Summary
Heterozygous mutations in the gene encoding CHD7, an ATP-dependent chromatin remodeler result in a complex constellation of congenital anomalies called CHARGE syndrome. Here we show that in humans and in Xenopus, CHD7 is essential for the formation of multipotent migratory neural crest cells, a transient cell population that is ectodermal in origin, but undergoes a major gene expression reprogramming to acquire a remarkably broad differentiation potential and ability to migrate throughout the body to give rise to bones, cartilages, nerves, and cardiac structures. We demonstrate that CHD7 function is essential for activation of core components of neural crest transcriptional circuitry, including Sox9, Twist and Slug. Moreover, the major features of CHARGE are recapitulated in Xenopus embryo by the downregulation of CHD7 levels or overexpression of its catalytically inactive ATP-ase mutant. We further show that in human multipotent neural crest cells, CHD7 associates with a BRG1-containing complex PBAF, and both factors co-occupy a neural crest-specific distal SOX9 enhancer, as well as a novel genomic element located upstream from TWIST1 gene and marked by H3K4me1. Furthermore, in the embryo CHD7 and PBAF act synergistically to promote neural crest gene expression and cell migration. Our work identifies an evolutionary conserved role for CHD7 in orchestrating neural crest gene expression programs, provides insights into the synergistic regulation of distal genomic elements by two distinct chromatin remodelers, and illuminates the patho-embryology of CHARGE syndrome.
Recent studies demonstrate that unique chromatin states are associated with retained or restricted differentiation potential. 1 During organismal development, cells gradually restrict their differentiation potential to produce specialized tissues and organs. One exception is germ cell formation, which is accompanied by reacquisition of the pluripotent state. Another major developmental reprogramming event occurs in vertebrate organisms during formation of the neural crest, when neural plate border territory cells that are ectodermal in origin undergo epithelial-to-mesenchymal transition (EMT) and acquire broad differentiation potential including ability to form derivatives typical of the mesoderm, such as bone, cartilage and smooth muscle 2,3. Although significant progress has been made in understanding chromatin modification that accompanies reprogramming in the germline 4,5, very little is known about mechanisms of chromatin regulation during neural crest formation.
One candidate chromatin modifier that may be involved in this process is CHD7. Human CHD7 is a large, 340 kD protein that belongs to the CHD family of ATP-dependent chromatin remodelers, distinguished by the presence of tandem chromodomains in addition to the DNA-dependent ATPase domain, which catalyzes nucleosome movement on DNA6,7. CHD7 is one of the four vertebrate homologs of the Drosophila trithorax group protein Kismet, a positive regulator of transcriptional elongation and an antagonist of Polycomb function in Drosophila8,9,10. CHD7 haploinsufficiency is a major cause of CHARGE syndrome, a sporadic, autosomal dominant disorder occurring with a prevalence of about 1 in 10,000 live births and characterized by malformations of the craniofacial structures, peripheral nervous system, ears, eyes and heart11,12. Heterozygous mutations within the CHD7 gene coding region account for about two-third of reported CHARGE cases12.
A large subset of congenital anomalies associated with the syndrome is consistent with defects in neural crest development and it was postulated almost a quarter of the century ago that CHARGE is a neurocristopathy 13. Nevertheless, this hypothesis was never experimentally tested and the mechanisms underlying CHARGE embryo-pathology and the relationship between disease phenotype and genotype remain poorly understood. We hypothesized that CHD7 is involved in orchestrating gene expression programs during neural crest formation and that aberrant execution of this process during human development results in CHARGE syndrome.
In vitro model of human multipotent migratory neural crest formation
To address the role of CHD7 in human neural crest formation, we first established an in vitro model for the efficient derivation of neural crest cells from hESCs. hESCs were differentiated in suspension to form neuroectodermal spheres composed of radial arrangements of neuroepithelial cells or “rosettes” (Figure 1A, panels a and b)14. Human neural rosettes were previously shown to give rise to neural crest cells 15. We developed a modified protocol that enriches for rosettes, and allows for the isolation of a relatively homogenous, multipotent neural crest cell population without the need for cell sorting. Between days 6 and 9 after induction of differentiation, 50-80% of the spheres spontaneously adhered to the culture dishes, and a population of stellate-morphology cells migrated away from the rosette clusters (Figure 1A, panels c and d). As determined by immunofluorescence analysis, these migratory cells expressed NESTIN, but lacked nuclear SOX2 localization suggesting they were distinct from neural precursors (Supplementary Figure 1).
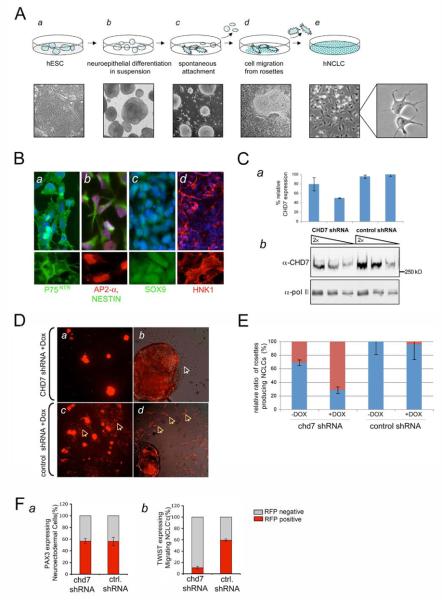
Figure 1. CHD7 knockdown disrupts formation of the migratory, multipotent Neural Crest Like Cell (NCLC) population derived from human embryonic stem cells.
(A) Schematic and morphological overview of the hNCLC derivation in vitro. H9 hESCs were collected by collagenase IV digestion and developed into compact spheres in neural induction medium (a and b). The neuroepithelial spheres spontaneously attached and formed rosette-like structures, with stellate morphology cells migrating away at 6 to 9 days of differentiation (c and d). Neural rosettes were manually removed and migratory cells formed a uniform population of stellate shaped cells on fibronectin-coated dishes (e). High magnification image showing representative morphology of the NCLCs (f).
(B) hNCLCs express early neural crest markers. hNCLCs were immunostained with antibodies recognizing: p75ntr (a, green), AP2-α (b, red), NESTIN (b, green), SOX9 (c, green) and cell surface marker HNK1 (d, red); (lower panels show single channel images). Nuclei were stained with DAPI (a-d, blue in merged channel images).
(C) Dox-inducible shRNA-mediated downregulation of CHD7 mRNA and protein levels.(a) CHD7 mRNA expression levels were analyzed by quantitative RT PCR of RNA samples prepared from hESCs infected with CHD7 shRNA or control shRNA lentiviruses and induced to differentiate as described in Figure 1A, either in the presence or in the absence of Dox. % of relative CHD7 expression at day 9 of differentiation is shown, with CHD7 expression in cells infected with a control shRNA lentivirus and cultured in the presence of Dox normalized to 100%. Error bars represent the standard deviation from two independent biological replicate experiments. (b) Extracts from shRNA lentivirus-infected cells differentiated in the presence of Dox, were normalized for protein concentration and analyzed by immunoblotting with α-CHD7 and α-RNA pol II antibodies. A two-fold dilution series is shown.
(D) Formation of the migratory hNCLC population from CHD7 shRNA and control shRNA-expressing neural rosettes. hESC were infected with appropriate lentiviral shRNA vectors and induced to differentiate in the presence of Dox. At day 9 of differentiation, shRNA-expressing neural rosettes were identified by RFP expression at 5X magnification (a and c) and 10X magnification (b and d; images contain merged fluorescence and bright-field). Yellow arrows (c and d) highlight areas where RFP-expressing hNCLCs migrate out of the rosettes. The white arrow depicts RFP-negative cells migrating out of an RFP positive rosette (b).
(E) Relative ratio of RFP expressing rosettes producing migratory cells. Migration defects as observed in C were quantified. The blue and red segments of the bar graph represent the percentage of rosettes from which hNCLCs did or did not, respectively, migrate out (N~200). Both red and non-red emigrating cells were included in the calculations. Error bars represent the standard deviations in two independent biological replicate experiments.
(F) PAX3 and TWIST1 expression in CHD7 shRNA-infected cells. hESCs were infected in parallel with CHD7 or control shRNAs and induced to differentiate in the presence of Dox. (a) Neuroectodermal cells (attached and floating rosettes) were collected as a single cell suspension and stained with α-PAX3 antibody. The number of PAX3/RFP double-positive and PAX3 positive/RFP negative cells was determined by cell counting (N >500) (b) In the same experiment, the hNCLCs were isolated by discarding the floating spheres and dissecting out the attached rosettes (>95% of isolated cells showed nuclear TWIST1 expression). The number of TWIST1/RFP double-positive and TWIST1 positive/RFP negative cells was determined by cell counting (N >500).
To determine whether the stellate cells migrating out of rosettes behave like neural crest cells, we removed the rosettes by dissection (Figure 1A, panel e) and immunostained the remaining migrating cells with markers characteristic of early neural crest such as SOX9, AP2-α, p75 and HNK1 16. At this stage, 95% of migrating cells expressed SOX9, p75 and AP2-α, whereas only 30% of these cells expressed HNK1, consistent with HNK1 marking only a subset of the early neural crest cells (Figure 1B) 17,18. Additionally, the isolated migratory cells preferentially proliferated on the fibronectin support (Supplementary Figure 2), in agreement with the adhesion and migration of neural crest cells on fibronectin in vivo 19. We further characterized these cells by showing that upon transplantation to the developing neural tube of a chick embryo, they properly migrate to the craniofacial mesenchyme and heart (Supplementary Figure 3). The observed gene expression characteristics, adhesion properties and in ovo migration patterns are consistent with an early neural crest cell identity, and thus we termed cells migrating out of attached neural rosettes human Neural Crest Like Cells (hNCLCs).
We also recapitulated the broad differentiation potential associated with neural crest cells in isolated hNCLCs, as they spontaneously differentiated into peripheral neurons, glia, melanocytes, adipocytes osteoblasts and chondrocytes upon growth factor withdrawal, and matured into lineage restricted Mesenchymal Progenitors (MPs) upon further amplification in the neural induction medium (for details see Supplementary Figure 4). Having established an in vitro model of human multipotent neural crest formation, we then asked whether CHD7 is important for neural crest formation and/or differentiation.
CHD7 is required for the formation of hNCLCs
CHD7 expression is upregulated in hNCLCs as compared to hESCs or hMPs (Supplementary Figure 5A). To determine whether CHD7 is important for hNCLCs specification, we downregulated CHD7 by transducing hESCs with a lentivirus encoding doxycycline (Dox) inducible short hairpin RNA (shRNA) targeting CHD7 mRNA. shRNA expression was linked to the expression of red fluorescent protein (RFP) (Supplementary Figure 5B). Infected cells were subsequently induced to form neural rosettes. Quantitative RT PCR and immunoblot analyses revealed a two-fold downregulation of CHD7 mRNA and protein levels in cells infected with CHD7 shRNA lentivirus in the presence of Dox, as compared to cells infected with a control non-targeting shRNA lentivirus (Figure 1C). Although we were unable to downregulate CHD7 below 50% of control levels, such two-fold reduction recapitulates the CHD7 dosage deficiency observed in CHARGE patients.
To analyze the role of CHD7 in formation of the hNCLC population, neural rosettes derived from hESC transduced with CHD7 or control shRNAs and treated with Dox were allowed to spontaneously attach. Formation and morphology of neural rosettes was not appreciably affected in cells expressing CHD7 shRNA (Supplementary Figure 5C and D). Although a total number of rosettes formed was unaffected by the CHD7 downregulation (Supplementary Figure 5E), rosettes expressing CHD7 shRNA attached less efficiently (compare panels a and c in Figure 1D). Upon attachment, control shRNA expressing rosettes gave rise to migratory hNCLCs (Figure 1D, see yellow arrows in panels c and d). However, this cell population was severely affected in rosettes expressing CHD7 shRNA (top panels). Upon bright-field illumination we observed some cells migrating from the CHD7 shRNA expressing rosettes, nevertheless these cells either lacked or emitted highly reduced levels of red fluorescence, indicating loss of RFP and thus of shRNA expression (see white arrow in panel b). Quantification of the migratory defect revealed a three-fold reduction in the number of rosettes forming hNCLCs in CHD7 shRNA treated cells relative to control shRNA treated cells (quantification included total red and non-red hNCLCs, Figure 1E).
Next, we assayed effects of CHD7 downregulation on the induction of PAX3- and TWIST1-positive cell populations during differentiation. PAX3 is involved in the establishing competence of the neural plate border territory for neural crest induction, whereas TWIST1 is a transcription factor critical for the formation of the multipotent migratory neural crest cells 2. Downregulation of CHD7 did not affect induction of PAX3, as evidenced by the equal representation of PAX3/RFP double positive cells in CHD7 shRNA and control shRNA-infected neuroectodermal populations (Figure 1F, panel a). In contrast, TWIST1/RFP double positive cells were significantly underrepresented in CHD7 shRNA vs control shRNA infected NCLCs obtained from the same neuroectodermal population (Figure 1E, panel b). Moreover, TWIST1/RFP double positive cells infected with CHD7 shRNA had significantly lower levels of RFP expression than controls, likely due to the strong selection against CHD7 downregulation in NCLCs. Similar results were obtained with additional shRNA targeting CHD7, indicating that the observed phenotype is not due to off-target effects (Supplementary Figure 6). Taken together, our data demonstrate that downregulation of CHD7 affects formation of the multipotent, migratory hNCLC population.
CHD7 is critical for neural crest cell migration in vivo and controls expression of genes encoding core neural crest transcriptional circuitry
Many fundamental mechanisms governing neural crest formation are conserved among vertebrates 2,20. To address whether CHD7 is important for neural crest migration in vivo, we established a migration assay using the Xenopus laevis embryo. CHD7 is conserved between humans and Xenopus (Supplementary Figure 7A), and during embryogenesis Xenopus CHD7 is expressed in the neural crest, neural and preplacodal ectodermal tissues (Figure 2A). A morpholino oligonucleotide targeting the 5′ UTR region of both non-allellic X. laevis CHD7 transcripts (CHD7 MO; Supplementary Figure 7B) was synthesized and shown to deplete CHD7 protein levels in the embryo upon injection (Figure 2B). CHD7 MO was then co-injected with mRNA encoding photo-activatable protein Kaede into both blastomeres of a two-cell stage X. laevis embryo. The anterior neural folds were exposed to UV, and cell migration to pharyngeal arches (PAs) was analyzed at the tailbud stage (for schematics of experimental design see Supplementary Figure 8A). Injection of CHD7 MO abolished PA cell migration (Figure 2C, compare panels b and c; quantified in Supplementary Figure 8B), a defect that was partially rescued by co-injecting human CHD7 mRNA with CHD7 MO (panel d).
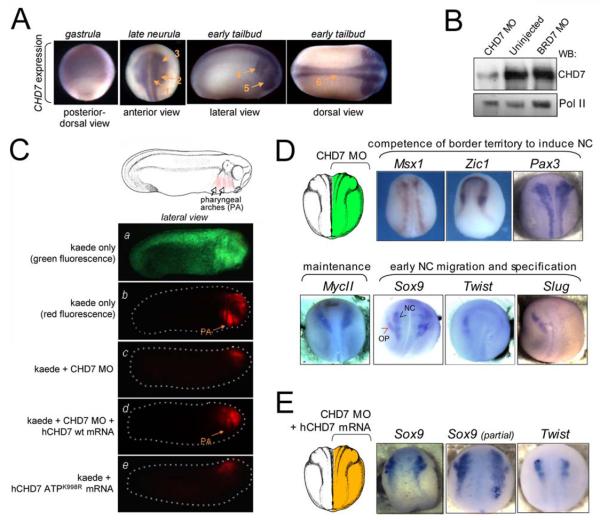
Figure 2. CHD7 and its ATP-ase function are required for neural crest migration in vivo.
(A) CHD7 mRNA expression during Xenopus embryogenesis. CHD7 expression was visualized by RNA in situ hybridization at indicated stages of development, showing diffuse pattern at the gastrula stage, but expression localized to the neural (arrow 1), neural crest (arrow 2) and preplacodal ectodermal (arrow 3) tissues at the late neurula stage. At the tailbud stages, CHD7 is expressed in the pharyngeal arches (arrow 4) and optic placode (arrow 5), as well as alongside the neural tube (arrow 6).
(B) Morpholino mediated knockdown of CHD7 protein levels in Xenopus laevis embryos. Embryos were injected on both sides at the two-cell stage with morpholino oligonucleotide (MO) targeting either CHD7 or BRD7 at 3.3 uM concentration. Nuclei were extracted from these as well as uninjected control embryos at neurula stage. Whole nuclear lysates were normalized for protein concentrations and analyzed by immunoblotting with α-CHD7xl and α-RNA pol II antibodies.
(C) Neural crest migration defect in CHD7 MO and hCHD7 ATPK998R injected embryos. Two-cell stage embryos were injected with mRNA encoding a photo-activatable protein Kaede alone (a and b), Kaede and CHD7 MO (3.3 uM) (c), Kaede, CHD7 MO, and hCHD7 wt mRNA (1 ng) (d), or Kaede and hCHD7 ATPK998R mRNA (9 ng) (e). At the neurula stage embryonic structures corresponding to a subset of the anterior neural and neural crest tissues were UV-irradiated to photo-convert Kaede protein from green to red fluorescence (schematics in Supplementary Figure 8A). Cell migration to the pharyngeal arches (PA) was assayed at the tailbud stage (orange arrows).
(D) Effect of CHD7 knockdown on expression of transcription factors involved in neural crest formation. Two cell-stage embryos were injected with CHD7 MO at 3.3 uM asymmetrically into a single blastomere and analyzed by whole mount RNA in situ hybridization at neurula stage to visualize expression patterns transcription factors controlling: neural plate border territory specification (Msx1, Zic1, Pax3), maintenance of the competent border (MycII) and early neural crest formation (Sox9, Twist, Slug). Sox9 is detected in two major expression domains: the neural crest (blue arrow) and prospective otic placode (red arrow).
(E) hCHD7 mRNA rescues defects in Sox9 and Twist expression. Two cell-stage embryos were co-injected with CHD7 MO (3.3 uM) and wild type full-length hCHD7 mRNA (1 ng) asymmetrically into a single blastomere. At late neurula stages the embryos were analyzed by whole mount RNA in situ hybridization. Representative examples of full and partial rescue are shown.
A point mutation of the conserved lysine residue within the catalytic ATPase domain of chromatin remodelers often produces a dominant negative protein variant as exemplified by K798R substitution in Brg121 We identified the corresponding conserved lysine residue in the ATPase domain of human CHD7 (K998). Overexpression of the CHD7 ATPaseK998R mutant mRNA in Xenopus embryo perturbed cell migration to PAs, although some migrating cells were still evident (Figure 2C, panel e; quantified in Supplementary Figure 8B). This result demonstrates that the intact ATPase domain is important for the role of CHD7 in neural crest migration.
Observed defects in cephalic crest migration may result from indirect effects on non-neural crest embryonic structures caused by the broad presence of morpholino throughout the embryo. For a more targeted disruption of CHD7 function, MO and/or hCHD7 mRNA was co-injected with a lineage tracer (mRNA encoding a fluorescent protein KikGR) into a single dorsal-animal (DA) blastomere of an eight cell stage embryo. The DA blastomere is predicted from the Xenopus embryonic fate maps to form dorsal anterior structures and thus gives rise to the anterior neural tube, neural crest and placodal tissues. The lineage label is clearly discernable along the pharyngeal arch tracts as well as parts of the neural tube and anterior head region. Injected embryos were subsequently scored for defects in the pharyngeal arch labeling pattern (we verified that all scored embryos were properly labeled in the neural tube and dorsal head regions). Injection of CHD7 MO resulted in absence of pharyngeal arch labeling, hCHD7 mRNA alone had no effect, and co-injection of CHD7 MO and hCHD7 mRNA resulted in partial labeling of pharyngeal arches (Supplementary Figure 8C). Although the PA labeling was not fully recovered by hCHD7 mRNA co-injection (possible reasons are discussed in Supplementary Figure 8 legend), the rescue was statistically significant (p<0.00042; Fisher's exact test for count data), further supporting specificity of the observed phenotype.
The effect of CHD7 downregulation on neural crest migration may result from interference with gene regulatory circuitry affecting one or multiple steps involved in neural crest development. To dissect the molecular processes controlled by CHD7, we performed whole-mount in situ RNA hybridization analyses of embryos injected into a single blastomere at the two-cell stage to examine the expression of transcription factors playing a critical role in: (i) establishing competence of the neural plate border territory to induce the neural crest (Pax3, Zic1 and Msx1), (ii) survival of neural crest cells (MycII), and (iii) formation of the multipotent, migratory neural crest (Sox9, Twist and Slug)2. Expression of Pax3, Zic1 and Msx1 was not appreciably affected by CHD7 knockdown (Figure 2D), indicating that the neural induction takes place and that the neural plate border territory is properly specified. Moreover, Pax3, Zic1 and Msx1 expression requires inductive signals from the underlying mesoderm and adjacent non-neural ectoderm2, therefore our results demonstrate that the ability of border territory to interpret signaling from mesoderm is not affected. Similarly, MycII expression was also unaffected, consistent with survival of the neural crest cells induced at the border territory.
In contrast, expression of core transcriptional circuitry for multipotent neural crest formation was severely affected by CHD7 knockdown (Figure 2D). For example, Sox9, a Sox-family transcriptional factor required for otic placode and neural crest specification showed diminished expression levels in both the neural crest and otic placode expression domains 22. Moreover, two critical neural crest and EMT regulators Twist and Slug (Snail2)2 were strongly downregulated on the CHD7-depleted side of the embryo. Defects in Sox9 and Twist expression were fully or partially rescued by co-injection of CHD7 mRNA along with morpholino (Figure 2E). Taken together, our results demonstrate that CHD7 controls gene expression programs for multipotent neural crest formation, but does not appear to be essential for the earliest inductive events at the neural plate border territory. These data are also in agreement with results obtained in the in vitro model of human multipotent neural crest formation, where TWIST1-positive, but not PAX3-positive cell population was affected by CHD7 downregulation (Figure 1F).
Disruption of CHD7 function in Xenopus results in defects that mimic major CHARGE features
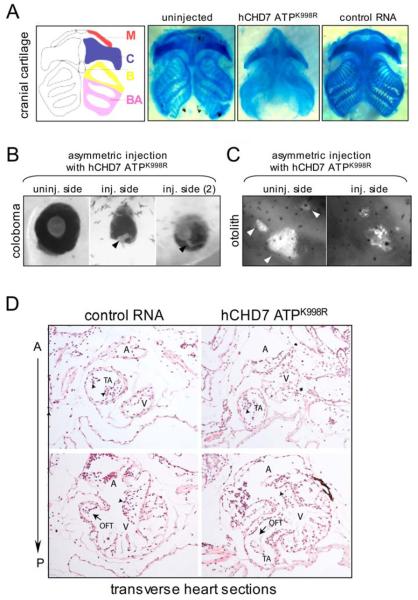
The main set of clinical criteria currently used for CHARGE diagnostics are: (i) ear abnormalities including abnormal semicircular canals, (ii) coloboma of the eye with or without microphtalmia, (iii) malformations of craniofacial structures including choanal atresia, and (iv) heart defects 12,23. Phenotypic analyses of CHD7 ATPaseK998R mRNA-injected tadpoles revealed defects consistent with those used to diagnose CHARGE: (i) missing or malformed otolith, a part of the vestibular system analogous to the human ear (Figure 3C), (ii) coloboma of the eye with or without microphtalmia (Figure 3B), (iii) malformations of craniofacial cartilage, including compression of ceratohyal cartilage, malformation of Meckel's cartilage, and collapsed branchial pouches (Figure 3A), and (iv) heart defects, including abnormal positioning of the truncus arteriosus and cardiac outflow tract, heart structures that receive developmental contribution from the neural crest (Figure 3D). Moreover, facial width compression and reduced eye distance observed in ATPaseK998R mRNA-expressing tadpoles may be indicative of the mild midline defects (Supplementary Figure 9); interestingly arhinencephaly is often associated with CHARGE syndrome12.
Figure 3. Overexpression of CHD7 ATPase mutant in Xenopus recapitulates CHARGE traits.
(A) Craniofacial defects in Xenopus embryos expressing ATPaseK998R mRNA. Two-cell stage embryos were symmetrically injected with 9 ng of either ATPaseK998R or control mRNA (Kaede) on both sides and allowed to develop to the late tadpole stage (stage 45) for analysis of craniofacial cartilage by Alcian Blue staining and dissection. M: Meckel's cartilage, C: ceratohyle, B: basohyl, BA: branchial arch.
(B) Coloboma of the eye with microphtalmia. Two-cell stage embryos were injected with ATPaseK998R mRNA asymmetrically into a single blastomere and allowed to develop to the late tadpole stage (stage 45). The eyes from uninjected and injected side were imaged under a stereomicroscope under the same magnification. Black arrows indicate the coloboma, a fissure that has not completely closed.
(C) Defects in the formation of the otolith, a structure analogous to human ear. Four cell embryos were injected in one of the dorsal blastomeres with ATPaseK998R mRNA and raised to early tadpole stages (stage 40-44?). The left and right otolith was imaged at the same magnification. White arrows indicate three parts of the properly developed otolith.
(D) The truncus arteriosus and cardiac outflow tract are abnormally positioned in ATPaseK998R expressing tadpoles. Transverse section through the stage 45 tadpoles expressing control mRNA (left panels) or ATPaseK998R mRNA (right panels). A: atrium, OFT: outflow tract, TA: truncus arteriosus, V: ventricle. Upper left section: The TA is located to the right and superior to the ventricle of the heart. Arrowheads indicate valves within the truncus arteriosus. Upper right section: Arrowhead indicating the TA in the ATPaseK998R mutant heart that is located to the right, but inferior to the ventricle of the heart. Lower left section: A relatively posterior transverse section of the control tadpole showing the normal connection of cardiac OFT to the ventricle of the heart. OFT is directly to the right and superiorly. Arrowheads indicate valves at the atrioventricular junction. Lower right section: Transverse section through a similar region of the ATPaseK998R expressing tadpole heart showing the aberrant orientation of cardiac OFT which is directed to the right and inferiorly.
Similar phenotypes were observed in tadpoles derived from embryos injected with CHD7 MO, however with MO injections we observed a strong dosage-sensitive response: injection of MO at 5 uM concentration caused late neurula stage lethality (Supplementary Figure 10A), injection at 3.3 uM resulted in partial loss of viability (Supplementary Figure 10B) with surviving late tadpoles exhibiting CHARGE-like phenotypes (Supplementary Figure 13 and not shown), and injection at 1.7 uM resulted in only very mild defects. Loss of viability associated with CHD7 MO injection was rescued by co-injection of CHD7 mRNA, suggesting that it was not caused by an intrinsic toxicity of the morpholino (Supplementary Figure 10). Observed eye coloboma and otolith defects suggest that in addition to neural crest, CHD7 is also important for development of placodal derivatives. Consistently, CHD7 is expressed in ectodermal placodes during embryogenesis (Figure 2A). Moreover, otic placode-specific expression of Sox9, as well as optic and otic placode-specific expression Pax2, a gene whose mutations results in ocular colobomas and hearing loss in humans24, are both affected by CHD7 knockdown (Figure 2D and Supplementary Figure 11). Taken together, our data indicate that the major features of CHARGE can be recapitulated by the downregulation of CHD7 levels or impairment of its ATP-ase activity. These observations underscore the validity of the mechanistic insights obtained in the Xenopus model for understanding CHARGE pathology.
In human neural crest cells CHD7 associates with BRG1-containing PBAF complex
We demonstrated that CHD7 is required for multipotent neural crest formation and expression of critical neural crest genes. To gain insight into molecular partners that cooperate with CHD7 to control neural crest gene expression we immunopurified CHD7-associated proteins from hNCLCs. Nuclear extracts prepared from hNCLC-enriched cell population derived from hESC were used as input for the anti-CHD7 and control antibody immunoaffinity purifications as schematically depicted in Figure 4A. We established that CHD7 antibody is able to deplete CHD7 from extract to near completion (Supplementary Figure 12). Due to challenges in obtaining large numbers of hESC-derived NCLCs for biochemical analysis, we performed parallel purifications from human teratocarcinoma NT2 cells that were differentiated to NCLCs (we verified that CHD7 expression, as well as neural and neural crest markers are induced during NT2 differentiation, not shown). Following immunopurification, the bound proteins were digested “in-solution” with trypsin, and the resultant complex mixture of peptides was separated and identified by liquid chromatography tandem mass spectrometry (LC-MS/MS). The major polypeptide detected in our MS/MS analysis was CHD7 (Figure 4A and Supplementary Table 1). In addition, multiple common subunits of BAF/PBAF complexes, PBAF-specific complex subunits, and Poly(ADP) ribose polymerase 1 (PARP1) were uniquely present in anti-CHD7 immunoprecipitates (Figure 4A). We recovered fewer peptides from hESC-derived NCLCs than from NT2-derived cells, most likely due to limiting amounts of the input material available for biochemical analysis.
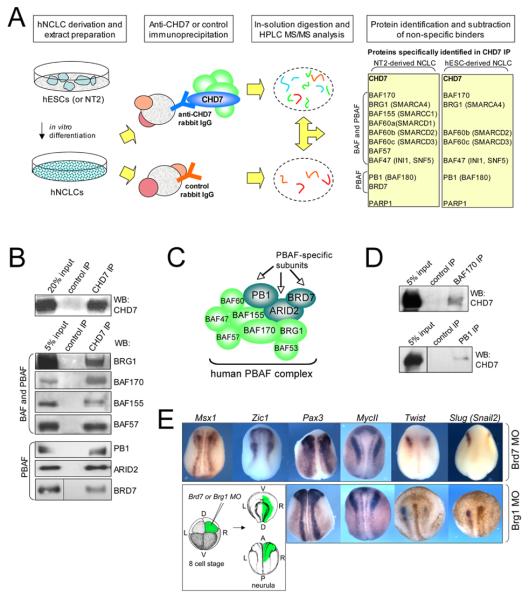
Figure 4. CHD7 interacts with the PBAF complex in neural crest cells.
(A) Identification of CHD7 protein partners in hNCLCs. A schematic representation of the purification process with polypeptides uniquely identified in CHD7 immunoaffinity purifications from NT2-derived or hESC-derived NCLCs shown on the left (see Methods for details).
(B) Confirmation of association between CHD7 and PBAF. Nuclear extracts from hESC-derived hNCLCs were used as input for immunoprecipitation analyses with α-CHD7 or control antibodies. Bound polypeptides were analyzed by immunoblotting with antibodies recognizing common BAF/PBAF subunits as PBAF specific subunits, as indicated.
(C) Schematic representation of the human PBAF complex. Arrows indicate unique PBAF subunits.
(D) CHD7 co-immunoprecipitates with PBAFcomplex. Nuclear extracts from hESC-derived hNCLCs were used as input for immunoprecipitation analyses with α-BAF170, α-PB1 or control antibodies. Bound polypeptides were analyzed by immunoblotting with α-CHD7 antibody.
(E) Effect of Brd7 and Brg1 MO injection on expression of transcription factors involved in neural crest formation. Eight cell-stage embryos were injected with either BRD7 MO (3.3 uM) or BRG1 MO (2.5uM) asymmetrically into a single dorsal anterior blastomere that gives rise to the dorsal-anterior part of the neurula embryo, as shown in the schematic. The late neurula stage embryos were analyzed by whole mount RNA in situ hybridization to visualize expression of Msx1, Zic1, Pax3, MycII, Twist and Slug.
PBAF and BAF are related, yet distinct chromatin remodeling complexes, which share multiple core complex components including the catalytic subunit BRG1, but are distinguished by the presence of ARID1A and ARID1B in BAF complex, and Polybromo (PB1, BAF180), ARID2 and a newly identified subunit BRD7 in PBAF complex25,26,27 (Figure 4C). Immunoblot analyses of the CHD7 immunoprecipitates with anti-BRG1, BAF170, BAF155, BAF57, PB1, and BRD7 antibodies confirmed association of CHD7 and PBAF complex components (Figure 4B). Moreover, ARID2, a PBAF-specific subunit that was not identified in our mass spectrometry analysis, also co-immunoprecipitated with CHD7 (Figure 4B). Reciprocal immunoprecipitation analyses with anti-BAF170 and anti-PB1 antibodies recovered CHD7 (Figure 4D), indicating that the presence of the PBAF complex in CHD7 immunoprecipitates is not an artifact of the antibody cross-reactivity. Although we did not recover BAF-specific subunits in our purification, at present we cannot exclude a possibility that CHD7 can also associate with BAF. Nevertheless, our results demonstrate that in human neural crest cells PBAF is a major CHD7-associated protein complex.
PBAF function is important for neural crest formation in Xenopus
Brg1, a catalytic engine of both BAF and PBAF complexes was implicated in neural crest formation in zebrafish 28, but whether this activity can be attributed to the function of BAF or PBAF is not known. Brg1-containing complexes have very broad roles in regulating early development 29, therefore to target PBAF function specifically in the dorsal-anterior structures, we injected morpholinos targeting Brg1 (Brg1 MO, described in 30) or a specific PBAF subunit Brd7 (Brd7 MO) into a single DA blastomere of eight cell stage embryos (Figure 4E). Importantly, although Brg1 is involved in neurogenesis, it is not essential for neural induction30. Next we examined expression of selected transcription factors involved in the neural crest formation. Similarly to what we observed for CHD7, downregulation of Brg1 or Brd7 does not affect expression of neural plate border specifiers or maintenance factor MycII, but leads to downregulation of core neural crest transcription factors, such as Twist and Slug (Figure 4E). Moreover, analysis of Brg1 and Brd7 morphants injected into a DA blastomere at the eight cell stage revealed significant phenotypic overlap with CHD7 morphants, including eye coloboma, otolith defects and craniofacial malformations (Supplementary Figure 13). Co-injection of BRD7 mRNA with Brd7 MO rescued eye and craniofacial defects (not shown; other traits were not scored), and the specificity of Brg1 MO was characterized previously 30. Moreover, Brd7 MO injection did not appreciably affect CHD7 levels (Figure 2B). Taken together, our results demonstrate that PBAF function is essential for neural crest development in Xenopus and suggest that CHD7 and PBAF cooperate in gene expression regulation during neural crest and placode formation.
Significant overlap between CHD7 and Brg1 genomic targets in mouse ESCs
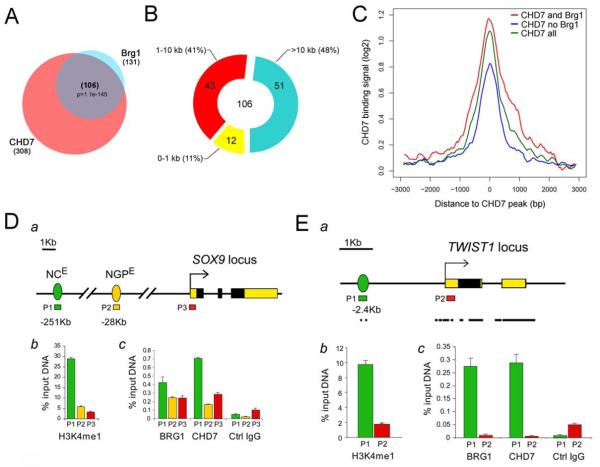
Next we wanted to determine whether CHD7 and PBAF can co-occupy same genomic targets. Although no genomic binding data are available for either CHD7, Brg1 or any of the PBAF components in neural crest cells and such analysis is challenging due to limiting availability of neural crest cells, CHD7 occupancy analysis over the ENCODE regions representing about 1% of the genome was recently reported for mouse ESCs and NPCs 31. This study demonstrated that CHD7 binding is cell type specific, and overlaps with a subset of regions marked by H3K4me1, a modification shown to be enriched at enhancer elements31,32. In addition, genome-wide Brg1 occupancy analysis in mouse ESCs was recently reported 33.To address whether Brg1 and CHD7 co-occupy gene targets in mouse ESCs, we used the reported datasets to identify Brg1 binding sites within the ENCODE regions and compared them with the CHD7-bound regions. As shown in Figure 5A, 106 out of 131 or 81% of Brg1 binding sites present within the ENCODE regions were also bound by CHD7, whereas 34% of CHD7 sites were also bound by Brg1. For a comparison, out of 126 Suz12 binding sites present within ENCODE regions, none were bound by CHD7 (not shown). Analysis of the distance of CHD7-Brg1 co-occupied regions from transcription start sites (TSS) revealed that 89% of co-occupied sites was located more than 1 kb away from the nearest TSS (Figure 5B). Furthermore, CHD7 binding footprints at sites co-occupied by Brg1 displayed stronger and broader signals than sites only bound by CHD7 (Figure 5C). Taken together, these observations are suggestive of the synergistic CHD7-Brg1 co-occupancy at distal regulatory elements. We thus hypothesized that in multipotent neural crest cells, CHD7 cooperates with Brg1 and PBAF to regulate enhancer elements controlling expression of critical neural crest genes.
Figure 5. Co-occupancy of CHD7 and BRG1 at distal elements.
(A) Overlap between CHD7 and Brg1 occupancy in mouse ESCs. Brg1 bound regions occurring within mouse ENCODE regions (n=131) were identified in the genome-wide dataset reported by 33, and compared to 308 CHD7-bound regions within mouse ENCODE regions reported by 31.
(B) Most CHD7-Brg1 sites are located more than 1 kb away from an annotated TSS. Distribution of CHD7-Brg1 co-occupied regions identified in A relative to annotated transcription start sites is shown.
(C) CHD7 sites co-occupied by Brg1 display stronger and broader binding signals. Mouse ESC CHD7 binding signal (log2, y-axis) footprints were generated around CHD7 bound regions for: all mouse CHD7 bound regions (green), regions bound by both CHD7 and Brg1 (red) or by CHD7 alone (blue) as indicated above. Chd7 ChIP-chip signals used in footprint generation were obtained from Gene Expression Omnibus (GEO) data set GSE14460 31.
(D) CHD7 and BRG1 co-occupy neural crest specific distal enhancer controlling SOX9 expression.(a) Schematic representation of the SOX9 locus showing the relative position of the primer sets (P1-3) used for ChIP-qPCR analyses. NCE: neural crest, and NGPE: notochord, gut and pancreas SOX9 distal enhancer elements identified by 34. (b) ChIP-qPCR analysis of H3K4me1 levels at indicated genomic regions. Y axis shows percent of input recovery; (c) ChIP-qPCR analysis of CHD7 and Brg1 occupancy at indicated genomic regions. Y axis shows percent of input recovery.
(E) CHD7 and BRG1 co-occupy a conserved distal element upstream from TWIST1 TSS.(a) Schematic representation of the TWIST1 locus showing the relative position of the primer sets (P1-2) used for ChIP-qPCR analyses. Conservation index for 31 eutherian mammals from ENSEMBL is shown at the bottom. (b) ChIP-qPCR analysis of H3K4me1 levels at indicated genomic regions. Y axis shows percent of input recovery; (c) ChIP-qPCR analysis of CHD7 and Brg1 occupancy at indicated genomic regions. Y axis shows percent of input recovery.
In hNCLCs CHD7 and BRG1 co-occupy distal elements located upstream from SOX9 and TWIST1 gene promoters
In Xenopus embryo Sox9 expression in the neural crest and otic placode domains is affected by CHD7 downregulation (Figure 2D). Interestingly, a conserved enhancer element located 251 kb upstream from human SOX9 gene (NCE) was shown to mediate expression specifically in the cranial neural crest and otic placode when assayed in the mouse embryo34 (Figure 5D, panel a). We used chromatin immunoprecipitation coupled to quantitative PCR (ChIP-qPCR) with anti-H3K4me1, anti-CHD7 and anti-BRG1 antibodies to test for the enrichment at the SOX9 NCE element in hNCLCs. As shown in Figure 5D, panel b, NCE region was marked by H3K4me1, a modification associated with active enhancers32, but H3K4me1 was not enriched at SOX9 TSS or another distal enhancer element located 28 kb upstream from TSS and shown to mediate expression in the notochord, gut and pancreas (NGPE). Both CHD7 and BRG1 were enriched at the SOX9 NCE (Figure 5D, panel c).
Unlike SOX9, distal elements controlling neural crest-specific expression of TWIST1 gene are not well understood. We have identified a genomic region located 2.4 kb upstream from the TWIST1 TSS that is evolutionarily conserved among eutherian mammals and highly marked by H3K4me1 in hNCLCs (Figure 5E, panels a and b). Next, we demonstrated that CHD7 and BRG1 both bound to this genomic region, but were not detected at the TWIST1 TSS. Taken together, our results demonstrate that in hNCLCs CHD7 and BRG1 co-occupy a known neural crest-specific enhancer controlling SOX9 expression, as well as novel genomic element located upstream from TWIST1 TSS and marked by the histone modification signature consistent with the enhancer identity.
CHD7 and PBAF act synergistically to promote neural crest gene expression and cell migration
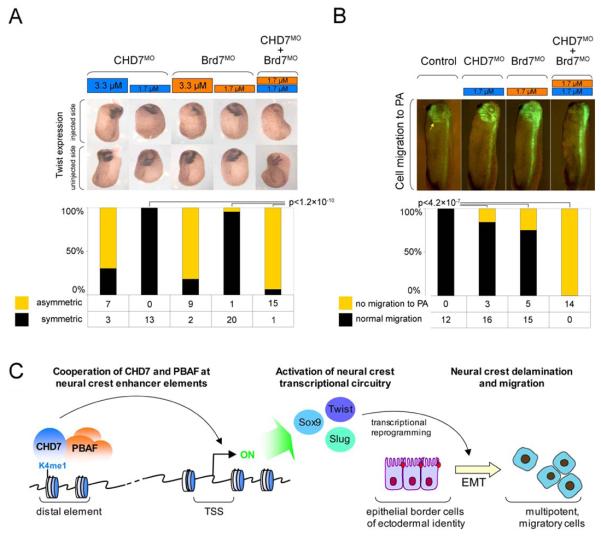
CHD7, Brg1 and Brd7 are important for Twist expression during neural crest migration in Xenopus (Figures 2D and 4E). To test whether CHD7 and PBAF synergistically regulate Twist expression in vivo, we took advantage of the dosage-sensitive effect of CHD7 and Brd7 MOs. Injection of CHD7 or Brd7 MO at 3.3 uM into a DA blastomere of an 8-cell stage embryo results in downregulation of Twist on the injected side, but a two-fold lower concentration of each morpholino (1.7 uM) has only a minor effect (Figure 6A; all injection volumes were kept constant). However, co-injection of both morpholinos at the 1.7 uM concentration results in a dramatic downregulation of Twist on the injected side (Figure 6A; p<1.2×10−10, Fisher's exact test for count data). These results indicate that CHD7 and Brd7 have synergistic effect on Twist gene expression. Next we asked whether CHD7 and Brd7 cooperate in promoting cephalic neural crest migration. Co-injection of KikGR fluorescent tracer and either CHD7 or Brd7 MO at 1.7 uM into a DA blastomere at the 8-cell stage had only a minor effect on PA labeling (Figure 6B). In contrast, simultaneous co-injection of both morpholinos at the same concentration resulted in lack of cell migration to PAs (Figure 6B). In sum, our results strongly suggest that CHD7 and PBAF act synergistically to promote neural crest gene expression and cell migration.
Figure 6. Cooperation between CHD7 and Brd7 in regulation of neural crest specific transcription and migration.
(A) Synergistic effect of CHD7 and Brd7 MOs on Twist expression. Twist in situ hybridization analysis of embryos injected into a single dorsal-anterior blastomere at the 8-cell stage with indicated doses of CHD7 and/or Brd7 MOs. Representative images are shown and asymmetry in Twist expression is quantified at the bottom. P values were calculated by Fisher's exact test for count data.
(B) Synergistic effect of CHD7 and Brd7 MOs on neural crest migration. Analysis of cell migration to PA in tadpoles derived from embryos co-injected into a single dorsal-anterior blastomere at the 8-cell stage with fluorescent lineage tracer (KikGR) and indicated doses of CHD7 and/or Brd7 MOs. Representative images are shown and migration to PA is quantified at the bottom. P values were calculated by Fisher's exact test for count data.
(C) Model of CHD7 function in neural crest formation. We propose that CHD7 and PBAF cooperatively regulate activity of enhancer elements controlling expression of critical neural crest transcription factors. Activation of core components of neural crest transcriptional circuitry by coordinate action of CHD7 and PBAF in turn allows for transcriptional reprogramming of ectodermal border territory cells leading to epithelial to mesenchymal transition (EMT), acquisition of multipotency and migratory potential characteristic of the early neural crest cells.
We propose that during formation of the multipotent neural crest, CHD7 and PBAF cooperatively regulate activity of enhancer elements controlling expression of critical neural crest transcription factors (Figure 6C). It remains on open question how occupancy of CHD7-PBAF complex at neural crest enhancers is regulated, but recognition of H3K4me1 by CHD7 tandem chromodomains 31 may contribute to this process, similarly to the way recognition of H3K4me3 by a PHD finger contributes to stabilization of the NURF remodeling complex at promoters 35. The activation of core components of neural crest transcriptional circuitry by coordinate action of CHD7 and PBAF in turn allows for transcriptional reprogramming leading to acquisition of multipotency and migratory potential characteristic of the early neural crest cells.
A large subset of developmental anomalies observed in CHARGE patients is likely a direct consequence of the defects in establishment of gene expression programs orchestrating neural crest formation and migration. Moreover, our results indicate that expression of placodal transcription factors such as Pax2 is also regulated by CHD7, which may account for ocular coloboma and inner ear defects in CHARGE patients. In addition to illuminating syndrome patho-embryology, our work identifies PBAF complex components such as BRD7 as potential candidates for novel genes affected in CHARGE patients without CHD7 mutations. Beyond relevance for CHARGE pathogenesis, this study suggests broader implications for the role of CHD7 in cell motility. Given that CHD7 function is essential for the expression of critical regulators of EMT and metastasis Twist and Slug, we predict that in addition to CHD7 loss-of-function causing CHARGE, CHD7 gain-of-function may play a role in tumor progression, particularly in cancers of the neural crest origin, such as neuroblastomas and melanomas.
Methods Summary
Human ESC culture and derivation of hNCLCs
WA-09 (H9; WiCell) cells were passaged as described.36 Briefly, H9 cells were grown on matrigel-coated dishes with irradiated mouse embryonic fibroblasts in medium supplemented with 8ng/ml bFGF. For neuroepithelial induction, MEF and matrigel depleted H9 cells were harvested by digestion with 1mg/ml collagenase IV for ~30-40 minutes, followed by PBS washes and gentle trituration. The hESC clusters were transferred to neural induction medium (NIM) 1:1 ratio of DMEM-F12 (Invitrogen #10565-018) and Neurobasal (21103-049) media with 0.5xN2, 0.5xB27, 5ug/ml Insulin, 20ng/ml bFGF, 20ng/ml EGF, Ix Penn Strep in uncoated polypropylene dishes. Spheres/rosettes were allowed to spontaneously attach, and between 6-9 days of differentiation, hNCLCs migrated from rosettes. Subsequently, rosettes were dissected away using a 28G needle under an inverted microscope. These P0 isolated NCLCs were passaged with accutase and replated on 1ug/ml fibronectin coated dishes for 10-15 days in NIM for maturation into MPs. P1 NCLCs were obtained by collecting migratory cells from the dissected and re-plated rosettes.
Methods
NCLC differentiation
P0 and P1 derived NCLCs were plated on 100ug/ml ornithine and 5ug/ml laminin coated coverslips in growth factor reduced medium (3:1 ratio of DMEMF12 and Neurobasal media supplemented with 0.25x B27, 0.5x glutamine and 1x antibiotic antimycotic mix). Medium was changed once a week. Cells were fixed and analyzed after 1, 4 or 7 weeks of differentiation. Differentiation of mature MPs (after 3-4 weeks of amplification) into specific lineages (adipocytes and osteoblasts) was carried out essentially as described.37
Immunocytochemistry
Cells were fixed in fresh 4% paraformaldehyde. For stainings with antibodies recognizing intracellular epitopes, phosphate buffered saline with 0.5 mg/ml BSA and 0.1% Triton X-100 was used for blocking and permeabilization. For cell surface stainings TritonX-100 was eliminated from the blocking buffer. The list of primary antibodies and dilutions used for staining is shown in the Supplementary Table 3. Appropriate Alexa 488, Rhodamine or Alexa 594, labeled secondary antibodies and/or DAPI counterstaining was used for visualization on a confocal (Leica TSC SP2) or inverted (Leica DMI 6000B) microscope.
RNA isolation and RT PCR
For Quantitative RT-PCR, total RNA was extracted using TRIZOL followed by turbo DNAse I treatment (Ambion) to avoid genomic contamination. Total RNA (1ug for each sample) was reverse transcribed (Superscript II, Invitrogen) and quantitative real time PCR analysis was performed using Roche Lightcycler 480 according to manufacturers recommendations. Primer sequences for each of the analyzed genes are listed in the Supplementary Table 4.
In ovo transplantation
Rosettes with migrating cells were labeled with 5uM CMFDA (a green fluorescent, viable dye, Molecular Probes) for 1 hr. The cells were washed several times with medium and cultured for 12 hr. Fertile white leghorn chicken eggs (Charles River) were incubated at 37 °C in a humidified incubator. For transplantation, the vitelline membrane of H&H stage 8/9 embryos was peeled and a small piece of the dorsal neural tube was dissected out with tungsten needles and replaced with either NCLCs that migrated out of the rosettes or a part of the rosette. 6 hrs later embryos in which the transplanted cells were still associated with the neural tube and not dispersed due to mechanical agitation were incubated further and imaged in ovo at 32 hrs post-transplantation using a fluorescent stereomicroscope.
Lentiviral infection, selection and quantification of the migration defect
pSM2c retroviral constructs encoding miRNA-resembling shRNAs targeting human CHD7 (RHS4529-NM_017780) or control, verified non-silencing shRNA (RHS4335) were purchased from Open Biosystems The MluI and XhoI fragment from pSM2c containing the hairpin was cloned into a Dox inducible lentiviral vector, TRIPZ (Open Biosystems # RHS4696-99634943).38,39 VSVG pseudotyped lentiviruses were produced in 293T cells as described previously.40 Undifferentiated hESC were infected in suspension for 2 hrs with a concentrated virus as described.36 Infected hESC were amplified for one week, replated on puromycin resistant MEF's and then selected with 2ug/ml puromycin for 10 days prior to differentiation. The infected pool populations were differentiated in the presence or absence of 1ug/ml doxycyclin. The attached spheres with the migrating cells were imaged on an inverted fluorescent microscope on day 9 of differentiation. For quantification of the migration defect, all spheres with red fluorescence were counted and the percentage of RFP expressing spheres that were attached and had migrating NCLCs was determined. For RT-PCR and immunoblot analyses, the predominantly RFP expressing spheres were separated on the second day of initiation of differentiation in the presence of doxycyclin and RNA and protein extracted at day 9 of differentiation.
Protein extraction, immunoprecipitation and immunoblotting
Nuclei were prepared from hESC and NT2 derivatives using the Dignam protocol and extracted with buffer containing 100mM Tris (pH 8.0), 300mM NaCl, 0.1% NP-40, 10% glycerol, 1mM EDTA, 1mM EGTA and 1mM DTT and freshly added 1mM PMSF and protease inhibitor cocktail (Roche).41 For immunoprecipitation, 0.5-2 mg of nuclear extracts with salt concentration adjusted to 250 mM NaCl, were incubated with specific antibodies and protein A-Dynal beads for 2hrs at 4 degrees. The bound proteins were captured with a magnet, washed in IP buffer and eluted in 0.5M NH4OH, 0.5mM EDTA. The eluted proteins were either used for mass spectrometric analysis or western blotting. Alternatively, proteins from the washed beads were eluted by boiling in Laemmli buffer. Extracts or IP eluates were separated by SDS PAGE and analyzed by immunoblottingblotting with antibodies listed in table S2.
For Xenopus extracts, ~30 staged embryos were washed twice in 0.1x MMR followed by one wash in Dignam buffer A. The embryos were dounced using loose pestle in 500ul of Dignam buffer A containing 2M sucrose. The homogenate was layered over a 2.05M sucrose containing-Dignam buffer A column and centrifuged for 1hr in a Beckman SW50.1 rotor. The pelleted nuclei were boiled in Laemmli buffer prior to separating proteins by SDS PAGE 42
HPLC/MS/MS analysis
HPLC/MS/MS analysis was performed in an integrated system that includes an Agilent 1100 series nanoflow liquid chromatography system and an LTQ two-dimensional ion trap mass spectrometer equipped with a nanoelectrospray ionization source. The eluate from each immunoprecipitation, either using a polyclonal anti-CHD7 antibody (ab31824) or control rabbit IgG, was lyophilized and then reconstituted in 30 to 50 μl of 50 mM ammonium biocarbonate and pH adjusted to 7-9. Trypsin (200ng of enzyme) digestion lasted 2 hours at 37° C. The resulting peptides were cleaned with C18 ZipTips (Millipore Corp.) according to the manufacturer's instructions. Subsequently, trypic peptides were separated in a capillary HPLC column (11 cm length × 75 μm inner diameter) packed in-house with Luna C18 resin (5 μm particle size, 100 Å pore diameter) (Phenomenex). Peptides were eluted from the column with a gradient of 2.0% to 90% buffer B (90% acetonitrile/9.95% water/0.05% acetic acid) in a 2 h LC/MS/MS analysis. Liquid chromatography tandem mass spectrometry was operated in a data-dependent mode such that the ten strongest ions in each MS scan were subjected to collisionally activated dissociation with a normalized collisionally activated dissociation energy of 35%. Database search was conducted with Mascot 2.1 and the NCBInr database. One missed cleavage was allowed for database search. The protein score is derived from the ions scores. More details can be found at http://www.matrixscience.com/help/interpretation_help.html.
Antibody production
To generate antibodies specifiaclly recognizing Xenopus CHD7 two rabbits were immunized (Covance) with a purified immunogen corresponding to the N terminal region of xl CHD7 amplified by the following primers Forward primer: 5′- ATGGCAGACCCTGGCATGATGAGTTTATTT -3′ and Reverse primer: 5′- GTAATTTCCCATCTGTTGAACATGCTGAGG -3′
Chromatin immunoprecipitation (ChIP) and ChIP-qPCR
ChIP of chromatin from NCLC enriched cells was performed according to Boyer et al 43. For each ChIP, 100 to 500 ug of chromatin was used. After overnight antibody-Dynal bead incubation, 5% of the pull down was taken for subsequent qPCR analysis using Roche Lightcycler 480. ChIP-qPCR signals of individual ChIP reaction was standardized to its own input qPCR signals as % input. Primer sequences are shown in Table S3.
Genomic data analyses
Chd7 31 or Brg1 33 binding regions in mouse ESC were as defined in the corresponding publications. For calculating the overlaps between Chd7 and Brg1, we first determined those Brg1 bound regions occurring within mouse ENCODE regions (n=131), since this binding data was generated for the whole genome. Subsequently, Chd7 regions were extended 500 bp on each side. At least 1bp overlap between the Chd7-extended and Brg1 regions was required to considered a region as co-occupied by both proteins (Brg1 regions were not extended since they are already quite large, with an median size of 6 kb). Chd7 regions being also bound by Brg1 were mapped with respect to protein coding gene transcription start sites, according to Refseq gene annotations based on Mm7 mouse genome. For creating binding signal footprints, mouse ESC Chd7 binding signal was obtained from from Gene Expression Omnibus (GEO) data set GSE14460 31 and analyzed using genomic Signal Aggregator (GSA)(http://zlab.bu.edu/GSA/44), considering a window size of +/− 3000 bp.
Morpholino oligonucleotide design, CHD7 mRNA cloning and mutagenesis
Translation-blocking morpholinos designed to Xenopus laevis CHD7 (CHD7 MO: 5′ - AACTCATCATGCCAGGGTCTGCCAT 3′) BRD7 (BRD7 MO: 5′- GCTTCTTATGTTTTTTCCCCATTGC -3′) and BRG1 (BRG1 MO: 5′-TCACTGCTAACCTGTCCCCGAATCC -3′) 45 were obtained from Gene Tools LLC (http://www.gene-tools.com) and resuspended in water at a concentration of 1mM. The CHD7 cDNA fragment (KIAA1416) (human CHD7) containing all the domains listed in figure S7 was obtained from Kazusa DNA Research Institute (KDRI). The missing 5′ and 3′ fragments were PCR-amplified from the human teratocarcinoma NT2 cell line using following primers: 5′ (primer 1: 5′ - cgcgtggcgcgccttatggcagatccaggaatgatgagtc – 3′; primer 2: 5′ - agtcgactctgtccaggttcggaattccctt – 3′)and 3′ (primer 1: 5′ - cgcgttcgatgtttgaccgccttctcact – 3′; primer 2: 5′ - agtcgaccgtacgctggaactggtactggttattcatcattttcattg – 3′). Amplified fragments were ligated to KIAA1416 via internal MfeI and NotI sites for the 5′ and 3′ fragments, respectively. The resulting full length human CHD7 transcript was cloned into pcDNA vector (Invitrogen) as a FLAG-His-CHD7 fusion transcript. 5′-capped and poly-A tailed mRNAs were transcribed from AvrII-linearized template using SP6 RNA polymerase. Catalytically inactive ATPase mutant CHD7 protein (CHD7K998R) was generated using a modified PCR-based site-directed mutagenesis strategy.46 Two sets of primers (Set 1: forward primer: 5′- gcaccaagggaattgactg - 3′, reverse primer: 5′- tcccaaacccatttcatctgctaaaatg - 3′; Set 2: forward primer: 5′- atccagtccattacatttctctatgag - 3′, reverse primer: 5′- gtgtgggttcgacgtaggag - 3′) containing the desired point mutation were used to PCR-amplify two fragments of CHD7 from the pcDNA vector containing wildtype human CHD7. The ligated product of the two fragments was then cloned into the pcDNA vector containing wildtype human CHD7, replacing the corresponding region of wildtype CHD7.
Xenopus embryology
Xenopus laevis embryos were staged according to standard procedures.47 To knockdown global xCHD7 expression levels, CHD7 MO was injected into both blastomeres of two-cell stage embryos at final concentrations of 1.7, 3.3, 4 and 5 μMin a 10nL volume. Embryo viability was scored throughout development to the tadpole stage (stage 35), whereupon embryos were scored for the penetrance of phenotypic traits. At the highest tolerated concentration of injected CHD7 MO (5μM), embryos were viable to the neurula stage (stage 16), after which their survival was severely perturbed. For rescuing the viability perturbation 1ng of FLAG-His-CHD7 mRNA was co-injected with the MO. For injections in one blastomere at the 8-cell stage, 2- cell stage embryos where the first cleavage plane clearly bisected the lightly pigmented future dorsal domain were separated and allowed to develop. 1.25 nl volume was injected into one dorsal blastomere at the 8-cell stage. For RNA whole mount in situ hybridization and phenotypic trait analysis at the tadpole stage embryos were injected at a CHD7 MO(3.3), BRD7 MO(3.3uM) or BRG1 MO(2.5uM) unless otherwise mentioned. 5′ capped and 3′tailed mRNA's were synthesized using the Ambion message machine ultra kit. Wildtype CHD7 and CHD7K998R templates were with AvrII and transcribed using SP6 RNA polymerase . A titer of CHD7 ATPaseK998R mRNA (ranging from 2.25ng to 9ng) co-injected with CHD7 MO (3.3μM) in a 10nL volume at the two-cell stage did not rescue the phenotypic traits observed in the CHD7 morphants. Overexpression of CHD7 ATPaseK998R mRNA alone produced phenotypes similar to those obtained with low doses of morpholino, indicating that CHD7 ATPaseK998R mRNA acts as a dominant negative. To score for the craniofacial and heart defects observed upon overexpression of CHD7 ATPaseK998R mRNA, two-cell stage embryos were injected in both blastomeres with 9ng CHD7K998R mRNA and developed to late stage tadpoles (beyond stage 45) along with uninjected controls or similar amounts of Kaeda mRNA injected controls . Tadpoles were imaged and scored or embdded in paraffin for serial sectioning prior to Hemotoxylin and Eosin staining
Whole mount in situ hybridization analysis
Staged embryos were fixed in MEMFA buffer for 1 hour, dehydrated in 100% methanol, and stored at −20°C prior to use. In situ hybridizations were performed according to the standard protocol.48 cDNA fragments corresponding to CHD7, Sox9, MycII and Twist genes were PCR amplified from stage 35 Xenopus laevis cDNA and cloned into pCS2+ vector . Primers are listed in table S4 Plasmids containing cDNA fragments for several genes were the kind gifts of Ellen Langer (Msx1), Kristen Kroll (Pax3) Julie Baker (Slug, Pax2), Jean-Pierre Saint-Jeannet (Zic1). Digoxigenin-labeled RNA probes were transcribed in vitro using SP6 or T7 RNA polymerase and DIG labeling mix (Roche).
In vivo migration assay
At the two-cell stage, embryos were injected in both blastomeres with 450pg photo-convertible Kaede mRNA transcribed with SP6 RNA polymerase from pCS2+ vector containing the open reading frame of the full length Kaede gene (a kind gift from Ilya Shestopalov and James K. Chen, Stanford University) in a 10nL volume. Embryos were monitored to the neurula stage (stage 17), and a subset of the anterior neural and neural crest tissues was UV-irradiated using a Leica DM4500 microscope with rectangular pinhole. Embryos were imaged at the late tailbud stage (stage 25) and positive migration to the pharyngeal arches was quantified. To assay for the perturbation of neural crest cell migration to the pharyngeal arches in an asymmetrical setup, two-cell embryos were injected in one blastomere with 3.3uM CHD7 MO or 9ng CHD7K998R mixed with 450pg Kaede mRNA in a 5nL volume. Embryos were monitored and UV-irradiated as described above. At the late tailbud stage (stage 25), asymmetry (defined as positive migration to the pharyngeal arches on the uninjected side and absence of positive migration to the pharyngeal arches on the injected side) was quantified.
Supplementary Material
Acknowledgements
The authors thank all laboratory members for input during the course of this work; S. Brugmann and A.C. Foley for sharing expertise in Xenopus and chick embryology, B.J. Chord and R. Rodriguez for assistance with H9 culture; J. Baker, B. Bedogni (M.B. Powell lab), Lena Ho (G. Crabtree lab), E. Langer (K. Kroll lab), and J.P. Saint-Jeannet a for reagents; A. Sanchez-Alvarado for cartilage staining protocol; G. Crump and S. Cox for sharing unpublished information, S. Brugmann, E. Duncan, Z. Ma, J. Peng, A.M. Ring, and R.A. Roth for comments on the manuscript. This work was supported by CIRM SEED RS1-00323, W.M. Keck Foundation Distinguished Young Scholar in Biomedical Research, and Searle Scholar grants for J.W.
Footnotes
Supplementary Information
Supplementary Figures 1-6 with legends and Supplementary Tables 1-4 are attached in a separate file accompanying the manuscript.
References
- 1.Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 3.Dupin E, Creuzet S, Le Douarin NM. The contribution of the neural crest to the vertebrate body. Adv Exp Med Biol. 2006;589:96–119. doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- 4.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Schaner CE, Kelly WG. Germline chromatin. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.73.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan S, et al. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 10.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 12.Sanlaville D, Verloes A. CHARGE syndrome: an update. Eur J Hum Genet. 2007;15:389–399. doi: 10.1038/sj.ejhg.5201778. [DOI] [PubMed] [Google Scholar]
- 13.Siebert JR, Graham JM, Jr., MacDonald C. Pathologic features of the CHARGE association: support for involvement of the neural crest. Teratology. 1985;31:331–336. doi: 10.1002/tera.1420310303. [DOI] [PubMed] [Google Scholar]
- 14.Bajpai R, et al. Molecular stages of rapid and uniform neuralization of human embryonic stem cells. Cell Death Differ. 2009;16:807–825. doi: 10.1038/cdd.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 16.Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- 17.Minarcik JC, Golden JA. AP-2 and HNK-1 define distinct populations of cranial neural crest cells. Orthod Craniofac Res. 2003;6:210–219. doi: 10.1046/j.1601-6335.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- 18.Del Barrio MG, Nieto MA. Relative expression of Slug, RhoB, and HNK-1 in the cranial neural crest of the early chicken embryo. Dev Dyn. 2004;229:136–139. doi: 10.1002/dvdy.10456. [DOI] [PubMed] [Google Scholar]
- 19.Tucker GC, Duband JL, Dufour S, Thiery JP. Cell-adhesion and substrate-adhesion molecules: their instructive roles in neural crest cell migration. Development. 1988;103(Suppl):81–94. doi: 10.1242/dev.103.Supplement.81. [DOI] [PubMed] [Google Scholar]
- 20.Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008;46:673–682. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- 21.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 22.Saint-Germain N, Lee YH, Zhang Y, Sargent TD, Saint-Jeannet JP. Specification of the otic placode depends on Sox9 function in Xenopus. Development. 2004;131:1755–1763. doi: 10.1242/dev.01066. [DOI] [PubMed] [Google Scholar]
- 23.Blake KD, Prasad C. CHARGE syndrome. Orphanet J Rare Dis. 2006;1:34. doi: 10.1186/1750-1172-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eccles MR, Schimmenti LA. Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clin Genet. 1999;56:1–9. doi: 10.1034/j.1399-0004.1999.560101.x. [DOI] [PubMed] [Google Scholar]
- 25.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eroglu B, Wang G, Tu N, Sun X, Mivechi NF. Critical role of Brg1 member of the SWI/SNF chromatin remodeling complex during neurogenesis and neural crest induction in zebrafish. Dev Dyn. 2006;235:2722–2735. doi: 10.1002/dvdy.20911. [DOI] [PubMed] [Google Scholar]
- 29.Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Seo S, et al. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnetz MP, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagheri-Fam S, et al. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–397. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 36.Loring JF, Wesselschmidt RL, Schwartz PH. Human stem cell manual : a laboratory guide. 1st edn Elsevier/Academic Press; 2007. [Google Scholar]
Methods references
- 37.Lee G, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 38.Stegmeier F, Hu G, Rickles R, Hannon G, Elledge S. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada T, Fujii H, Mitsuya H, Nienhuis AW. Targeted and highly efficient gene transfer into CD4+ cells by a recombinant human immunodeficiency virus retroviral vector. J Clin Invest. 1991;88(3):1043–1047. doi: 10.1172/JCI115365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zufferey R, Nagy D, Mandel R, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 41.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu K, Gurdon JB. A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc Natl Acad Sci U S A. 1999;96:6791–6796. doi: 10.1073/pnas.96.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 46.Higuchi R, Krummel B, Saiki R. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) Garland Publishing Inc; 1994. [Google Scholar]
- 48.Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.