Abstract
Songbirds share some essential traits but are extraordinarily diverse, allowing comparative analyses aimed at identifying specific genotype–phenotype associations. This diversity encompasses traits like vocal communication and complex social behaviors that are of great interest to humans, but that are not well represented in other accessible research organisms. Many songbirds are readily observable in nature and thus afford unique insight into the links between environment and organism. The distinctive organization of the songbird brain will facilitate analysis of genomic links to brain and behavior. Access to the zebra finch genome sequence will, therefore, prompt new questions and provide the ability to answer those questions.
Introduction
Songbirds are the dominant members of the largest order of birds on earth, the Passeriformes. Over half of the around 10,000 extant bird species are passerines, and most of these are further classified as songbirds because they share an additional trait: the ability to communicate via learned vocalizations. Songbirds are social in sophisticated ways, displaying features, such as monogamy and cultural inheritance, that we often identify as important in human life. They are adaptive, both physiologically, responding immediately to environmental cues, and evolutionarily, inhabiting diverse ecological niches. And they are diverse, yet related in a lineage about as old as the mammalian radiation. These qualities have made songbirds attractive subjects for research in ecology, evolution, and the neurobiology of behavior.
Now, songbirds are about to assume a new role in genomic research. The zebra finch (Figure 1) was chosen as the second bird species [1] for whole genome sequencing by the National Human Genome Research Institute. The first draft assembly of the zebra finch genome was recently released (http://www.songbirdgenome.org) and provides a new vantage point for interpreting the evolution of the vertebrate genome. In addition, the zebra finch genome will be a springboard for research in all songbird species, allowing application of genome-level tools and perspectives to a range of biological questions not well represented in the traditional genetic model organisms. Here, we introduce some of the most compelling features of songbird biology such as the neural control of singing, the complex social lives of songbirds, and the ability of songbirds to adapt to their environments, with special emphasis on areas where the new zebra finch genome sequence may have transformative impact.
Figure 1. Zebra finches.

A zebra finch male (center) flanked by two females. Zebra finches are highly social, maintaining lifelong monogamous pair bonds in large colonies.
The Songbird Radiation
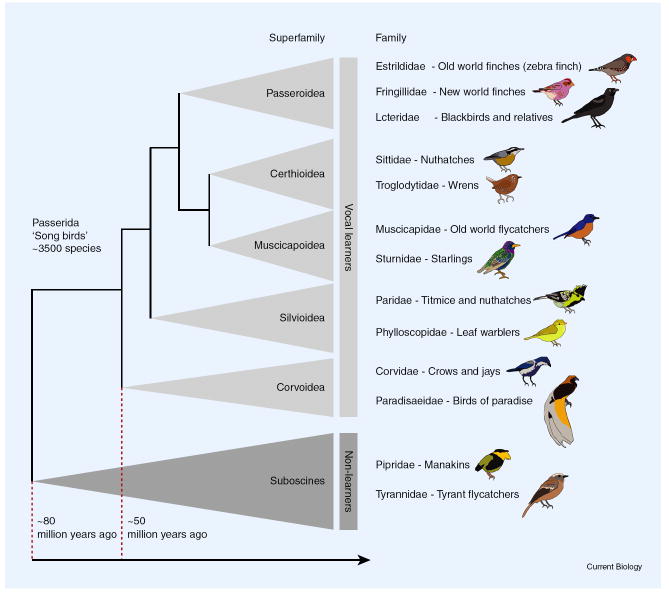
Passeriformes diverged from the rest of birds about 80 million years ago (Figure 2). The phylogenetic placement of passerines is still highly uncertain, and most recently, a novel hypothesis that places parrots as the closest relatives of passerines was proposed [2]. This relationship may have implications for the genetic and neurobiological characteristics associated with the evolution of vocal learning and plasticity, discussed below. The radiation of oscine (suborder Passeri) passerine birds began only about 65 million years ago following a split of two major passerine lineages, the oscines and suboscines (Figure 2) [3], at about the same time when non-avian dinosaurs went extinct. Even within the oscines, precise taxonomic relationships are still debated [3]. All oscines are, however, believed to have arisen in Australia, New Zealand and New Guinea. There are two major oscine subgroups: the corvids (Corvoidea), such as crows and jays, and the passerids (Passerida).
Figure 2. Overview of songbird phylogeny.
Note the position of family Estrildidae (which includes the zebra finch) as one of the ‘vocal learner’ families, in contrast to the ‘non-learner’ suboscines. Some families and species from each superfamily are listed as representatives (in total there are 122 families within the five major superfamilies of the Passerida). Drawings of a male of one species contained within the representative families are included (drawings not to scale). Estrildidae: zebra finch (Taeniopygia guttata); Fringillidae: purple finch (Carpodacus purpureus); Icteridae: Brewer’s blackbird (Euphagus cyanocephalus); Sittidae: red-breasted nuthatch (Sitta canadensis); Troglodytidae: winter wren (Troglodytes troglodytes); Muscicapidae: vivid niltava (Niltava vivida); Sturnidae: European starling (Sturnus vulgaris); Paridae: yellow-cheeked tit (Parus spilonotus); Phylloscopidae: Tickell’s leaf warbler (Phylloscopus affinis); Corvidae: western scrub jay (Aphelocoma californica); Paradisaeidae: lesser bird-of-paradise (Paradisaea minor); Pipridae: golden-collared manakin (Manacus vitellinus); Tyrannidae: brown-backed chat-tyrant (Ochthoeca fumicolor).
Passerida is a large group of ~ 3500 species, including the birds commonly considered songbirds, and is the group to which the zebra finch belongs. The term ‘zebra finch’ actually refers to two subspecies of zebra finches present in the wild. One, Taeniopygia guttata castanotis, ranges across essentially the entire extent of Australia, and is the progenitor of all common laboratory populations of zebra finch. The other, T. guttata guttata, is found among the islands of the Lesser Sundas archipelago of eastern Indonesia [4]. The two subspecies differ in body size, bill color, plumage and vocal characteristics [5], and are also genetically differentiated [6].
In general, songbirds share several characteristics, such as excellent color vision [7], a high metabolic rate (body temperature ~ 40°C), and a small body size. The zebra finch, for example, weighs a modest 10–15 grams. Songbirds are altricial and require parental care after hatching. Most species mature in less than 1–2 years, and the zebra finch matures in only 3–4 months. The key trait that has made oscines like the zebra finch attractive research subjects is that they learn their vocalizations — unlike most other avian groups, in which vocalizations are innate. Outside of birds, there is strong evidence for vocal learning in humans and cetaceans [8], and more limited evidence that some species of bats [9] and elephants [10] can also learn vocalizations. Although vocal learning may be a rare trait, among birds it appears to have evolved independently three times, in songbirds, hummingbirds, and parrots. A fourth possible case of vocal learning has been suggested in three-wattled bellbirds, Procnias tricarunculata [11], a member of the sub-oscines, which are believed not to learn vocalizations (Figure 2). The repeated evolution of vocal learning presents a unique opportunity to identify important commonalities and differences in genetic control, developmental timing, and neurophysiology.
The Genome and the Brain
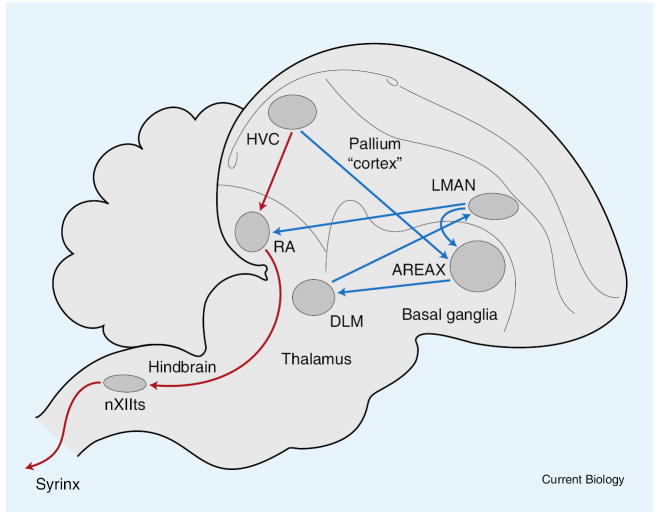
In songbirds, learned vocal communication is physically associated with evolution of a specialized vocal control apparatus, the syrinx, and a dedicated neural system. The syrinx, a bipartite voice box, allows songbirds to produce complex sounds, in some species even two different sounds simultaneously [12]. The neural song control system is composed of a set of distinct brain nuclei organized in two major interconnected pathways (Figure 3). The posterior branch is responsible for direct control of the syringeal muscles and respiratory centers and is necessary for song production. The anterior branch is more clearly associated with song learning and vocal plasticity. Comparative analyses suggest that the posterior circuit evolved from a ‘general motor pathway’ and the anterior circuit evolved from a ‘cortical-basal ganglia’ pathway present in non-oscine birds [13]. The anterior pathway in particular has strong anatomical and functional parallels to basal ganglia circuits involved in trial-and-error learning in mammals [14]. These parallels led to a recent revision of avian brain nomenclature to reflect homologies among avian and mammalian brains [15]. The discrete telencephalic song control nuclei are found only in oscines, although recent molecular analyses have indicated striking parallels in neuroanatomy and gene expression patterns between parrots and hummingbirds, and the oscine song control system [16-19].
Figure 3. Core elements of the ‘song circuit’.
Several brain centers responsible for the production of learned vocal signals in songbirds are shown. Red lines indicate the motor output pathway, descending from the nucleus known as HVC (used as a proper name) to the robust nucleus of the arcopallium (RA). Blue lines indicate an anterior forebrain loop needed for song learning, which begins with projections from a distinct set of neurons in HVC to a specialized region within the basal ganglia known as Area X. Area X projects to the medial dorsolateral nucleus of the anterior thalamus (DLM), which projects back up to the lateral portion of the magnocellular nucleus of the anterior nidopallium (LMAN). LMAN projects back onto neurons in RA that also receive input from HVC; this projection also sends collaterals to Area X.
The distinctive nuclear organization of the song control system has practical advantages for neurobiological research because its functions may be interpreted in the context of specific circuit models and behavioral outputs. The concentration of specific functions in discrete bounded brain areas has undoubtedly helped researchers recognize how behavioral change is associated with observable physical changes in particular brain areas. These changes include the shifting of song control nuclei boundaries across the seasons of breeding and molting [20,21], changes in the cell size and dendritic branching patterns [21,22], the ongoing recruitment of new neurons into the song nuclei [23] that is also influenced by the seasonal status of singing behavior [24], and the sex-specific development of the motor pathway [25].
Furthermore, it has been relatively easy to visualize distinguishing molecular features of song nuclei to show how genomic and neural control systems are integrated. Some of the earliest studies described the presence of steroid hormone binding sites [26] and recently this description has advanced even to the level of small fatty acids [27]. Expression of several genes, including the immediate early gene zenk (zif268, egr-1, ngfi-a, krox24) is activated by both the sound of birdsong [28] and the act of singing [29,30], but in different components of the song control system. Numerous genes have been identified that are differentially regulated within the song nuclei (reviewed in [31]), and with the application of microarray technologies (e.g., [32-34]) their number is increasing rapidly. By providing the ability to capture even more genetic diversity and expression changes, the zebra finch genome will further our understanding of how this song control system is dynamically regulated in real time by biologically relevant behaviors. For example, it may soon be possible to identify the genes that regulate the neural plasticity required for song learning, either during one sensitive period in development as in the case of zebra finches, or every year in the case of seasonal learners such as canaries. With the zebra finch genome, it should be possible to identify not only the genes that change in their expression across conditions, but also the regulatory elements that control them. This would enable analysis of gene–gene interactions that influence the development, function and evolution of brain systems for vocal communication.
Songbird Vocalizations
Singing behavior, the hallmark of songbirds, is easy to observe, record, analyze, and manipulate [35-37]. Hence singing may represent a beachhead for both mechanistic studies of gene function in behavior and for evolutionary studies of variation and natural selection on behavioral phenotypes. Songbirds produce different kinds of vocalizations for different purposes. Here, we approach all learned, modulated vocalizations produced by songbirds as different manifestations of a unique communicative ability – the ‘singing’ of songbirds. In some contexts, ‘song’ specifically connotes the particular stereotyped vocalization that is most closely associated with courtship and territorial defense, whereas other vocalizations are often referred to as ‘calls’. In many temperate species, primarily males sing. If considered across all songbird species, however, the ability of males and females to sing ranges from only one sex singing to equally-prolific males and females singing duets together.
The zebra finch has a limited vocal repertoire: only the males sing, and each male learns to produce one song that lasts only a few seconds. Each individual male, however, sings a unique song that is rhythmic and harmonically complex (Figure 4), and the structure of which is maintained across the bird’s lifetime. Therefore, each male’s song is probably essential for the individual recognition that stabilizes mate-pairing, family-building, and the social structure of the colony. Further, while only male zebra finches produce “song”, both sexes produce about a dozen other calls used in a range of contexts, including nest building, conflict, sexual behavior, and close localization [4]. Thus, in spite of the limited song repertoire of the male zebra finch, vocalizations still make a major contribution to the complex social behaviors that zebra finches display.
Figure 4. Spectrograms of songs from several songbird species.
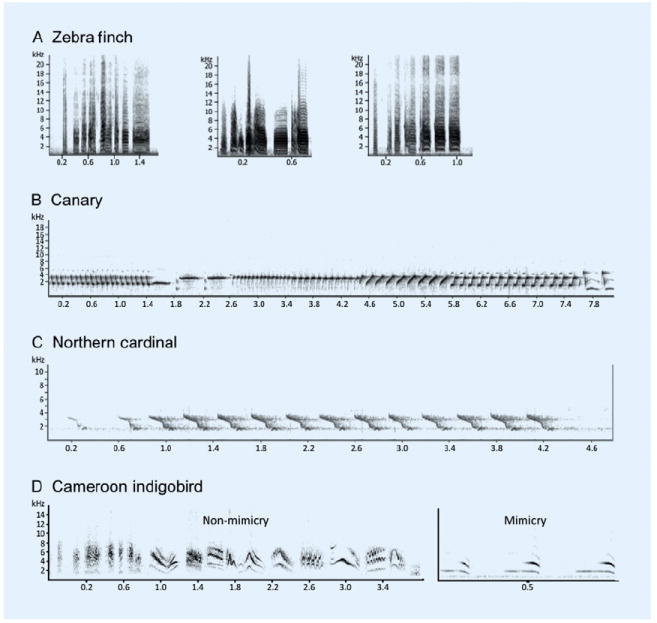
(A) Songs of three unrelated male zebra finches. Each zebra finch sings a unique song, observable by the distinct structure of each song element, or syllable, and by the ordering of those syllables. Note the typical ‘harmonic stack’ structure to the songs (visible as alternating black and white horizontal bars), individual variation in syllable structure, number of syllables, and total duration of song. (B) Canary song featuring pure-tone like syllables, syllable repetition, a multitude of different syllables, and a long song duration. (C) Tonal whistles of the northern cardinal, a species in which both sexes sing. (D) Song sparrow song highlighting syllable diversity; tones and harmonic stacks are visible. Elements can be repeated, and there are several types of syllables that comprise the song. (D) Songs of brood parasitic Cameroon indigobirds. There are two types of song: ‘non-mimicry’ songs, which are indigobird-specific, and ‘mimicry’ songs, which have syllables that are copied from the host species song. Note that the conspecific non-mimicry song is more complex than the mimicry song; it is composed of several distinct syllables types, many with rapid frequency modulation, and is longer in duration than the mimicry song.
Still, other species are much more prolific singers (Figure 4; reviewed in [38]). The northern mockingbird Mimus polyglottos repertoire consists of 100–200 different songs, requiring half an hour of continuous singing to complete. The winter wren Trogolodytes trogolodytes produces complex songs that last up to 20 seconds per bout. The hermit thrush Cathartus guttatus sings long sequences of different songs, each one in the series chosen apparently for contrast to the previous one. The brown thrasher Toxostoma rufum coordinates the two sides of his syrinx to sing in couplets, and produces them literally by the thousands: in one example a single bird produced 4653 couplets in one two-hour period.
Although many songbird vocalizations are undoubtedly reflexive, some vocalizations clearly have semantic content and flexibility. For example, black-capped chickadees, Poecile atricapilla, produce mobbing calls when challenged by a predator, and the acoustic structure of the mobbing call is varied to communicate the size of the predator [39]. Their “chick-a-dee-dee-dee” call is used in many different ways with different meanings, and has been likened to human language [40]. European starlings, Sturnus vulgaris, can form open-ended song categories [41] and may use grammatical rules in processing sequences of song elements [42].
Apart from the intrinsic interest and its significance for evolutionary biology, birdsong also has an immediate application in biomedicine as it represents the best and perhaps only viable animal model of complex human-like vocal communication [43-45]. Thirteen percent of Americans (roughly 36.5 million people) struggle with communicative disorders; stuttering, for example, affects Americans with a prevalence of 1% and has a strong genetic component, e.g., [46]. The zebra finch specifically has been proposed as an animal model for the study of stuttering [47]. A number of other heritable syndromes have major effects on vocal communication systems including autism, Parkinson’s disease and Fragile X syndrome [48]. Using genome sequence as a bridge, it may become possible to use songbirds to test specific molecular hypotheses and therapies for these human disorders. Indeed, enticing parallels between expression patterns of a gene called FoxP2, implicated in speech-related disorders in humans, have been described in the brains of humans and songbirds [49].
The Interaction of Nature and Nurture
Songbirds provide some of the best demonstrations in non-human animals of how early social experience influences behavioral development. The underlying mechanisms are not fully understood, but almost surely involve the deployment of different gene products according to social experience and developmental state, and may also involve epigenetic modification of the genome (e.g., [50]). Here again, singing provides a specific focal point for behavioral analysis. Because songbirds are altricial, singing behavior and its controlling neural circuit do not develop until well after the bird has hatched. Thus, singing behavior can be modified by social experience, and changes in the song control system are easy to observe against the backdrop of an otherwise relatively mature brain. This is a major advantage for uncovering genomic elements that may contribute to song.
The ontogeny of song has been studied most thoroughly in lab-reared zebra finches [51,52]. In zebra finches, developmental song learning begins around 30 days after hatching, when the bird begins to form a sensory memory of a tutor’s song. Both the anterior forebrain pathway and the auditory forebrain, which responds to song stimulation in adult birds, have been implicated in this process [53,54]. Between days 25 and 30 in male zebra finches, the motor pathway develops and birds begin to produce immature song-like sounds. Over the next month, young birds practice their vocal performance, and it comes to resemble closely the acoustic structure of the tutor song that they memorized. By sexual maturation, at around 90 days of age, song learning is complete and birds produce a stereotyped song that will be their trademark throughout life. In the zebra finch, the ability to memorize a tutor song ceases before sexual maturation. However, the timing of song development is subject to at least some modification based on social experience; birds reared in isolation can incorporate elements of tutor songs they eventually hear as adults, unlike birds who were reared in their normal social environment [54,55].
The boundaries of the song learning period are also apparently sensitive to genetic factors, as some species retain the ability to incorporate new elements into song during adulthood. Among domesticated birds, the canary Serinus canaria is an ‘open learner’ because it may add and subtract new song elements each breeding season [56]. Open learning is even more clearly evident in some wild species such as the northern mockingbird Mimus plyglottos, who learns new songs each year and readily mimics sounds of other species (or even jet planes and washing machines) [38].
Brood parasitic species illustrate the complex interplay between genetic and experiential components of song learning. About 25 songbird species are obligate brood parasites and parasitic behavior appears to have evolved independently twice in passerines [57]. A familiar North American example is the brown-headed cowbird, Molothrus ater. Cowbirds parasitize more than 200 different host species, laying eggs in the host species nests so that young cowbirds are raised without exposure to their own species-typical behavior. It is not until fledging that cowbirds join a flock of other cowbirds. Then, the male cowbird learns his species-specific song from social interactions within the flock [58-60], guided by behavioral responses from females to whom they sing [61]. In other words, a male develops a species-specific song that is partly influenced by conspecific females even though the early auditory experience of both birds was in the nests of other species. Another group of brood parasites, the African indigobirds (Vidua sp.), actually incorporate elements of host song into their song repertoire (Figure 4) [62].
Genomics of Sociality
Song is fundamentally a social behavior. Birds produce different vocalizations in different social contexts and they modify their responses to songs they hear based on their social interactions with the singer. Songbird species vary on multiple axes of sociality, for example, from colonial to territorial.
In territorial species, singing reveals exquisite sensitivity to the context of social interactions. A classic example is the song sparrow Melospiza melodia in western North America. Each male learns the songs of his neighbors and uses them in a subtle process of recognition, aggression and deference. Early in the breeding season, when territorial competition is intense, he responds to a specific neighbor by singing back the same song his neighbor just sang (‘type matching’), which acts as an aggressive signal. Later, once territorial boundaries are established, he responds by singing back a different element of that neighbor’s particular song repertoire. This ‘repertoire matching’ implies recognition and reduces aggression. Significantly, he never responds to his neighbor with a song that is not in both birds’ repertoire [63,64], a clear demonstration that birds can control the type of vocal behavior to appropriately suit the specific social context.
In contrast, zebra finches are colonial, with no known territorial function for song. Instead, song is used by zebra finches on a daily basis to recognize mates, kin and potentially other members of the colony [4]. Mate recognition is important for maintenance of the male–female pair bond, which is considered monogamous and stable throughout life. Fledglings recognize their parents by their vocalizations and preferentially associate with their siblings, who tend to sing similar songs [65]. Zebra finches are aware of more than just their mates and family members — they also appear to be acute observers of broader social contexts. A female zebra finch may be influenced by the choices of other females when selecting her mate [66]. Males pay attention to the mating status of other conspecific pairs, and use this information to control their behavior towards their own female partner. This sensitivity to social relationships is comparable to that demonstrated by social mammals such as primates [67].
Amongst the family of estrildid finches (Estrildidae), to which zebra finches belong, there is variation in social organization structures that may have genetic correlates [68-70]. For example, zebra finches and spice finches, Lonchura punctulata, are gregarious and colonial (living in groups of at least 100 individuals). At the other pole, the violet-eared waxbill, Uraeginthus granatina, and the melba finch, Pytilia melba, are highly territorial. The Angolan blue waxbill, U. angolensis, is an intermediate social phenotype; these birds are social during the breeding season, nest in a semi-colonial manner, and form small feeding flocks. Among these species a number of molecular measures, including neuropeptide receptors and immediate early gene responses, differ in specific brain areas associated with control of social behavior. These differences correlate with the degree of sociality displayed by the species. The zebra finch genome sequence may, therefore, serve as a common reference and nucleation point for comparative studies of genome and social structure within a single family.
The genome has already been linked to songbird sociality at multiple levels. Song perception [28] and song production [29,30] are associated with dynamic changes in gene expression in the brain that differ with the social and perceptual context in which the experience occurs [71-75]. Hence, social context influences the readout of the genome. Conversely, species differences in social behavior are likely to arise from genomic differences. Perhaps the starkest example is seen in the white-throated sparrow, Zonotrichia albicollis, in which there are two distinct morphs that differ in both plumage and behavior. Birds of the “tan” morph invest more in parental care, whereas the “white” morph invests more in territorial and sexual aggression [76]. These differences are correlated with a pair of nested chromosomal inversions spanning 98 megabases that appears to be around two million years old [77,78]. The inversion polymorphism is maintained in the population through disassortative mating (each morph prefers to breed with the opposite morph) and through the effects of recombination suppression within the inversion. Analysis of the chromosomal inversion may lead to identification of gene sequences that differ in the two morphs, and that correlate with both the plumage and behavioral differences. The zebra finch genome has already been an invaluable reference for targeted analysis of specific chromosomal domains identified in the white-throated sparrow [78], and will enable the identification of biologically-relevant genomic changes in other passerines.
Adaptation to Natural and Artificial Environments
Songbirds are exquisitely sensitive to their environments, and are even used as sentinels for environmental integrity on both local and global scales (e.g., [79]). If individual songbirds are notably sensitive to their environments, as a group they are also notably adaptable to them. The relative ease with which laboratory and field biology can be integrated in songbirds has made them major subjects for study of how environmental factors influence physiology and behavior, ultimately through effects at the level of the genome (Figure 5). For example, in western North America are different populations of the white-crowned sparrow, Zonotrichia leucophrys. The populations differ in corticosteroid-binding globulin proteins and in stress-induced free corticosteroid levels, adaptations hypothesized to maximize reproductive success for different breeding season durations [80]. Another classic example is the rapid evolution of species-specific beak morphologies in Darwin’s finches (Geospiza sp.), now explained in part through variations in expression of calmodulin and bone morphogenic proteins (reviewed in [81]).
Figure 5. Genome–brain–behavior relationships.
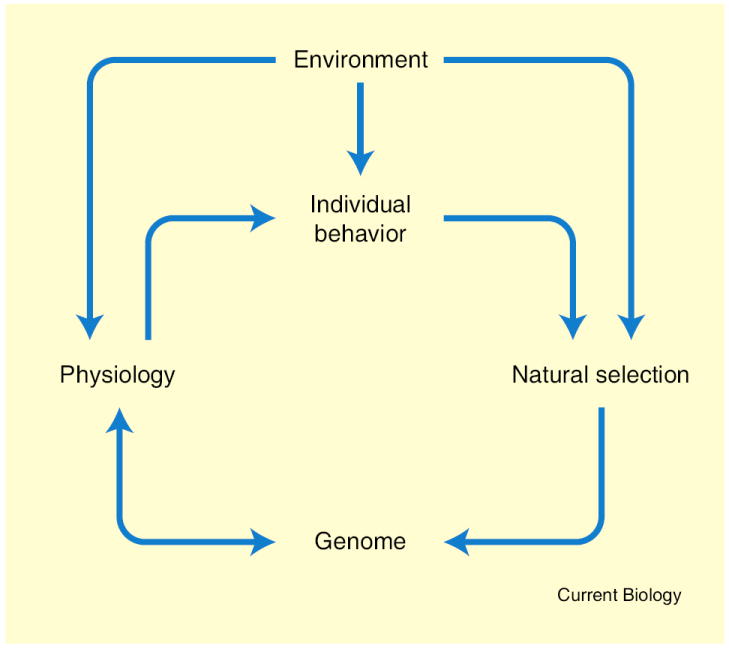
The genome influences behavior of individuals indirectly by encoding proteins and RNAs, which are components of cells and circuits, which make up the neural systems responsible for behavioral control. Individual behavior is subject to natural selection, leading over generations to the establishment of gene sequences associated with successful behavioral phenotypes. Environment affects behavior in physiological time via the responses of neural systems, and it affects the genome in evolutionary time as a component of natural selection. Physiological processes can lead to epigenetic modifications of the genome. An individual’s environment includes the behavior of other interacting individuals. Social organization emerges out of the behavioral interactions of individuals.
The zebra finch genome will greatly facilitate studies aimed at linking genetic and phenotypic changes in birds. For example, zebra finches have been artificially selected for high or low corticosterone responses to a mild stressor [82]. Over the first four generations of the experiment, the high lines demonstrated a significant realized heritability of about 20%. Ironically, selection for birds with low responses to stress was confounded by the apparent adaptation even of the control lines to the housing environment (with concomitant reduction of control line stress responses). Among captive zebra finches basal metabolic rate also appears to be heritable and hence subject to natural selection [83,84]. Song is itself a quantifiable behavior; thus, a genetic basis for song learning abilities may be identified with approaches such as selective breeding.
Status and Future of Songbird Genomics
An initial assembly and annotation of the zebra genome draft sequence has recently been released, with characterization of the preliminary assembly ongoing. The zebra finch genome comprises about 1.2 billion base pairs organized into 7 macrochromosome pairs, 32 pairs of microchromosomes, and the Z and W sex chromosomes. The genome sequence was derived from the DNA of a single male zebra finch from an aviary maintained at the University of California, Los Angeles. Like the chicken, the zebra finch has a more compact genome than mammals, supporting the conclusion that birds generally have the smallest and least variable (in terms of size) genomes among vertebrates (e.g., [85]). Analysis of the zebra finch genome assembly will test generalizations about avian genomes derived from the chicken genome assembly.
The primary genome assembly will be complemented by other sequencing resources already developed or under development in the research community. These include several BAC libraries and EST sequencing projects, which will provide information for annotating the genic organization of the genome. Three independent groups initiated EST databases of sequences expressed in the zebra finch brain and the sequences from all three sources have now been merged into a single master database (http://www.uiuc.edu/goto/songbird). New cDNA (house finch Carpodacus mexicanus [86]), cosmid (red-winged blackbirds [87]), BAC (Emu [88]), and small-insert genomic libraries (e.g., estrildid finches, Poephila spp. [89], and the red-backed fairywrens, Malurus melanocephalus [90]) have also been recently reported.
In addition to a primary draft of the genome itself, the genome sequencing project will provide or stimulate the development of other resources that will be useful for comparative genomics. Even though the genome of only a single bird is being sequenced, it will still provide a collection of mapped single nucleotide polymorphisms (SNPs) because of diploidy. To date, the genetic structure of songbird populations has been studied predominantly by the use of the mitochondrial DNA and microsatellite sequences, but these approaches are inherently limited [91,92]. Zebra finch genomic resources have already facilitated the development of large panels of nuclear markers for population genetics and systematics (e.g, [6,93]), a change that will promote the use of powerful new statistical approaches to population and phylogenetics (reviewed in [94]).
The genome sequence will also be useful as a template for production of comprehensive DNA microarrays that can be used for both gene expression studies and for comparative genomic hybridizations (CGH) to test for changes in gene copy number and genome organization. Initial results applying zebra finch cDNA microarrays to studies in other species are promising [95-97], although careful interpretation of cross-hybridization experiments is of course critical [98]. An even more promising approach will be to use sequence data obtained directly from other species using the new generation of sequencing technologies [99]. Soon, it will be possible to map direct sequencing results onto the zebra finch genome assembly, a clear advance in the ability to investigate all songbirds in a more powerful way than ever before.
Conclusion
Songbirds have long been of interest to taxonomists for their diversity, to ethologists for their complex behaviors, and to lay people for their natural beauty and engaging vocalizations. Neurobiologists have made great progress elucidating the brain circuits underlying vocal communication in songbirds. Assembly of the zebra finch genome now brings a new set of questions, perspectives and tools, making songbirds even more compelling as models for probing the links between gene, brain, behavior, environment and evolution.
Acknowledgments
We thank Margaret Ferris, Lynn Huynh, Dan Janes, Chris Organ, and Kirstin Replogle for helpful discussions and comments on the manuscript. Supported by NIH R01 045264, T32DC006612, and F32NS055413.
References
- 1.Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MAM, Delany ME, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 2.Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 3.Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci USA. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zann RA. The Zebra Finch: a Synthesis of Field and Laboratory Studies. Oxford University Press; 1996. [Google Scholar]
- 5.Clayton NS. Assortative mating in zebra finch subspecies, Taeniopygia-guttata-guttata and T-g-castanotis. Philos Trans R Soc Lond B. 1990;330:351–370. [Google Scholar]
- 6.Balakrishnan C, Edwards S. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata) Genetics. 2009;181:1–16. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart NS. Variations in cone photoreceptor abundance and the visual ecology of birds. J Comp Physiol. 2001;187:685–697. doi: 10.1007/s00359-001-0240-3. [DOI] [PubMed] [Google Scholar]
- 8.Noad MJ, Cato DH, Bryden MM, Jenner MN, Jenner KCS. Cultural revolution in whale songs. Nature. 2000;408:537–537. doi: 10.1038/35046199. [DOI] [PubMed] [Google Scholar]
- 9.Boughman JW. Vocal learning by greater spear-nosed bats. Proc Roy Soc B. 1998;265:227–233. doi: 10.1098/rspb.1998.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole JH, Tyack PL, Stoeger-Horwath AS, Watwood S. Animal behaviour: Elephants are capable of vocal learning. Nature. 2005;434:455–456. doi: 10.1038/434455a. [DOI] [PubMed] [Google Scholar]
- 11.Saranathan V, Hamilton D, Powell GVN, Kroodsma DE, Prum RO. Genetic evidence supports song learning in the three-wattled bellbird Procnias tricarunculata (Cotingidae) Mol Ecol. 2007;16:3689–3702. doi: 10.1111/j.1365-294X.2007.03415.x. [DOI] [PubMed] [Google Scholar]
- 12.Suthers RA. Contributions to birdsong from the left and right sides of an intact syrinx. Nature. 1990;347:473–477. [Google Scholar]
- 13.Farries MA. The oscine song system considered in the context of the avian brain: Lessons learned from comparative neurobiology. Brain Behav Evol. 2001;58:80–100. doi: 10.1159/000047263. [DOI] [PubMed] [Google Scholar]
- 14.Doupe AJ, Perkel DJ, Reiner A, Stern EA. Birdbrains could teach basal ganglia research a new song. Trends Neurosci. 2005;28:353–363. doi: 10.1016/j.tins.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, et al. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gahr M. Neural song control system of hummingbirds: Comparison to swifts, vocal learning (songbirds) and nonlearning (suboscines) passerines, and vocal learning (budgerigars) and nonlearning (dove, owl, gull, quail, chicken) nonpasserines. J Comp Neurol. 2000;426:182–196. [PubMed] [Google Scholar]
- 17.Jarvis ED, Mello CV. Molecular mapping of brain areas involved in parrot vocal communication. J Comp Neurol. 2000;419:1–31. doi: 10.1002/(sici)1096-9861(20000327)419:1<1::aid-cne1>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis ED, Ribeiro S, da Silva ML, Ventura D, Vielliard J, Mello CV. Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature. 2000;406:628–632. doi: 10.1038/35020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wada K, Sakaguchi H, Jarvis ED, Hagiwara M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol. 2004;476:44–64. doi: 10.1002/cne.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nottebohm F. A brain for all seasons – Cyclical anatomical changes in song control. Science. 1981;214:1368–1370. doi: 10.1126/science.7313697. [DOI] [PubMed] [Google Scholar]
- 21.Tramontin AD, Smith GT, Breuner CW, Brenowitz EA. Seasonal plasticity and sexual dimorphism in the avian song control system: Stereological measurement of neuron density and number. J Comp Neurol. 1998;396:186–192. doi: 10.1002/(sici)1096-9861(19980629)396:2<186::aid-cne4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 22.AlvarezBuylla A, Kirn JR. Birth, migration, incorporation, and death of vocal control neurons in adult songbirds. J Neurobiol. 1997;33:585–601. [PubMed] [Google Scholar]
- 23.Goldman SA, Nottebohm F. Neuronoal production, migration, and differentiation in a vocal control nucleus of the adult female canary brain. Proc Natl Acad Sci USA. 1983;80:2390–2394. doi: 10.1073/pnas.80.8.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirn J, Oloughlin B, Kasparian S, Nottebohm F. Cell-death and neuronal recruitment in the high vocal center of adult male canaries are temporally related to changes in song. Proc Natl Acad Sci USA. 1994;91:7844–7848. doi: 10.1073/pnas.91.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konishi M, Akutagawa E. Neuronal growth, atrophy and death in a sexually dimorphic song nucleus in the zebra finch brain. Nature. 1985;315:145–147. doi: 10.1038/315145a0. [DOI] [PubMed] [Google Scholar]
- 26.Arnold AP, Nottebohm F, Pfaff DW. Hormone concentrating cells in vocal control and other areas of brain of zebra finch (Poephila-guttata) J Comp Neurol. 1976;165:487–511. doi: 10.1002/cne.901650406. [DOI] [PubMed] [Google Scholar]
- 27.Amaya KR, Monroe EB, Sweedler JV, Clayton DF. Lipid imaging in the zebra finch brain with secondary ion mass spectrometry. Internat J Mass Spect. 2007;260:121–127. [Google Scholar]
- 28.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene-expression in the songbird forebrain. Proc Natl Acad Sci USA. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, Clayton DF. Localized changes in immediate-early gene regulation during sensory and motor learning in zebra finches. Neuron. 1997;19:1049–1059. doi: 10.1016/s0896-6273(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 30.Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci USA. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clayton D. Molecular neurobiology of birdsong. In: Blaustein J, editor. Handbook of Neurochemistry and Molecular Neurobiology. Vol. 21. New York: Kluwer; 2007. [Google Scholar]
- 32.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: neurochemical specializations of the oscine song system. PLoS One. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.London SE, Dong S, Replogle K, Clayton DF. Developmental shifts in gene expression in the auditory forebrain during the sensitive period for song learning. Dev Neurobiol. 2009;69:437–450. doi: 10.1002/dneu.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong S, Replogle KA, Hasadsri L, Imai B, Yau P, Rodriguez-Zas S, Southey BR, Sweedler J, Clayton DF. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci USA. 2009;106:11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beecher MD, Brenowitz EA. Functional aspects of song learning in songbirds. Trends Ecol Evol. 2005;20:143–149. doi: 10.1016/j.tree.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Brenowitz EA, Beecher MD. Song learning in birds: diversity and plasticity, opportunities and challenges. Trends Neurosci. 2005;28:127–132. doi: 10.1016/j.tins.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Catchpole CK, Slater PJB. Bird Song: Biological Themes and Variations. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- 38.Kroodsma D. The Singing Life of Birds: The Art and Science of Listening to Birdsong (Houghton Mifflin) 2005 [Google Scholar]
- 39.Templeton CN, Greene E, Davis K. Allometry of alarm calls: Black-capped chickadees encode information about predator size. Science. 2005;308:1934–1937. doi: 10.1126/science.1108841. [DOI] [PubMed] [Google Scholar]
- 40.Freeberg TM, Lucas JR. Receivers respond differently to chick-a-dee calls varying in note composition in Carolina chickadees, Poecile carolinensis. Anim Behav. 2002;63:837–845. [Google Scholar]
- 41.Chaiken M, Gentner TQ, Hulse SH. Effects of social interaction on the development of starling song and the perception of these effects by conspecifics. J Comp Psychol. 1997;111:379–392. doi: 10.1037/0735-7036.111.4.379. [DOI] [PubMed] [Google Scholar]
- 42.Gentner TQ, Fenn KM, Margoliash D, Nusbaum HC. Recursive syntactic pattern learning by songbirds. Nature. 2006;440:1204–1207. doi: 10.1038/nature04675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marler P. Animal communication signals. Science. 1967;157:769–774. doi: 10.1126/science.157.3790.769. [DOI] [PubMed] [Google Scholar]
- 44.Doupe AJ, Kuhl PK. Birdsong and human speech: Common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- 45.Kuhl PK. Human speech and birdsong: Communication and the social brain. Proc Natl Acad Sci USA. 2003;100:9645–9646. doi: 10.1073/pnas.1733998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansson H. Childhood stuttering: Incidence and development. J Fluency Disord. 2000;25:47–57. [Google Scholar]
- 47.Rosenfield DB, Viswanath NS, Helekar SA. An animal model for stuttering-related part-word repetitions. J Fluency Disorders. 2000;25:171–171. [Google Scholar]
- 48.Roberts J, Steven LH, Malkin C, Barnes E, Skinner M, Hennon EA, Anderson K. A comparison of phonological skills of boys with fragile X syndrome and Down syndrome. J Speech Lang Hear Res. 2005;48:980–995. doi: 10.1044/1092-4388(2005/067). [DOI] [PubMed] [Google Scholar]
- 49.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver ICG, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein A mediates epigenetic programming: Altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tchernichovski O, Mitra P, Lints T, Nottebohm F. Dynamics of the vocal imitation process: how a zebra finch learns its song. Science. 2001;291:2564–2569. doi: 10.1126/science.1058522. [DOI] [PubMed] [Google Scholar]
- 52.Bolhuis JJ, Gahr M. Neural mechanisms of birdsong memory. Nature Rev Neurosci. 2006;7:347–357. doi: 10.1038/nrn1904. [DOI] [PubMed] [Google Scholar]
- 53.London SE, Clayton DF. Functional identification of sensory mechanisms required for developmental song learning. Nat Neurosci. 2008;11:579–586. doi: 10.1038/nn.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eales LA. Song learning in femal-raised zebra finches – Another look at the sensitive phase. Anim Behav. 1987;35:1356–1365. [Google Scholar]
- 55.Slater PJB, Jones A, Tencate C. Can lack of experience delay the end of the sensitive phase for song learning. Netherlands J Zool. 1993;43:80–90. [Google Scholar]
- 56.Nottebohm F, Nottebohm ME, Crane L. Developmental and seasonal changes in canary song and their relation to changes in the anatomy of song-control nuclei. Behav Neural Biol. 1986;46:445–471. doi: 10.1016/s0163-1047(86)90485-1. [DOI] [PubMed] [Google Scholar]
- 57.Sorenson MD, Payne RB. Molecular genetic perspectives on avian brood parasitism. Integrative Comparative Biol. 2002;42:388–400. doi: 10.1093/icb/42.2.388. [DOI] [PubMed] [Google Scholar]
- 58.Freeberg TM. The cultural transmission of courtship patterns in cowbirds, Molothrus ater. Anim Behav. 1998;56:1063–1073. doi: 10.1006/anbe.1998.0870. [DOI] [PubMed] [Google Scholar]
- 59.Freeberg TM, King AP, West MJ. Cultural transmission of vocal traditions in cowbirds (Molothrus ater) influences courtship patterns and mate preferences. J Comp Psychol. 2001;115:201–211. doi: 10.1037/0735-7036.115.2.201. [DOI] [PubMed] [Google Scholar]
- 60.Miller JL, Freed-Brown SG, White DJ, King AP, West MJ. Developmental origins of sociality in brown-headed cowbirds (Molothrus ater) J Comp Psychol. 2006;120:229–238. doi: 10.1037/0735-7036.120.3.229. [DOI] [PubMed] [Google Scholar]
- 61.West MJ, King AP. Femal visual displays affect the development of male song in the cowbird. Nature. 1988;334:244–246. doi: 10.1038/334244a0. [DOI] [PubMed] [Google Scholar]
- 62.Payne RB, Payne LL, Woods JL. Song learning in brood-parasitic indigobirds Vidua chalybeata: song mimicry of the host species. Anim Behav. 1998;55:1537–1553. doi: 10.1006/anbe.1997.0701. [DOI] [PubMed] [Google Scholar]
- 63.Beecher MD, Stoddard PK, Campbell SE, Horning CL. Repertoire matching between neighbouring song sparrows. Anim Behav. 1996;51:917–923. doi: 10.1006/anbe.1999.1276. [DOI] [PubMed] [Google Scholar]
- 64.Beecher MD, Campbell SE, Burt JM, Hill CE, Nordby JC. Song-type matching between neighbouring song sparrows. Anim Behav. 2000;59:21–27. doi: 10.1006/anbe.1999.1276. [DOI] [PubMed] [Google Scholar]
- 65.Burley N, Minor C, Strachan C. Social preference of zebra finches for siblings, cousins and nonkin. Anim Behav. 1990;39:775–784. [Google Scholar]
- 66.Brown GR, Fawcett TW. Sexual selection: Copycat mating in birds. Curr Biol. 2005;15:R626–R628. doi: 10.1016/j.cub.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Vignal C, Mathevon N, Mottin S. Audience drives male songbird response to partner’s voice. Nature. 2004;430:448–451. doi: 10.1038/nature02645. [DOI] [PubMed] [Google Scholar]
- 68.Goodson JL, Wang YW. Valence-sensitive neurons exhibit divergent functional profiles in gregarious and asocial species. Proc Natl Acad Sci USA. 2006;103:17013–17017. doi: 10.1073/pnas.0606278103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goodson JL, Evans AK, Lindberg L, Allen CD. Neuro-evolutionary patterning of sociality. Proc Royal Soc B. 2005;272:227–235. doi: 10.1098/rspb.2004.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodson JL, Evans AK, Wang Y. Neuropeptide binding reflects convergent and divergent evolution in species-typical group sizes. Horm Behav. 2006;50:223–236. doi: 10.1016/j.yhbeh.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: Context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 72.Kruse AA, Stripling R, Clayton DF. Context-specific habituation of the zenk gene response to song in adult zebra finches. Neurobiol Learn Mem. 2004;82:99–108. doi: 10.1016/j.nlm.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Heimovics SA, Riters LV. Immediate early gene activity in song control nuclei and brain areas regulating motivation relates positively to singing behavior during, but not outside of, a breeding context. J Neurobiol. 2005;65:207–224. doi: 10.1002/neu.20181. [DOI] [PubMed] [Google Scholar]
- 74.Heimovics SA, Riters LV. ZENK labeling within social behavior brain regions reveals breeding context-dependent patterns of neural activity associated with song in male European starlings (Sturnus vulgaris) Behav Brain Res. 2007;176:333–343. doi: 10.1016/j.bbr.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vignal C, Andru J, Mathevon N. Social context modulates behavioural and brain immediate early gene responses to sound in male songbird. Eur J Neurosci. 2005;22:949–955. doi: 10.1111/j.1460-9568.2005.04254.x. [DOI] [PubMed] [Google Scholar]
- 76.Tuttle EM. Alternative reproductive strategies in the white-throated sparrow: behavioral and genetic evidence. Behav Ecol. 2003;14:425–432. [Google Scholar]
- 77.Thorneycroft H. Chromosomal polymorphism in the white-throated sparrow, Zonotrichia albicollis. Science. 1966;154:1571–1572. doi: 10.1126/science.154.3756.1571. [DOI] [PubMed] [Google Scholar]
- 78.Thomas JW, Caceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearragement and suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hames RS, Rosenberg KV, Lowe JD, Barker SE, Dhondt AA. Adverse effects of acid rain on the distribution of the Wood Thrush Hylocichla mustelina in North America. Proc Natl Acad Sci USA. 2002;99:11235–11240. doi: 10.1073/pnas.172700199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breuner CW, Orchinik M, Hahn TP, Meddle SL, Moore IT, Owen-Ashley NT, Sperry TS, Wingfield JC. Differential mechanisms for regulation of the stress response across latitudinal gradients. Am J Physiol. 2003;285:R594–R600. doi: 10.1152/ajpregu.00748.2002. [DOI] [PubMed] [Google Scholar]
- 81.Schneider RA. How to tweak a beak: molecular techniques for studying the evolution of size and shape in Darwin’s finches and other birds. Bioessays. 2007;29:1–6. doi: 10.1002/bies.20517. [DOI] [PubMed] [Google Scholar]
- 82.Evans MR, Roberts ML, Buchanan KL, Goldsmith AR. Heritability of corticosterone response and changes in life history traits during selection in the zebra finch. J Evol Biol. 2006;19:343–352. doi: 10.1111/j.1420-9101.2005.01034.x. [DOI] [PubMed] [Google Scholar]
- 83.Ronning B, Moe B, Bech C. Long-term repeatability makes basal metabolic rate a likely heritable trait in the zebra finch Taeniopygia guttata. J Exp Biol. 2005;208:4663–4669. doi: 10.1242/jeb.01941. [DOI] [PubMed] [Google Scholar]
- 84.Ronning B, Jensen H, Moe B, Bech C. Basal metabolic rate: heritability and genetic correlations with morphological traits in the zebra finch. J Evol Biol. 2007;20:1815–1822. doi: 10.1111/j.1420-9101.2007.01384.x. [DOI] [PubMed] [Google Scholar]
- 85.Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV. Origin of avian genome size and structure in non-avian dinosaurs. Nature. 2007;446:180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- 86.Wang ZS, Farmer K, Hill GE, Edwards SV. A cDNA macro-array approach to parasite-induced gene expression changes in a songbird host: genetic response of house finches to experimental infection by Mycoplasma gallisepticum. Mol Ecol. 2006;15:1263–1273. doi: 10.1111/j.1365-294X.2005.02753.x. [DOI] [PubMed] [Google Scholar]
- 87.Gasper JS, Shiina T, Inoko H, Edwards SV. Songbird genomics: Analysis of 45 kb upstream of a polymorphic Mhc class II gene in red-winged blackbirds (Agelaius phoeniceus) Genomics. 2001;75:26–34. doi: 10.1006/geno.2001.6596. [DOI] [PubMed] [Google Scholar]
- 88.Janes D, Ezaz T, Graves JM, Edwards S. Recombination and nucleotide diversity in the sex chromosal pseudoautosomal region of the emu, Dromaius novaehollandiae. J Hered. 2009;100:125–136. doi: 10.1093/jhered/esn065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jennings WB, Edwards SV. Speciational history of Australian grass finches (Poephila) inferred from thirty gene trees. Evolution. 2005;59:2033–2047. [PubMed] [Google Scholar]
- 90.Lee JY, Edwards SV. Divergence across the carpentarian barrier: Statistical phylogeography of the red-backed fairywren (Malurus melanocephalus) Evolution. 2008;62:3117–3134. doi: 10.1111/j.1558-5646.2008.00543.x. [DOI] [PubMed] [Google Scholar]
- 91.Edwards S, Bensch S. Looking forwards or looking backwards in avian phylogeography? Mol Ecol. 2009;18:2930–2933. doi: 10.1111/j.1365-294X.2009.04270.x. [DOI] [PubMed] [Google Scholar]
- 92.Edwards SV, Kingan SB, Calkins JD, Balakrishnan CN, Jennings WB, Swanson WJ, Sorenson MD. Speciation in birds: Genes, geography, and sexual selection. Proc Natl Acad Sci USA. 2005;102:6550–6557. doi: 10.1073/pnas.0501846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Backstroem N, Fagerberg S, Ellegren H. Genomics of natural bird populations: a gene-based set of reference markers evenly spread across the avian genome. Mol Ecol. 2008;17:964–980. doi: 10.1111/j.1365-294X.2007.03551.x. [DOI] [PubMed] [Google Scholar]
- 94.Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- 95.Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, et al. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Naurin S, Bensch S, Hansson B, Johansson T, Clayton DF, Albrekt AS, Von Schantz T, Hasselquist D. A microarray for large-scale genomic and transcriptional analyses of the zebra finch (Taeniopygia guttata) and other passerines. Mol Ecol Resources. 2008;8:275–281. doi: 10.1111/j.1471-8286.2007.01979.x. [DOI] [PubMed] [Google Scholar]
- 97.Romanov MN, Dodgson JB. Cross-species overgo hybridization and comparative physical mapping within avian genomes. An Gene. 2006;37:397–399. doi: 10.1111/j.1365-2052.2006.01463.x. [DOI] [PubMed] [Google Scholar]
- 98.Bar-Or C, Czosnek H, Koltai H. Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends Genet. 2007;23:200–207. doi: 10.1016/j.tig.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 99.Bonneaud C, Burnside J, Edwards SV. High-speed developments in avian genomics. Bioscience. 2008;58:587–595. [Google Scholar]