Abstract
Deoxyribozymes are promising biotechnological tools. In a recent JACS article Mokany et al. reported on the design of multi-component deoxyribozyme (MNAzyme) sensors based on 10–23 and 8–17 DNA enzymes. The sensors can detect down to 5 pM of a specific nucleic acid. The versatility of MNAzyme platform allows the design of catalytic cascades for signal amplification. This work is a step forward to PCR-free molecular diagnostics.
The discovery of catalytic DNA molecules, known also as deoxyribozymes, DNA enzymes or DNAzymes, by Breaker and Joyce in 1994 has introduced a new versatile scaffold for the design of variety of biotechnological tools (reviewed by Schlosser and Li, 2009 and by Baum and Silverman, 2008). Sharing the advantages of biocompatibility and simplicity of structural prediction and modification with ribozymes, DNA enzymes are more stable and have lower cost of chemical synthesis than their RNA counterparts. DNAzymes have a great, though not completely explored, potential for sensing a variety of analytes (Liu et al., 2009) mainly due to the possibility of catalytic signal amplification.
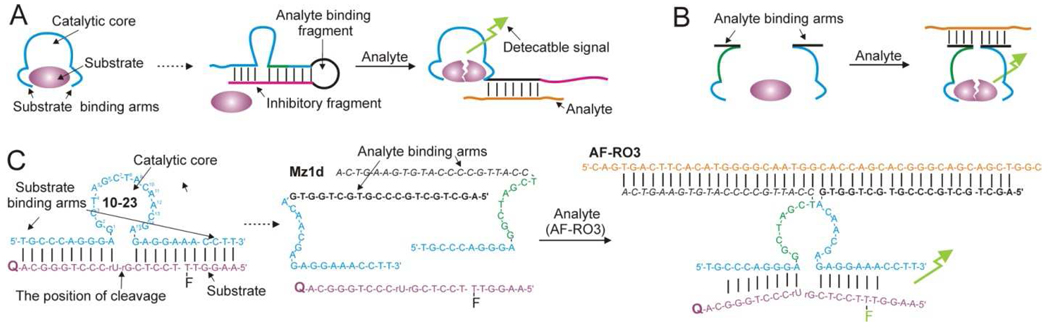
Among all possible applications of DNAzyme-based sensors the detection of specific RNA/DNA sequences is of particularly interest because deoxyribozymes can be easily tailored to recognize these types of analytes by Watson-Crick base pairing. On the other hand there is a need for the PCR-free assays in straightforward formats that do not require highly trained personnel and such expensive equipment as real time PCR thermocyclers. Indeed, PCR is an ultimate in terms of sensitivity, but relatively complex, expensive and sensitive to contaminations. Furthermore, microarrays of deoxyribozyme sensors may enable simultaneous analysis of hundreds of thousands of analytes, and thus revolutionize molecular diagnostics. Nucleic acid analysis, therefore, is the field, in which the enormous potential of deoxyribozyme-based sensors can be put into practice. Recently reported MNAzymes technology (Mokany et al. 2009) takes advantage of catalytically efficient DNAzymes 10–23 and 8–17 (Santoro and Joyce, 1998) to create efficient sensors for specific nucleic acid sequences. Selectivity and sensitivity are two major characteristics that determine the efficiency of any sensor. Choice of the proper design is a crucial step toward creating an efficient molecular sensor. Many DNAzymes share common architecture (Fig. 1A, left) with a substrate binding domain and a catalytic core (cyan), which are essential to bind and process the substrate. One strategy for the sensor design uses an inhibitory fragment (magenta in Fig. 1A, middle) connected to the catalytically essential nucleotides via an analyte-binding fragment (black). This fragment hybridizes to the essential nucleotides, thus preventing their interaction with the substrate. Hybridization of the analyte (orange) to the inhibitory fragment liberates the substrate-processing nucleotides and restores the catalytic activity. Active enzyme cleaves multiple substrates, thus generating a detectable signal. This mechanism most closely resembles intrasteric regulation of natural enzymes. In this design the analyte competes with the deoxyribozyme’s essential nucleotides for binding to the inhibitory fragment. This competition may lead either to high background reaction in the absence of the analyte if the inhibited sensor structure is unstable or to the reduced sensitivity if this structure is too stable. The optimization of the length and the sequence of the inhibitory fragment can turn to be a tedious task. In addition, such sensors are not always selective enough to distinguish analytes that differ by only one nucleotide.
Figure 1. Design of deoxyribozyme sensors for nucleic acid analysis.
A: Allosterically regulated deoxyribozyme sensors (e.g. Stojanovic et al., 2003). B: Binary deoxyribozyme sensors (e.g. Kolpashchikov, 2007 and Kolpashchikov, 2008). C: One of the MNAzyme sensors reported by Mokany et al., 2009. The enzymatic core of DNAzyme 10–23 was divided between T8 and A9 , and the analyte binding arms (black sequences) were connected to T8 and A9. The presence of oligonucleotide AF-RO3 led to the formation of the catalytically active associate (right), which cleaved the fluorogenic substrate.
An alternative strategy takes advantage of splitting the deoxyribozyme structure into two fragments (subunits) shown in Figure 1B (left). Each fragment consists of the essential substrate binding and processing nucleotides and a fragment complementary to the analyte (analyte binding arm). Hybridization of the two subunits to the abutting positions of an analyte re-unites the catalytically active core leading to the substrate transformation followed by generation of the detectable signal (Fig 1B, right). In this approach the degree of the sensor association can be fine-tuned simply by varying the concentration of the two sensor’s strands, thus completely eliminating the undesired background activity in the absence of the analyte. Moreover, the binary design enables great selectivity because each of the two short probe-analyte hybrids is more perceptive to a single base mis-paring than one long stable hybrid formed by the allosterically regulated construct shown in Figure 1A.
It has been shown that the split architecture is adoptable for deoxyribozymes, which contain either inessential for catalysis stem-loop fragments (Kolpashchikov, 2007) or G-quartets (Kolpashchikov, 2008). The present work demonstrates more general applicability of the binary design: Mokany and colleagues elegantly split catalytic domains of deoxyribozymes 10–23 and 8–17 to form multicomponent DNAzymes (MNAzymes). It was demonstrated that the MNAzymes can be used for sensitive nucleic acid detection in isothermal assay or in a PCR format. For example, a 10–23-derived MNAzyme, Mz1 (Fig. 1C), which demonstrated the highest analyte-dependent activation, detected as low as 5 pM DNA analyte. This limit of detection (LoD) is among the lowest reported so far for homogeneous fluorescent assays.
The LoD for enzyme-based sensors depends strongly on the catalytic efficiency of the parent enzymes. Deoxyribozyme 10–23 is the most catalytically active DNA known so far with kcat/Km of ~ 109 M−1 min−1 approaching the efficiency of the “catalytically perfect” enzymes. Despite optimal binary design, the LoD of 10–23-based sensor does not exceed picomolar range. This detection limit is not low enough to sense ~10−13–10−16 M viral RNA found in the blood samples of infected individuals (Steininger C. et al., 1994). Deoxyribozyme cascades, in which the signal of a deoxyribozyme sensor is amplified by downstream reactions, can help to achieve the desired sensitivity even without PCR amplification. Such cascades were introduced by Levy et al. in 2003 and now, in an original design, reported by Mokany and colleagues. It is to be demonstrated whether DNAzyme-based cascades can improve the LoDs by 1–4 orders of magnitude to enable applications of deoxyribozyme technology in molecular diagnostics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Baum DA, Silverman SK. Cell Mol. Life Sci. 2008;65:2156–2174. doi: 10.1007/s00018-008-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpashchikov DM. Chembiochem. 2007;8:2039–2042. doi: 10.1002/cbic.200700384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpashchikov DM. J. Am. Chem. Soc. 2008;130:2934–2935. doi: 10.1021/ja711192e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Ellington AD. Proc. Natl. Acad. Sci. USA. 2003;100:6416–6421. doi: 10.1073/pnas.1130145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokany E, Bone SM, Young PE, Doan TB, Todd AV. J. Am. Chem. Soc. 2009 doi: 10.1021/ja9076777. 10.1021/ja9076777 M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Steininger C, Kundi M, Jatzko G, Kiss H, Lischka A, Holzmann H. J. Infect. Dis. 2003;187:345–351. doi: 10.1086/367704. [DOI] [PubMed] [Google Scholar]