoral human breast tissues. Cancer Genet Cytogenet. 1992;59(1):26–29. doi: 10.1016/0165-4608(92)90152-x. [DOI] [PubMed] [Google Scholar]
- 31.Soini Y, Vakkala M, Kahlos K, Paakko P, Kinnula V. MnSOD expression is less frequent in tumour cells of invasive breast carcinomas than in in situ carcinomas or non-neoplastic breast epithelial cells. J Pathol. 2001;195(2):156–162. doi: 10.1002/path.946. [DOI] [PubMed] [Google Scholar]
- 32.Baker AM, Oberley LW, Cohen MB. Expression of antioxidant enzymes in human prostatic adenocarcinoma. Prostate. 1997;32(4):229–233. doi: 10.1002/(sici)1097-0045(19970901)32:4<229::aid-pros1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 33.Bostwick DG, Alexander EE, Singh R, Shan A, Qian J, Santella RM, Oberley LW, Yan T, Zhong W, Jiang X, et al. Antioxidant enzyme expression and reactive oxygen species damage in prostatic intraepithelial neoplasia and cancer. Cancer. 2000;89(1):123–134. [PubMed] [Google Scholar]
- 34.Salzman R, Kankova K, Pacal L, Tomandl J, Horakova Z, Kostrica R. Increased activity of superoxide dismutase in advanced stages of head and neck squamous cell carcinoma with locoregional metastases. Neoplasma. 2007;54(4):321–325. [PubMed] [Google Scholar]
- 35.Nelson KK, Melendez JA. Mitochondrial redox control of matrix metalloproteinases. Free Radic Biol Med. 2004;37(6):768–784. doi: 10.1016/j.freeradbiomed.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Nelson KK, Ranganathan AC, Mansouri J, Rodriguez AM, Providence KM, Rutter JL, Pumiglia K, Bennett JA, Melendez JA. Elevated sod2 activity augments matrix metalloproteinase expression: evidence for the involvement of endogenous hydrogen peroxide in regulating metastasis. Clin Cancer Res. 2003;9(1):424–432. [PubMed] [Google Scholar]
- 37.Yang JQ, Zhao W, Duan H, Robbins ME, Buettner GR, Oberley LW, Domann FE. v-Ha-RaS oncogene upregulates the 92-kDa type IV collagenase (MMP-9) gene by increasing cellular superoxide production and activating NF-kappaB. Free Radic Biol Med. 2001;31(4):520–529. doi: 10.1016/s0891-5849(01)00613-x. [DOI] [PubMed] [Google Scholar]
- 38.Vairaktaris E, Yapijakis C, Derka S, Serefoglou Z, Vassiliou S, Nkenke E, Ragos V, Vylliotis A, Spyridonidou S, Tsigris C, et al. Association of matrix metalloproteinase-1 (-1607 1G/2G) polymorphism with increased risk for oral squamous cell carcinoma. Anticancer Res. 2007;27(1A):459–464. [PubMed] [Google Scholar]
- 39.O-Charoenrat P, Leksrisakul P, Sangruchi S. A functional polymorphism in the matrix metalloproteinase-1 gene promoter is associated with susceptibility and aggressiveness of head and neck cancer. Int J Cancer. 2006;118(10):2548–2553. doi: 10.1002/ijc.21644. [DOI] [PubMed] [Google Scholar]
- 40.Cao ZG, Li CZ. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter enhances oral squamous cell carcinoma susceptibility in a Chinese population. Oral Oncol. 2006;42(1):32–38. doi: 10.1016/j.oraloncology.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Lin SC, Chung MY, Huang JW, Shieh TM, Liu CJ, Chang KW. Correlation between functional genotypes in the matrix metalloproteinases-1 promoter and risk of oral squamous cell carcinomas. J Oral Pathol Med. 2004;33(6):323–326. doi: 10.1111/j.1600-0714.2004.00214.x. [DOI] [PubMed] [Google Scholar]


