Summary
The outer membrane protein A (OmpA) of Escherichia coli is a well-known model for protein targeting and protein folding. Wild-type OmpA, isolated either from cytoplasmic inclusion bodies or from outer membranes, forms narrow pores of ~80 pS in planar lipid bilayers at room temperature. The pores are well-structured with narrow conductance range when OmpA is isolated using lithium dodecyl sulfate (LDS) or RapiGest surfactant but display irregular conductance when OmpA is isolated with urea or guanidine hydrochloride. Previous studies have shown that serine residues S163 and S167 of the sorting signal of OmpA (residues 163–169), i.e. the essential sequence for outer membrane incorporation, are covalently modified by oligomers of (R)-3-hydroxybutyrate (cOHB). Here we find that single mutants S163 and S167 of OmpA, which still contain cOHB on one serine of the sorting signal, form narrow pores in planar lipid bilayers at room temperature with lower and more irregular conductance than wild-type OmpA, whereas double mutants S163:S167 and S163:V166 of OmpA, with no cOHB on the sorting signal, are unable to form stable pores in planar lipid bilayers. Our results indicate that modification of serines in the sorting signal of OmpA by cOHB in the cytoplasm enables OmpA to incorporate into lipid bilayers at room temperature as a narrow pore. They further suggest that cOHB modification may be an important factor in protein targeting and protein folding.
Keywords: OmpA, oligo-(R)-3-hydroxybutyrate (OHB), complexed OHB (cOHB), outer membrane sorting, sorting signal, bilayer incorporation, pore formation, protein modification, amphipathic polymer
1. Introduction
Outer membrane protein A (OmpA) is a major outer membrane protein of Escherichia coli. The process by which OmpA and other outer membrane proteins are directed to and incorporated into the outer membrane is known as outer membrane sorting. OmpA precursors are synthesized in the cytoplasm with a 21-residue signal sequence at the amino terminal. They are guided by the Sec translocon into and through the cytoplasmic membrane into the periplasm in a largely unfolded state. The signal sequence is cleaved and the proteins, assisted by periplasmic chaperones and lipopolysaccharides, are folded and inserted into the outer membrane [1–4].
The mechanism by which periplasmic OmpA inserts into the outer membrane is not well understood. A segment of the 325 residue outer membrane protein OmpA, known as the sorting signal, has been shown to be critical to outer membrane incorporation. Klose et al. [5] used immunoelectron microscopy to examine a series of overlapping deletions of OmpA and defined a region between residues 154–180 as essential for outer membrane association; fragments missing this region were located in the periplasm. The critical region for outer membrane assembly was narrowed by Freudl et al. [6] to the eighth β-strand, residues 160–170. Klose et al [7] found that the double mutant G160V; L162R was not defective in membrane assembly. Accordingly, these studies indicate that the putative sorting signal is contained within residues 163–170 (SLGVSYRF) of the mature protein.
Xian et al. [8] showed by Western blot immunoassay and chemical assay that the serine residues on fragment 162–174 of OmpA (LSLGVSYRFGQGE) are covalently modified by oligo-R-3-hydroxybutyrates (cOHB). The presence of cOHB on fragment 162–174 of wild-type OmpA and the absence of cOHB on the same fragment of double mutant S163G:S167G were confirmed by MALDI-MS. The sorting signal of OmpA is modified by cOHB in protein isolated either from inclusion bodies or from outer membranes which suggests that this modification occurs in the cytoplasm.
cOHB-modified proteins are found in prokaryotes and also in eukaryotes [9–11]. The physical properties of these oligomers - highly flexible, amphiphilic molecules which are insoluble in water but soluble in lipids [12–14] - suggest that they may act as chaperones to facilitate the incorporation of hydrophilic polypeptides into bilayers. Here we examine the effect of cOHB modification of the sorting signal on bilayer incorporation and pore formation by OmpA.
2. Materials and methods
2.1. Purification of OmpA from outer membranes
Whole OmpA (325 residues) was extracted from the outer membranes of JM109 by a modification of the method of Sugawara and Nikaido [15,16]. Briefly, stationary-phase cells were collected by centrifugation and frozen at −20 °C overnight. The pellet was then suspended in 20 mM tris(hydroxymethyl) aminomethane (Tris)-HCl, pH 7.8, 5 mM ethylenediamine tetraacetic acid (EDTA), 0.1 mM PMSF (phenylmethylsulfonyl fluoride), 0.5 mg/ml lysozyme and disintegrated by ultrasonication (Branson). Mg2+ (10 mM) and DNase (0.1 mg/ml) were added and unbroken cells were removed by centrifugation at 5000 rpm for 20 min at 4 °C (Sorvall GSA rotor). Crude outer membrane fractions were recovered by centrifugation at 12,000 rpm for 40 min at 4 °C (Sorvall SS-34 rotor). Outer membranes were suspended in 0.3% lithium dodecyl sulfate (LDS), 5 mM EDTA in 20 mM Hepes, adjusted to pH 7.5 with KOH, to a final protein concentration of 2 mg/ml. After 1hr with mild mixing at 4 °C, the suspension was centrifuged at 35,000 rpm (Beckman Type 50.2 TI rotor) for 45 min. The pellet was resuspended in 0.5% LDS in the same solvent, gently mixed for 1 hr at 4 °C, and centrifuged in the same manner. The pellet from the second extraction was resuspended in 2% LDS, 20 mM Hepes, pH 7.5, and gently mixed at 4 °C for > 1hr. The pellet from this extraction was discarded and the supernatant containing soluble OmpA was loaded onto a column of Sephacryl S-300 (1.6 ×60 cm, HiPrep, Pharmacia) which had been equilibrated with 0.05% LDS, 0.4 M LiCl, 20 mM Hepes, pH 7.5. Fractions were eluted with the same solvent and examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). OmpA-rich fractions were combined and concentrated using Centricon-10 (Amicon). Further purification when necessary was performed on a column of Superdex 75 (10/300) using the same solvent.
Alternatively, OmpA was extracted using 8M urea as described by Hong et al. [17] or with 6M guanidine hydrochloride (GuHCl) using the same procedure and then purified by Sephacryl S-300 chromatography as above.
2.2 Expression and purification of OmpA from inclusion bodies
His-OmpA was overexpressed in E. coli BL21(DE3)pLysS cells (Novagen) containing the pET-45b(+)-His-OmpA plasmid, and was grown in LB medium supplemented with 100 μg/ml ampicillin at 37 °C with aeration to an OD600 of ~ 0.6. Protein expression was induced by the addition of 0.5 mM isopropyl-1-thio-3-D-galactopyranoside (IPTG), and the cells were cultured at 37 °C for 2 hrs before harvesting by centrifugation. Cells were disintegrated by ultrasonication as above and inclusion bodies were collected by centrifugation at 15,000 rpm for 30 min (Sorvall SS-34 rotor). His-OmpA was extracted from inclusion bodies with 0.1% RapiGest (Waters), pH 7.4, or alternatively with 8M urea, pH 7.4 or with 6M GuHCl, pH 7.4, and bound to Ni-agarose beads in the presence of 20 mM immidazole, pH 7.8. The beads were washed with 25 mM immidazole, pH 7.8, 1 mM dodecylmaltoside (DDM) and His-OmpA was eluted with 300 mM immidazole, pH 6.8, 1 mM DDM. His-OmpA was also extracted from inclusion bodies obtained by overexpression of the pET-45b(+)-His-OmpA plasmid in E. coli UH 203 OmpA-minus cells using the procedure described above for outer membranes, i.e. isolated with LDS and purified by Sephacryl-S-300 chromatography.
2.3. Reconstitution of M-OmpA and I-OmpA
M-OmpA and I-OmpA preparations were concentrated to ~ 1 mg/ml by centrifugal filtration using 10K Centricon filters. Buffer substitution was then performed 5X with 20 mM n-octyl tetraethylene glycol monoether (C8E4) in 20 mM KHepes, pH 7.4 using the same filters. The concentrate was then diluted with the C8E4 solution to 0.1 mg/ml. This solution (1 μl) was added to the cis side of a planar bilayer formed with synthetic diphytanoylphosphatidylcholine (DPhPC) (Avanti Polar lipids).
2.4. Western blot assays for cOHB
In the Western blot assay for cOHB, 5 μg of OmpA was added to Laemmli loading buffer The sample was heated in a boiling water bath for 5 min, cooled, loaded on a 12% SDS-PAGE and proteins were separated by electrophoresis. The gel was transferred to a supported nitrocellulose or PVDF membrane (Bio-Rad) in Tris glycine SDS buffer (Bio-Rad), 20% methanol, using a Mini Trans-Blot electrophoretic cell (Bio-Rad) or Gene Blotter (Idea Scientific Co.). The membrane was blocked with 1.25% gelatin (electrophoresis grade; Bio-Rad) in Tris-buffered saline, pH 7.5, 0.1% Tween-20. Primary incubation was with polyclonal anti-OHB IgG in blocking buffer produced in rabbits to a synthetic 8mer of OHB (courtesy of D. Seebach, ETH Zürich) conjugated to gelatin (Metabolix Inc.) and purified by protein A chromatography. The second antibody was goat anti-rabbit alkaline phosphatase conjugate (Bio-Rad) in the same buffer. Color development was performed with 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium (NBT) (Bio-Rad).
2.5. Planar lipid bilayer studies
Planar lipid bilayers were formed from a solution of DPhPC in n-decane (Aldrich) at a concentration of ~ 17 mg/ml. The solution was used to paint a bilayer in an aperture of ~150 μm diameter between aqueous solutions of 1M KCl in 20 mM Hepes, pH 7.4 in a Delrin cup (Warner Instruments). All salts were ultrapure (Aldrich). After the bilayer was formed, a solution of OmpA in C8E4 (1 μl of 0.1 mg/ml) was added to the cis compartment.
Unitary currents were recorded with an integrating patch clamp amplifier (Axopatch 200A). The trans solution (voltage command side) was connected to a CV 201A head stage input and the cis solution was held at virtual ground via a pair of matched Ag-AgCl electrodes. Currents through the voltage-clamped bilayers were low-pass filtered at 10 kHz and recorded after digitization through an analog to digital converter (Digidata 1322A, Axon Instr.). Data were filtered through an 8 pole Bessel filter (9021 PF, Frequency Devices) and digitized at 1 kHz using pClamp 9.0 software (Axon Instr.). Single-channel conductance events were identified and analyzed by using Clampfit9 software (Axon Instr.). The data were averaged from > 10 independent recordings.
3. Results
3.1. Planar bilayer observations of wild-type OmpA isolated from outer membranes and from cytoplasmic inclusion bodies
Whole OmpA (325 residues) was extracted from outer membranes (M-OmpA) and His-tagged whole OmpA was extracted from inclusion bodies (I-OmpA) using several different solubilizing agents. M-OmpA was isolated using either lithium dodecyl sulfate (LDS) or urea or GuHCl and purified by size-exclusion chromatography using LDS (see Methods). I-OmpA was extracted using either RapiGest surfactant or urea or GuHCl and purified by Ni-agarose chromatography using DDM (see Methods). I-OmpA was also extracted with LDS but this preparation did not bind to Ni-agarose beads; consequently, I-OmpA extracted with LDS was purified using size-exclusion chromatography in the same manner as M-OmpA. The purified proteins all displayed the same electrophoretic migration on SDS-PAGE (Fig. 1 lanes 1–4). Unheated proteins migrated at ~ 30 kDa and proteins heated in 2% SDS before loading migrated at ~ 35 kDa.
Figure 1.
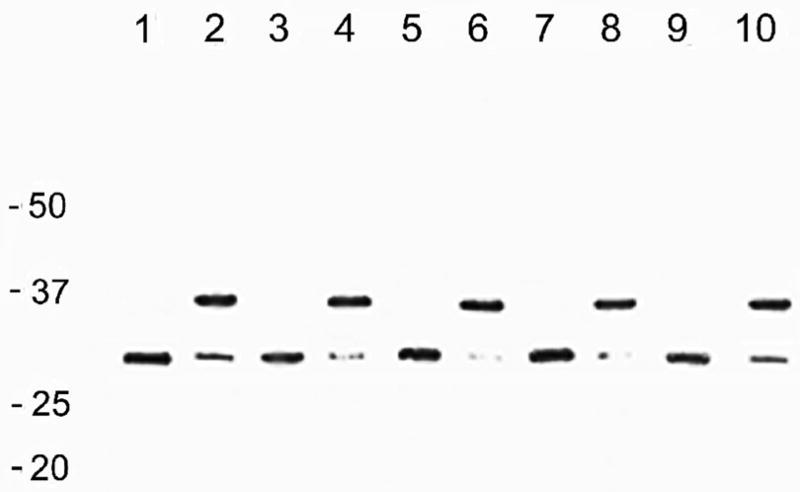
Electrophoretic migration of representative wild-type and mutant OmpA proteins on 12% SDS PAGE gels. Samples were loaded without heating (U) and after heating in 2% SDS in a boiling water bath for 3 minutes before loading (H). Lanes 1,2 – M-OmpA U, H; lanes 3,4 – I-OmpA U, H; lanes 5,6 – S163G U, H; lanes 7,8 – S167G U, H; lanes 9,10 – S163G:S167G U, H.
The purified M-OmpA and I-OmpA proteins were reconstituted into C8E4 micelles, the solubilizing agent was removed by repeated filtration and the proteins were then each incorporated into planar lipid bilayers of DPhPC at room temperature (see Methods). The conductance range and open time of pores formed by the various preparations were then compared.
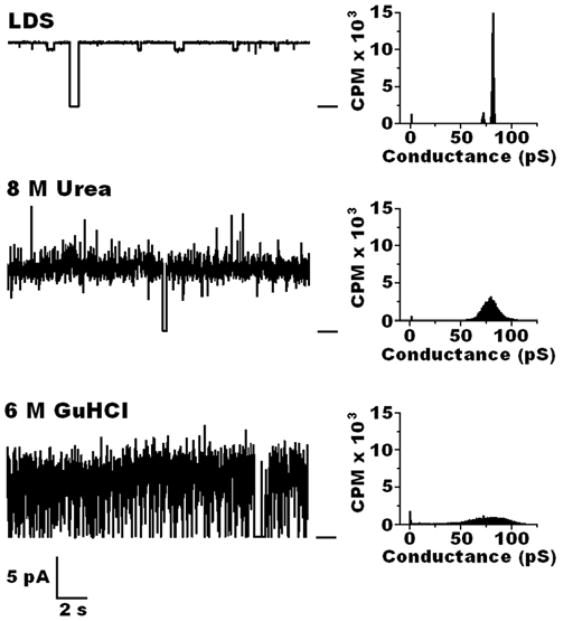
As shown in Fig. 2, M-OmpA extracted with LDS formed well-structured pores with an open time of 0.94 and a narrow conductance range of 80 ± 2 pS. M-OmpA extracted with urea had similar long open time of 0.92 and the same average conductance but with a significantly wider range of 80 ± 10 pS. M-OmpA isolated with GuHCl had a lower open time of 0.70 and a wide conductance range of 80 ± 17 pS.
Figure 2.
Left: Representative current traces of OmpA isolated from outer membranes of E. coli JM109 cells (M-OmpA), purified by chromatography on Sephacryl 300, reconstituted into C8E4 micelles, and incorporated into planar lipid bilayers of DPhPC at room temperature between aqueous solutions of 1M KCl in 20 mM Hepes, pH 7.4. Bar at right of the traces indicates the closed state of the channel. Clamping potential was +100 mV. Right: Histograms taken from records of ~ 10 minutes at room temperature.
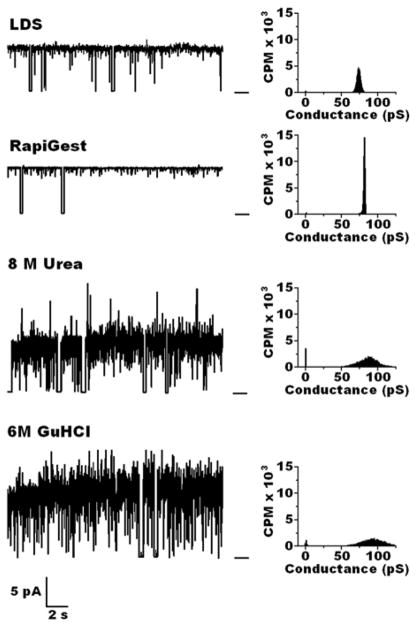
Pore configuration of I-OmpA was also affected by the solubilizing agent used in protein purification (Fig. 3). When isolated using LDS or the surfactant RapiGest, I-OmpA formed well-structured narrow pores which displayed long open times of 0.90 and 0.94 and conductances of 78 ± 3 pS and 80 ± 2 pS, respectively. I-OmpA isolated with urea formed irregular pores with a shorter open time of 0.78 and wide conductance range of 84 ± 18 pS. Pores formed by I-OmpA isolated with GuHCl had a similar open time of 0.79 but were even more irregular with conductance range of 84 ± 28 pS. Further incubation in C8E4 micelles or DPhPC did not improve the pore structure of M-OmpA or I-OmpA isolated using urea or GuHCl.
Figure 3.
Left: Representative current traces of OmpA isolated from cytoplasmic inclusion bodies of E. coli BL21(DE3)pLysS cells (I-OmpA), purified by Ni agarose chromatography, reconstituted into C8E4 micelles, and incorporated into planar lipid bilayers of DPhPC at room temperature between aqueous solutions of 1M KCl in 20 mM Hepes, pH 7.4. Bar at right of the traces indicates the closed state of the channel. Clamping potential was +100 mV. Right: Histograms taken from records of ~ 10 minutes at room temperature.
3.2. Planar lipid bilayer observations of OmpA sorting signal mutants S163G and S167G
Previous studies showed that cOHB is conjugated to residues S163 and S167 of the sorting signal (residues 163–169) in OmpA isolated from cytoplasmic inclusion bodies [8]. Each of the single mutants, S163G and S167G, displays positive reactions to anti-OHB IgG, which indicates that the remaining serine residue is conjugated to cOHB.
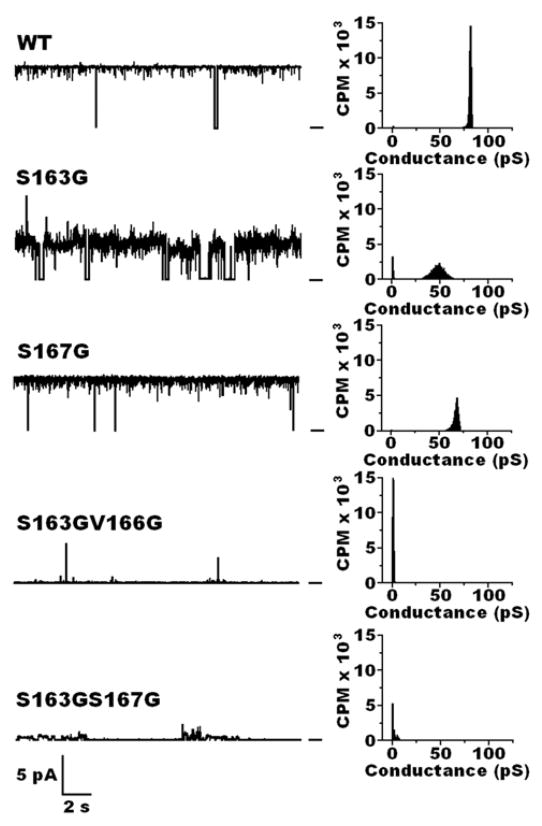
The effect of this partial loss of cOHB on the sorting signal on bilayer incorporation and pore formation by OmpA was examined. His-tagged OmpA carrying either the mutation S163G or alternatively S167G was overexpressedin E. coli BL 21 cells containing the pET-45b(+)-His gene. Each His-OmpA mutant was then purified from inclusion bodies using RapiGest (see Methods). The purified mutant proteins displayed the same pattern of electrophoretic migration on SDS-PAGE as the wild-type (Fig. 1 lanes 5–8). Each protein was reconstituted into C8E4 micelles and incorporated into planar lipid bilayers of DPhPC. As shown in Fig. 4, S167G formed narrow pores in planar lipid bilayers at room temperature with long open time of 0.94 but with significantly lower conductance, 67 ± 4 pS. S163G displayed a substantially lower open time of 0.80, with even lower conductance and wider conductance range, 49 ± 7 pS.
Figure 4.
Left: Representative current traces of OmpA wild-type and mutant proteins isolated from cytoplasmic inclusion bodies of E. coli BL21(DE3)pLysS cells, purified by Ni agarose chromatography, reconstituted into C8E4 micelles, and incorporated into planar lipid bilayers of DPhPC at room temperature between aqueous solutions of 1M KCl in 20 mM Hepes, pH 7.4. Bar at right of the traces indicates the closed state of the channel. Clamping potential was +100 mV. Right: Histograms taken from records of ~ 10 minutes at room temperature.
3.3. Planar lipid bilayer observations of cOHB-minus OmpA sorting signal mutants S163G:S167G, S163G:V166G
Previous studies showed that the double mutants S163G:S167G and S163G:V166G of OmpA, isolated from cytoplasmic inclusion bodies, lack cOHB on the sorting signal (residues 162–174) [8]. Each double mutant was overexpressed and purified from inclusion bodies of E. coli UH203 as above. The double mutants displayed the same pattern of electrophoretic migration on SDS-PAGE as the wild-type and single-mutants (Fig. 1 lanes 9,10). The mutants were each reconstituted into C8E4 micelles and multiple attempts were then made to incorporate each mutant protein into planar lipid bilayers of DPhPC at room temperature as above and again after incubation for > 2 hrs at 40 °C. More than 20 separate observations were made of two or more separate preparations of each mutant. In all cases, only trial insertions were observed with no stable pore formation (Fig. 4).
4. Discussion
Here we have examined the influence of purification protocols on the configuration of pores formed by wild-type E. coli OmpA in planar lipid bilayers at room temperature. Whole OmpA, extracted from outer membranes or from cytoplasmic inclusion bodies, incorporates into planar lipid bilayers forming narrow pores at room temperature (Figs. 2,3). The average conductance is not significantly influenced by the solubilizing agent used to extract the OmpA protein; however, the pore structure, as estimated by the conductance range and the open time, is notably affected. OmpA extracted from outer membranes with LDS and OmpA isolated from inclusion bodies with LDS or RapiGest forms well-structured pores in planar lipid bilayers of DPhPC at room temperature with a narrow conductance range and high open time (Figs. 2,3, Table 1). When urea or GuHCl, were used to extract OmpA, the imperfect refolding of the protein is evident in the conductance irregularity and in some cases lower open time (Figs. 2,3, Table 1).
Table 1.
Conductance and open time (Po) for OmpA isolated from cytoplasmic inclusion bodies (IB) or from outer membranes (OM) using different solubilizing agents.
| Extraction Agent | Conductance, pS | PO | |
|---|---|---|---|
| IB | LDS | 78 ± 3 | 0.90 |
| RapiGest | 80 ± 2 | 0.94 | |
| 8 M Urea | 84 ± 18 | 0.78 | |
| 6 M GuHCl | 84 ± 28 | 0.79 | |
| OM | LDS | 80 ± 2 | 0.94 |
| 8 M Urea | 80 ± 10 | 0.92 | |
| 6 M GuHCl | 80 ± 17 | 0.70 |
Problems in refolding suggested by the irregular conductance may be due to the increased complexity of refolding from the unfolded state obtained with urea and GuHCl. In the case of urea, carbamylation may contribute to this problem [18,19]. OmpA has 17 lysines and 2 cysteines which are potential targets for carbamylation. Regardless of the purity of the urea employed, a significant concentration of isocyanate will form on standing, even at the neutral pH and room temperatures used in this study. Carbamylation presents a more serious problem at the alkaline pH and elevated temperatures which have been used in many studies of OmpA folding [17,20–23].
We have also examined the effect of cOHB modification of the sorting signal on the bilayer incorporation and pore structure of OmpA. We find that single serine mutants S163G and S167G, which still contain some cOHB on the sorting signal [8] are able to form narrow pores in planar lipid bilayers (Fig. 4). The loss of cOHB on S167 results in significantly lower conductance while loss of cOHB on S163 appears to narrow the pore to a greater extent and reduces both conductance and open time. The double mutants S163G:S167G and L162G:S167G, which are without cOHB on the sorting signal [8], display only trial insertions into the bilayer which indicate that the proteins are associated with the bilayer and may even lie within it, but are unable to form stable pores (Fig. 4).
The wild-type and mutant OmpA proteins display the same electrophoretic mobility on SDS-PAGE. They all exhibit ‘heat modification’, i.e. the proteins migrate at ~ 35 kDa after heating in SDS whereas unheated proteins migrate at ~ 30 kDa (Fig. 1). It is widely believed that the native state of OmpA can be recognized by its mobility on SDS gels; however, our studies indicate that only the completely unfolded protein migrates at 35 kDa; all other forms have an apparent molecular weight of ~30 kDa.
R-3-hydroxybutyrate monomers are water-soluble, but oligomers of ≥4 units are water-insoluble [24,25]. The modification of serines of the eighth beta strand by short chains of cOHB increases the hydrophobicity of this segment and may thereby assist in targeting the protein to the bilayer and/or in folding within the bilayer to form a pore. The flexibility and amphiphilic structure of cOHB [12–14] enable it to adapt its conformation to the environment, i.e. cOHB may turn its hydrophilic ester oxygens inward and hydrophobic methyl groups outward as the protein moves from aqueous medium to bilayer and reverse this orientation as the protein moves from bilayer into aqueous medium. Our studies indicate that cOHB modification plays a critical role in bilayer incorporation and pore formation by OmpA and further suggest that cOHB may be an important agent in the bilayer incorporation and performance of other membrane proteins.
Acknowledgments
We thank NIH Grant GM 054090 and Metabolix Inc. (Cambridge, MA), for support of this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- 2.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 5.Klose M, Schwarz H, MacIntyre S, Freudl R, Eschbach M, Henning U. Internal deletions in the gene for an Escherichia coli outer membrane protein define an area possibly important for recognition of the outer membrane by this polypeptide. J Biol Chem. 1988;263:13291–13296. [PubMed] [Google Scholar]
- 6.Freudl R, Klose M, Henning U. Export and sorting of the Escherichia coli outer membrane protein OmpA. J Bioenerg Biomembr. 1990;22:441–449. doi: 10.1007/BF00763176. [DOI] [PubMed] [Google Scholar]
- 7.Klose M, MacIntyre S, Schwarz H, Henning U. The influence of amino substitutions within the mature part of an Escherichia coli outer membrane protein (OmpA) on assembly of the polypeptide into its membrane. J Biol Chem. 1988;263:13297–13302. [PubMed] [Google Scholar]
- 8.Xian M, Fuerst MM, Shabalin Y, Reusch RN. Sorting signal of Escherichia coli OmpA is modified by oligo-(R)-3-hydroxybutyrate. 2007;1768:2660–2666. doi: 10.1016/j.bbamem.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang R, Reusch RN. Poly(3-hydroxybutyrate) is associated with specific proteins in the cytoplasm and membranes of Escherichia coli. J Biol Chem. 1996;271:22196–22202. doi: 10.1074/jbc.271.36.22196. [DOI] [PubMed] [Google Scholar]
- 10.Seebach D, Brunner A, Bűrger HM, Schneider H, Reusch RN. Isolation and 1H-NMR spectroscopic identification of poly(3-hydroxybutanoate) from prokaryotic and eukaryotic organisms. Eur J Biochem. 1994;224:317–328. doi: 10.1111/j.1432-1033.1994.00317.x. [DOI] [PubMed] [Google Scholar]
- 11.Reusch RN. Low molecular weight complexed poly(3-hydroxybutyrate): a dynamic and versatile molecule in vivo. Can J Microbiol. 1995;41:50–54. doi: 10.1139/m95-167. [DOI] [PubMed] [Google Scholar]
- 12.Rueping M, Dietrich A, Buschmann V, Fritz MG, Sauer M, Seebach D. On the structure of poly(3-hydroxybutanoic acid) in solution and in phospholipid bilayers: Circular dichroism and fluorescence spectroscopy with oligo(3-hydroxybutanoic acid) derivatives. Macromol. 2001;34:7042–7048. [Google Scholar]
- 13.Waser P, Rueping M, Seebach D. On the solution structure of PHB: Preparation and NMR analysis of isotopically labeled oligo [(R)-3-hydroxybutanoic acids] (OHBs) Helv Chim Acta. 2001;84:1821–1845. [Google Scholar]
- 14.Gee PJ, Hamprecht FA, Schuler LD, Gunsteren WF, Duchardt E, Schwalbe H, Albert M, Seebach D. A molecular-dynamics simulation study of the conformational preferences of oligo(3-hydroxyalkanoic acids) in chloroform solution. Helv Chim Acta. 2002;85:618–632. [Google Scholar]
- 15.Sugawara E, Nikaido H. Pore-forming activity of OmpA protein of Escherichia coli. J Biol Chem. 1992;267:2507–2511. [PubMed] [Google Scholar]
- 16.Sugawara E, Nikaido H. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J Biol Chem. 1994;269:17981–17987. [PubMed] [Google Scholar]
- 17.Hong H, Szabo G, Tamm LK. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat Chem Biol. 2006;2:627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 18.Hagel P, Gerding JJ, Fieggen W, Bloemendal H. Cyanate formation in solutions of urea. I. Calculation of cyanate concentrations at different temperature and pH. 1971;243:366–373. doi: 10.1016/0005-2795(71)90003-1. [DOI] [PubMed] [Google Scholar]
- 19.Stark GR, Stein WH, Moore S. Reactions of the cyanate present in aqueous urea with amino acids and proteins. J Biol Chem. 1980;235:3177–3181. [Google Scholar]
- 20.Surrey T, Jähnig F. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc Natl Acad Sci USA. 1992;89:7457–7461. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinschmidt JH, Tamm LK. Folding intermediates of a β-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. [DOI] [PubMed] [Google Scholar]
- 22.Arora A, Rinehart D, Szabo G, Tamm LK. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J Biol Chem. 2000;275:1594–1600. doi: 10.1074/jbc.275.3.1594. [DOI] [PubMed] [Google Scholar]
- 23.Kim JE, Arjara G, Richards JH, Gray HB, Winkler JR. Probing folded and unfolded states of outer membrane protein A using steady-state and time-resolved tryptophan fluorescence. J Phys Chem. 2006;110:17656–17662. doi: 10.1021/jp061991r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jendrossek D, Hendrick R. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol. 2002;56:403–422. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]