Abstract
Chemoreceptors are crucial components in the bacterial sensory systems that mediate chemotaxis. Chemotactic responses exhibit exquisite sensitivity, extensive dynamic range and precise adaptation. The mechanisms that mediate these high-performance functions involve not only actions of individual proteins but also interactions among clusters of components, localized in extensive patches of thousands of molecules. Recently, these patches have been imaged in native cells, important features of chemoreceptor structure and on–off switching have been identified, and new insights have been gained into the structural basis and functional consequences of higher order interactions among sensory components. These new data suggest multiple levels of molecular interactions, each of which contribute specific functional features and together create a sophisticated signaling device.
High-performance signaling in bacterial chemotaxis
The high-performance chemotaxis signaling system of Escherichia coli (Box 1) involves a limited number of components but notable sophistication [1–4]. This system has become a paradigm for molecular characterization of biological signaling mechanisms. Transmembrane chemoreceptors, known as methyl-accepting chemotaxis proteins (MCPs), direct cell locomotion by regulating the histidine kinase CheA; CheA phosphorylates a response regulator, which in turn controls the rotational direction of the flagellar motor. Chemotactic sensitivity and the range of signal detection are regulated by adaptational modification of chemoreceptors via reversible glutamyl methylation. The resulting interplay between motor control and sensory adaptation produces directed motile behavior. E. coli chemoreceptors are the most extensively studied representatives of the MCP super-family, central components of homologous systems that mediate tactic responses [5,6] across the phylogenetic diversity of bacteria and archaea [7].In E. coli chemotaxis proteins cluster in membrane-associated patches [8]. Interaction within patches is thought to contribute to notable features of the signaling system: high sensitivity, wide dynamic range, extensive cooperativity and precise adaptation. Delineating the molecular mechanisms underlying these features will require knowledge of structures of the components, complexes and higher order arrays. In addition it will require definition of organization at the multiple levels of interaction among signaling components and an understanding of the changes in components and their interactions that mediate signaling at each level. In the past few years, notable progress has been made toward acquiring the information and insights necessary for understanding these molecular mechanisms. This review describes that progress. Here we follow the lead of most investigators in the area, and do not distinguish between studies of first cousins E. coli and Salmonella enterica serovar Typhimurium. Thus, references to E. coli can be taken to apply to both species.
High-performance features suggest functional arrays of interacting sensory components
The E. coli chemotaxis signaling pathway can amplify stimuli at least 50-fold (i.e. a 1% change in receptor occupancy elicits a 50% change in the rotational bias of the flagellar motors [9], a minimal estimate because receptors act in intermixed arrays [10]). Elegant measurements of CheA kinase activity in vivo using fluorescence resonance energy transfer (FRET) showed that much of this amplification (36-fold) occurs at the signaling complex [10]. In many signaling systems the mechanism of gain involves enzyme activation, generating many downstream signaling molecules from an occupancy change at one receptor. However, for E. coli chemoreceptors, attractant occupancy inhibits the activity of the associated kinase (Box 1). Thus, a 36-fold gain indicates that one receptor molecule can control the activity of three dozen kinase molecules, implying a functional network that links one receptor to multiple copies of the kinase. In addition, receptors are coupled to one another. For instance, the response to attractant stimulation is cooperative; Hill coefficients as high as 10 have been observed [11,12]. Strikingly, the presence of heterologous receptors or homologous receptors in different states of adaptational modification alters cooperativity and/or sensitivity to stimulation, as well as the level of kinase activation [11–14]. These properties imply that receptors operate as allosteric arrays with as many as several dozen in the cooperative unit [2,11]. Finally, precise adaptation, which is the exact resetting of signaling and behavior to the ground state after stimulation, involves cooperation between neighboring receptors [15–17]. Thus, the features of signal gain, cooperativity, inter-receptor influences and sensory adaptation all imply that multiple sensory components interact in communicating arrays.
Box 1. Two conformational states of chemoreceptor signaling.
Chemoreceptors mediate gradient-tracking behavior through ternary signaling complexes that contain receptors, CheA (a histidine autokinase) and CheW (a protein that couples CheA activity to receptor control) (Box 1. Figure I). The major features of the chemotaxis system [1–4] can be explained qualitatively by the notion that signaling complexes are in equilibrium between two conformational states. The ‘kinase-on’ receptor conformation stimulates the CheA autophosphorylation rate several hundred-fold, whereas the ‘kinase-off’ conformation deactivates CheA autophosphorylation. Net kinase activity reflects the proportion of complexes in the two states. Chemical stimuli elicit motor responses by shifting the equilibrium. An increase in attractant concentration shifts receptors to the ‘kinaseoff’ state; an increase in repellent concentration (or a decrease in attractant concentration) shifts receptors to the ‘kinase-on’ state.
Shifts in the kinase-on–kinase-off equilibrium modulate the flux of CheA phosphoryl groups to two response regulators, CheY for motor control, and CheB for sensory adaptation. In motor control, phospho-CheY binds to the flagellar rotary motor, enhancing the probability of clockwise (CW) rotation, which causes random directional changes; counter-clockwise (CCW) rotation, the default behavior, produces forward swimming. Phosphatase CheZ hydrolyzes phospho-CheY, ensuring a short duration in CW flagellar rotation so that phospho-CheY levels closely track receptor-modulated CheA activity.
Cells swimming through spatial chemical gradients monitor temporal changes in chemoeffector concentrations by means of a sensory adaptation system that records recently encountered chemical conditions in the form of reversible chemoreceptor methylation at four to six glutamyl residues in the adaptation region of the receptor's kinase control module. Hence, chemoreceptors are methyl-accepting chemotaxis proteins (MCPs). The modifications are catalyzed by two MCP-specific enzymes, methyltransferase CheR and methylesterase CheB. Two receptor-modification sites are synthesized as glutamines, which are functional mimics of methyl glutamates, and are deamidated by CheB to create methyl-accepting glutamates. The adaptation enzymes continuously update the methylation record using two feedback mechanisms: (i) activation of CheB by CheA-mediated phosphorylation and (ii) opposite propensities for the two modifications in the two receptor conformations. For example, the ‘kinase-off’ signaling conformation has high attractant affinity, high propensity for methylation and low propensity for demethylation. The ‘kinase-on’ conformation has low attractant affinity, low methylation propensity and high demethylation propensity. Because rates of modification are slow on the time-scale of binding an attractant or a repellent, the extent of receptor methylation provides a record of the recent chemical past with which to make comparisons to current chemoeffector levels, reflected in the extent of receptor ligand occupancy.
Qualitatively, a two-state model explains the major features of the chemotaxis system, but quantitative and modeling studies indicate that additional complexity is present that extends the dynamic range of the system beyond that possible for a simple two-state model [13,17,61].
Patches provide the physical organization in which functional arrays can form
Membrane-associated patches of ternary signaling complexes, containing chemoreceptors, the CheA histidine kinase and the coupling protein, CheW, were first detected by immunogold labeling of fixed, sectioned cells [18], later by imaging of fluorescently tagged chemotaxis proteins in living cells [8,19] and most recently by electron tomography of intact cells in vitreous ice [20]. All three methods detect no more than a few patches per cell, with most (~80%) located at a cell pole, although not at any particular position along the polar membrane. Non-polar, lateral patches are distributed at future division sites [21]. There is no evidence for functional differences between polar and lateral patches. Patches in the polar area are mobile within the curvature of the pole but lateral patches appear fixed [21]. Polar localization is a separate phenomenon from formation of focused patches. For instance, receptors form polar caps in the absence of CheA and CheW, but require these proteins to form distinct patches [22], and some receptors form only polar caps in the absence of adaptational modification [23].
Recent cryo-electron microscopy and tomography have provided the most revealing views of signaling complexes in patches [20]. The patches contain closely packed, needle-like receptors extending from the membrane with a layer of CheA and CheW at their cytoplasmic, membrane-distal tips (Figure 1). These images provide striking documentation of localized patches of chemosensory components with defined boundaries. Resolution is not yet sufficient to determine the degree to which components are packed in regular arrays. Patches are generally circular or ellipsoid and they have varying sizes, with an average diameter ~250 nm. A circular 250 nm patch could contain ~40% of the total of ~7500 receptor dimers present in the cell [24] and would occupy ~1% of the cell surface.
Figure 1.
A patch of membrane-embedded chemoreceptors in a signaling complex near the pole of an intact cell. The emerging technique of cryo-electron tomography can provide images of the detailed internal structure of intact cells, including macromolecular complexes, without the need for fixation, staining or other potentially perturbing treatments. Application of this technique to motile and chemotactic E. coli cells [20] reveals distinct patches of striations, usually near a pole, extending perpendicular to the cytoplasmic membrane. These patches occur in cells containing chemoreceptors, the histidine kinase CheA and the coupling protein CheW, but not in cells lacking any one of these proteins. Immuno-electron microscopy demonstrates that the striations consist of chemoreceptors and that a thin line of density parallel to the membrane at the membrane-distal end of the striations contains CheA and CheW. (a) A 5 nm tomographic slice of the region near a pole of an intact, chemotactically wild-type E. coli cell [20]. The slice is oriented essentially normal to the two membranes (indicated by labeled arrows) that surround this Gram-negative cell – the cytoplasmic membrane, which is the cell's permeability barrier, and the outer membrane, which is a penetration barrier that creates the periplasm between the two membranes. A patch of chemoreceptors, CheA and CheW is visible along part of the cytoplasmic membrane. The boundaries of the patch are marked by white arrows. (b) Schematic of the tomograph in (a) showing the membranes (outer membrane, dark gray; cytoplasmic membrane, light gray), periplasm (light blue) and cytoplasm (yellow). The boundaries of the patch are marked by white arrows as in part (a). Chemoreceptors (red) and the layer of CheA and CheW (blue) are indicated by labeled arrows. Panel (a) is derived from the same original image from which Figure 2a of Zhang et al. [20] was made. We thank S. Subramaniam for that original image. Part (b) is based on Figure 1c of the same paper. Adapted, with permission, from [20].
Interactions in these patches could enable receptors to control multiple kinases and multiple receptors to influence one other. However, researchers are only now beginning to identify patterns and mechanisms of higher order interactions among receptors. The first level at which insights have been gained has been characterization of chemoreceptor trimers-of-homodimers.
Chemoreceptor homodimers interact to form trimers-of-dimers
A fragment representing most of the cytoplasmic domain of the E. coli chemoreceptor Tsr (taxis to serine and repellents) crystallized as three extended, homodimeric four-helix bundles interacting at their helical hairpin tips [25]. Cysteine-directed cross-linking studies indicated that this trimer-of-dimers interaction occurs in the full-length receptor in vivo (Box 2). The principal trimer contact residues are identical in all five E. coli receptors [Tsr, Tar (taxis to aspartate and repellents), Trg (taxis to ribose and galactose), Tap (taxis to dipeptides) and Aer (taxis to oxygen)]. These contacts allow different receptors to form mixed trimers, the composition of which reflects their relative cellular abundance [26] (Box 2).
Recent studies of receptors with trimer contact lesions suggest that trimers-of-dimers are important in receptor function. Ames et al. [27] characterized amino acid replacements at 11 trimer contact residues in the serine receptor, Tsr. Receptors with proline introduced were defective in ternary complex assembly, kinase activation and patch formation. These loss-of-function (null) pheno-types are consistent with disruption of helical structure at the protein interaction tip. By contrast, receptors containing an alanine or tryptophan at a trimer contact formed patches. Some activated CheA kinase; others did not. Some alanine-replacement mutants even regained serine receptor function when co-expressed with wild-type aspartate (Tar) receptors. By contrast, all tryptophan-replacement mutants blocked the function of co-expressed Tar. These rescue and epistasis effects suggest that Tsr and Tar might function in mixed complexes. Tsr molecules with rescuable defects might be conformationally repaired by the presence of wild-type Tar in the complex, whereas Tsr molecules with epistatic defects might impose an aberrant conformation on the entire complex. In support of this idea, some epistatic Tsr defects were functionally rescued by Tar mutants harboring a complementary mutational change in the trimer contact region [28]. The Tar alterations suppressed Tsr defects in an allele-specific fashion, a hallmark of conformational suppression through direct protein–protein interactions.
Receptors form trimers-of-dimers in the absence of other chemotaxis proteins [26]. However, trimers assembled in the absence of either CheA or CheW exchange members with recently made receptor molecules, whereas trimers formed in the presence of both CheA and CheW do not undergo such exchanges [29]. Thus, CheA and CheW stabilize trimer arrangements, most likely through binding interactions within and/or between trimer units. The stoichiometries of receptor, CheA and CheW molecules during ternary complex assembly have profound effects on the resultant structure and function of the array. For example, an excess of CheW interferes with trimer formation, probably through binding interactions that mask the trimer contact surfaces on receptor molecules [29]. Similarly, CheW and CheA compete for binding sites on receptor molecules [30,31].
Macroscopic receptor patches are most likely imperfect arrays of receptor trimers-of-dimers. The fundamental unit of receptor signaling could be a single trimer with its associated CheA and CheW partners, or could be several trimers linked through shared CheA and CheW signaling proteins. The threefold symmetry of receptor trimers is the simplest geometry that could produce 2D arrays [32]. However, owing to stochastic assembly, the array probably contains gaps that could determine the effective size of the cooperative signaling unit.
Chemoreceptors and other chemotaxis proteins cluster in membrane-associated patches in all nine phylogenetically diverse species that have been examined [33].However, there is little experimental evidence that addresses whether chemoreceptors in these bacteria also have a trimer-based architecture. Sequence comparisons of >2000 chemoreceptors from 152 species revealed high conservation for 10 of 11 principal trimer contact residues across all families [7]. The one exceptional site (residue 373 in Tsr) is a phenylalanine in the E. coli chemoreceptor family and a polar, usually charged, residue in all others. This difference could conceivably destabilize the trimer interface in other chemoreceptors, and indeed a cytoplasmic fragment of a Thermotoga maritima receptor that carries a glutamic acid at this position crystallized not as trimers-of-dimers but rather as ‘hedgerows’, lines of parallel receptors interacting along their long axes [34]. It should be possible to elucidate the higher-order organization of receptors in other bacteria through in vivo cross-linking approaches such as those used in E. coli.
Architecture of the receptor–CheA–CheW signaling complex
The structure of ternary chemotaxis signaling complexes and the mechanism(s) of kinase control are poorly understood but the bulk of the data on E. coli receptors suggests that the ternary complex involves dimers interacting as a trimer. A working architectural model for this ternary complex, based on the probable docking surfaces of each component, has been proposed [35]. Electron microscopic image analysis of particles assembled from a soluble receptor fragment, CheA and CheW also shows trimers, but with significantly altered geometry in which the three dimers are not arranged in a threefold symmetry [36,37]. The hedgerow arrangement in crystals of an archaeal receptor fragment suggests that archaeal CheA and CheW might associate with separated dimers [34]. Whether the structural diversity implied by these in vitro studies occurs in vivo remains an open question.
Correlating receptor–receptor interactions and function
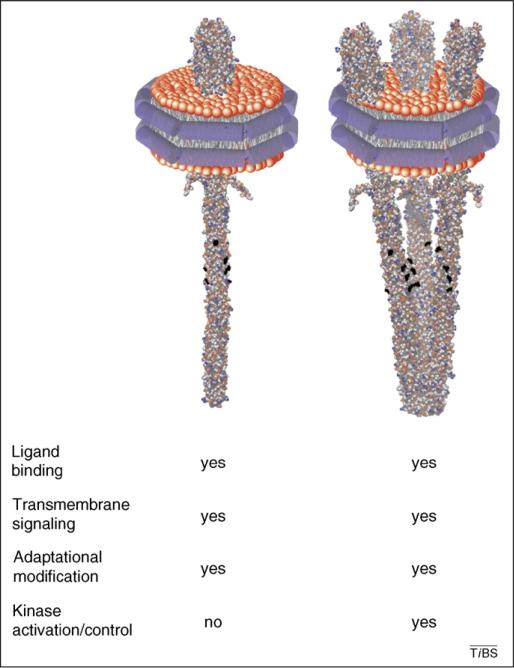
Which activities are inherent in individual chemoreceptor dimers and which require interactions between dimers? Individual, membrane-inserted dimers can be isolated from interaction with other receptors by placing them in Nanodiscs (Figure 2), which are defined-size, water-soluble plugs of lipid bilayer surrounded by a protein annulus [38,39]. These isolated dimers are efficiently methylated and the rate of methylation is augmented by attractant binding, demonstrating that individual dimers not only bind ligand, but are capable of transmitting ligand-induced conformational changes to the methylation region of the receptor [40]. Thus, transmembrane signaling occurs within single receptor dimers and does not require higher-order interactions among dimers. By contrast, Nanodisc-segregated dimers were ineffective at kinase activation, which required more than one dimer per disc [40]. Peak kinase activation occurred in preparations averaging almost three dimers per disc, and this activation was inhibited by ligand, as it is in intact cells [10]. An optimum for ligand-controlled kinase activation at approximately three dimers per disc is tantalizing because it correlates activation with potential formation of trimers-of-dimers, although these results cannot rule out activation by other oligomer sizes. In any case, it is notable that isolated, Nanodisc-embedded dimers (in the absence of receptor partners) bind ligand, are effective substrates for covalent modification and generate trans-membrane signals. Thus, a mechanistic understanding of high-performance features of chemoreceptors and the chemotaxis sensory system begins with characterization of the chemoreceptor homodimer.
Figure 2.
Functional activities of Nanodisc-embedded chemoreceptors. Nanodiscs are soluble, nanoscale (~10 nm diameter) particles of lipid bilayer surrounded by an annulus of amphiphilic membrane scaffold protein [39]. They form spontaneously when detergent is removed from mixtures of detergent-solubilized lipid and scaffold protein. If the preparation mixture contains detergent-solubilized membrane protein, the protein is incorporated into Nanodiscs. A Nanodisc-embedded protein is in a lipid bilayer and thus probably exists in its native state, but it is segregated from other membrane proteins. Chemoreceptors can be incorporated into Nanodiscs [40]. Preparations with different average number of chemoreceptor dimers per disc can be prepared by manipulating the input ratio of receptor to scaffold protein. These Nanodisc preparations enabled determination of chemoreceptor activity as a function of the number of receptor dimers per disc, that is the number of dimers that could potentially interact. The figure (a modified version of Figure 4 from Boldog et al. [40]), presents schematics of Nanodisc-embedded chemoreceptors in the onedimer per disc (left) or three-dimers per disc (right) state (shown as a trimer-of-dimers) and indicates the receptor activities exhibited in the two different conditions. Preparations of one dimer per Nanodisc were effectively methylated, and initial rates of this modification were doubled by the presence of a saturating concentration of an attractant ligand. These observations indicated that individual chemoreceptor dimers reconstituted in Nanodiscs were substrates for adaptational modification and coupled ligand-binding to methylation propensity. However, preparations of one chemoreceptor dimer per disc were ineffective at activating the chemotaxis kinase CheA. Nanodiscs with three dimers per disc exhibited a relatively sharp maximum of kinase activation, and activation was eliminated by the addition of a saturating concentration of an attractant ligand [40]. These observations indicated that functionally relevant kinase activation required the combined action of more than one chemoreceptor and were consistent with the functional importance of three dimers, for instance as a trimer-of-dimers. Adapted, with permission, from [40].
Chemoreceptor homodimers: structure
The helical, intertwined chemoreceptor homodimer contains three operational modules, each with a distinct structure and signaling mechanism: transmembrane sensing, signal conversion and kinase control (Box 3). Electron microscopy [41] reveals E. coli receptor dimers as ~300Å needle-like proteins oriented approximately normal to the membrane. The needle-like shape and dimensions imply that the helical receptor sub-domains for which detailed structural information is available are arranged end-toend (Box 3).
The signal-conversion module is a HAMP domain (found in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins and phosphatases [42]). A recent advance comes from nuclear magnetic resonance (NMR) characterization of an isolated HAMP domain from an archaeal, hyperthermophilic, transmembrane protein of unknown function [43] (Box 3). HAMP domains, characterized by two amphiphilic helices joined by a linker [44], occur in many bacterial signaling proteins [42,45]. Typically, HAMP domains are located between transmembrane and cytoplasmic signaling regions of known or putative transmembrane receptors, consistent with a role in converting ligand-induced conformational changes into kinase-controlling signals. In the recently solved HAMP structure each subunit contributes two amphiphilic helices (AS-1, AS-2), joined by a connector, to form a homodimeric, parallel, four-helix bundle [43]. The sequence locations, and the exposed and buried faces of these helices, are the same as identified previously by cysteine-scanning of an intact chemoreceptor [44], and the length of the domain is close to that deduced by analysis with electron microscopy [41]. Thus, at least in one signaling state, the chemoreceptor signal-conversion module might resemble the HAMP NMR structure, which could connect to adjoining receptor modules through extensions of its helical segments. However, the proposed HAMP architecture and its proposed connections to other modules have yet to be tested in a native, full-length chemoreceptor.
Chemoreceptor structure appears to be conserved across the diversity of bacteria and archaea. For instance, the kinase control modules of chemoreceptors from E. coli and from the evolutionarily distant species, Thermotoga maritima, are both anti-parallel, four-helix bundles formed by the dimerization of helical hairpins [25,34]. Impressively, recent analysis of >2000 kinase control module sequences from ~150 species revealed an orderly pattern of sequence relatedness [7]. Seven major receptor classes were distinguished by pairs of heptad (seven-residue) insertions or deletions symmetrically placed on each side of the membrane-distal hairpin turn, thus preserving register and packing of the coiled-coil. The analysis identified the protein-interaction region (Box 3)asfour paired, highly conserved heptads bracketing the membrane-distal hairpin turn. It identified the adaptation region as 8–10 paired heptads adjacent to the HAMP module. In addition it suggested the existence of a ‘flexible bundle’ region of 5–8 paired heptads between the protein-interaction and adaptation regions. The flexible bundle region appears to contain two separate heptad sets, with knobs-into-holes packing skewed in one direction in one set and in the other direction in the other set. The two sets center around glycine residues conserved among chemoreceptors and postulated to be a ‘glycine hinge’ important for on–off signaling [7,46] (Box 3, Figure II). The skewed sets of heptads are well placed to facilitate bending of the four-helix bundle at the hinge. The two available X-ray structures of chemoreceptor cytoplasmic fragments, from two distantly related species, are indeed bent at the position of the glycine hinge [25,34,46], and substitution of larger residues for hinge glycines appears to lock the receptor in the on- or off-state [46]. Finally, in vivo studies of fluorescently-tagged receptors showed that dimers move closer together on repellent stimulation [47] and further apart on attractant binding [48], movements that could depend on bending at the glycine hinge.
Figure II.
The glycine hinge of the kinase control module. (a) Atomic model of the kinase control module [25] illustrating the supercoiling of the two subunits (light and dark blue) and the location of the glycine hinge (box) that allows the four-helix bundle to bend ~10° [46]. (b) An expanded view of the hinge shows the helical backbones and highlights the six glycines of the hinge, residues G340, G341 and G439 in each subunit.
Chemoreceptor homodimers: conformational signaling
Chemoreceptors couple ligand recognition to kinase control, linking the ends of the extended protein by conformational, transmembrane signaling. In most respects, the receptor can be considered a two-state, on–off switch in which different inputs shift the equilibrium between two signaling states by triggering conformational changes in the three modules (Boxes 1 and 3).
Signaling in the transmembrane sensing module
The conformational signal induced by attractant binding in the transmembrane sensing module has been demonstrated by multiple, independent lines of evidence to be a piston-like sliding of the signaling helix towards the cytoplasm [49]. Increased receptor methylation in the kinase control module reverses the functional consequences of attractant binding, mediating precise adaptation. A recent disulfide cross-linking study in vivo using cysteine reporter positions in the signaling helix near the periplasm-membrane boundary showed that this functional reversal corresponds to a physical reversal of the conformational change in the transmembrane sensing module [50]. Attractant binding at the periplasmic binding site and covalent modification in the cytoplasm appear to drive the signaling helix piston in opposite directions (Figure 3). These findings support the two-state equilibrium notion in which ligand binding and adaptational modification move the signaling helix between its two piston positions, shifting the equilibrium between kinase-off and kinase-on. The opposing piston displacements generated by attractant binding and adaptational modification provide a simple, structural explanation for the opposite effects of these inputs on kinase activity. The results also imply that the HAMP module converts signals bi-directionally.
Box 2 In vivo evidence for receptor trimers-of-dimers.
The X-ray crystal structure of most of the kinase control module of the E. coli serine receptor, Tsr [25], revealed that the helical hairpin tips of three kinase control modules can pack together in a trimer-of-dimers arrangement (Box 2. Figure I). The residues promoting dimer–dimer interactions in the trimer are highly conserved throughout the MCP super-family [7] and are identical in the five E. coli receptors. These structural features suggested that trimer formation might be important for receptor function and that E. coli receptors of different types might form mixed trimers-of-dimers in vivo. Two cysteine-directed cross-linking approaches, guided by the Tsr crystal structure, have provided strong support for both of these ideas [3,26,62].
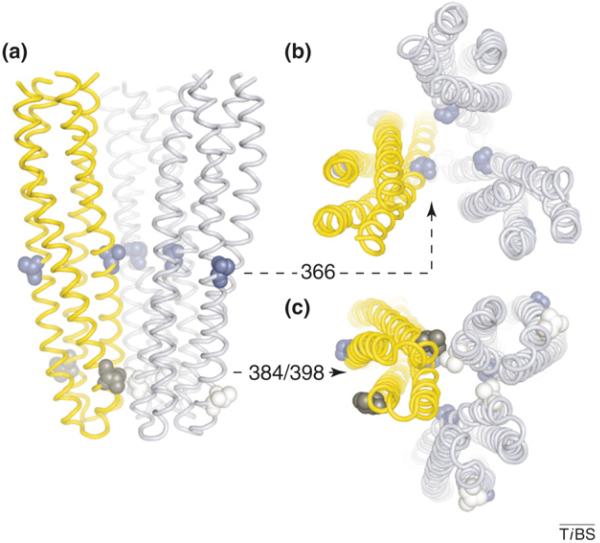
In one approach, cysteines were introduced into Tsr and the aspartate receptor (Tar) at residues predicted to interact closely only in the trimer-of-dimers structure, and not in dimers (see residues 384 and 398 in Figure I). The cysteine-substituted receptors, which retained normal signaling function, were expressed separately and together in cells under conditions that promoted disulfide formation in the cytoplasm. Because Tar and Tsr subunits do not form heterodimers [63], disulfide bonds between the receptors must form between dimers. As predicted by the trimer structure, Tsr molecules with one reporter cysteine efficiently formed disulfide cross-links with Tar molecules bearing the other cysteine, whereas neither cross-linked efficiently to itself.
A second cross-linking test for in vivo trimer formation used a cysteine reporter just membrane proximal to the putative trimer contact region (see residues 366 in Figure I). The trimer structure predicts that the cysteine in the inward-facing subunits of the dimers would have a trigonal spacing matching that of the reactive groups in TMEA [tris-(2-maleimidoethyl)amide], a commercially available tri-functional thiol-reactive cross-linking reagent. Indeed, TMEA treatments produced two- and three-subunit cross-linked products consistent with capture of the axial subunits in trimers-of-dimers [26]. Moreover, cells containing two different receptors with this cysteine replacement yielded mixed TMEA cross-linked products in proportions that reflected the relative abundance of each receptor type in the population.
Amino acid substitutions within receptors, particularly proline replacements, at principal trimer contact residues abolished both trimer-diagnostic signals, demonstrating that the cross-linking products probably form through specific structural interactions, rather than random collisional encounters.
Box 3. Functional architecture of the chemoreceptor dimer.
The receptor dimer contains three operational modules: transmembrane sensing, signal conversion and kinase control, each defined by distinct structural and functional features (Figure I). The ‘transmembrane sensing module’, a combination of the traditional periplasmic and transmembrane domains, is a dimer of two anti-parallel four-helix bundles in which two helices from each bundle extend across the membrane creating a membrane-spanning, four-helix bundle [49,51,64]. The ligand binds at periplasmic sites in the interface between subunits. The module signals attractant occupancy by a piston-like sliding of one helix toward the cytoplasm [49]. This signaling helix extends from the ligand-binding site across the membrane and connects to the ‘signal conversion’ module. This module is a HAMP (present in histidine kinases, adenylyl kinases, methyl-accepting chemotaxis proteins and phosphatases [42]) domain and the signaling helix connects specifically to its N-terminal (AS-1) helix. The parallel four-helix structure shown is a hypothetical model, based on a recent structure of a HAMP fragment from an atypical, archaeal protein of unknown function [43], which has not yet been tested in a full-length chemoreceptor.
The ‘kinase control module’ is a continuous four-helix, anti-parallel coiled-coil containing two helices from each subunit, with a hairpin turn at its membrane-distal end. The adaptation region contains sites of adaptational modification, specifically surface glutamates, or glutamines deamidated to become glutamates, located midway along the ~250Å coiled-coil (Figure I). A flexible region contains a conserved glycine hinge that enables the four-helix bundle to bend [46], as illustrated by the crystal structure of the Tsr kinase control module [25] (Figure II). The hinge consists of six glycine residues in a plane transecting the bundle, allowing the long axis of the bundle to bend ~10°. Substitution of the larger alanine side-chain at Gly 340 locked the receptor in the kinase-activating state, whereas alanine substitution at Gly 341 or 439 inhibited kinase docking or activation [46], suggesting that the hinge is crucial for ‘on–off’ switching, and perhaps also for kinase docking. A region of high sequence conservation bracketing the hairpin turn, termed the protein interaction region (Figure I) directly binds and regulates CheA kinase. Conformational signals in the kinase control module are transmitted by rearrangements of helix–helix contacts at the subunit interface (see text for details). The carboxyl terminus of some receptors carries a conserved pentapeptide sequence (NWETF or NWESF in the single-letter amino acid code) that interacts with the two enzymes of adaptational modification (CheR and CheB) and enhances the rates of the reactions they catalyze [69,70].
The sequence and 3D structure of kinase control modules are conserved across the diversity of bacteria and archaea [7]. Sequence conservation is more subtle among HAMP modules [42,45] but 3D structure is probably shared for the approximately two-thirds of all chemoreceptors that contain the module [43]. There is substantial variation in the sequence and in the deduced 3D organization among transmembrane sensing modules, but the anti-parallel four-helix bundle seems to be the most common structure [65] and this structure can be conserved even with minimal sequence identity [66].
Figure 3.
Conformational signaling in chemoreceptor dimers. The left-hand schematic, labeled ’Attractant response’ shows the conformational changes that convey an informational signal from one end of the chemoreceptor to the other. The right-hand schematic, labeled ’Sensory adaptation’, shows how covalent modification of an attractant-occupied receptor (methylation of specific glutamyl residues) reverses the ligand-induced conformational changes and thus mediates sensory adaptation. For clarity, a single receptor dimer is shown, embedded in the cytoplasmic membrane (light gray rectangle). As seen in the left-hand schematic, binding of an attractant chemoeffector (gray circle) to the transmembrane sensing module (green) initiates downward displacement of the signaling helix (see arrow heads on the signaling helix of the transmembrane sensing module) [49]. This movement generates an as-yet undefined conformational change in the signal conversion module (gray), which shifts its signaling state. This shift in turn weakens subunit interactions (symbolized by heavy arrows pointing away from the interface of subunit interaction) in the kinase control module (blue), resulting in increased flexibility, twisting and/or bending of the receptor molecule [46,56–58]. These changes, either directly or through effects on the trimer [47,48,60], deactivate coupled CheA kinase molecules leading to a counter-clockwise motor response. The same conformational changes also increase the propensity of the kinase control module for methylation and decrease its propensity for demethylation. The ligand-induced kinase inhibition decreases the level of active, phosphorylated demethylating enzyme CheB, thereby further increasing the number of methylated adaptation sites (black circles in the kinase control domain) at the expense of demethylated adaptation sites (white circles in the kinase control domain). Increased methylation terminates the motor response by reversing the attractant-triggered conformational changes [50]. Methylation strengthens subunit interactions (symbolized by heavy arrows pointing toward the interface of subunit interaction) in the kinase control domain, reducing its flexibility and activating coupled CheA kinases [58]. The reduced dynamic motion and strengthened subunit interactions of the kinase control domain also reverse the signaling state of the signal conversion module and thus cause upward movement of the signaling helix, reversing the conformational change of the transmembrane sensing module and influencing the conformation of the ligand-binding site [50]. Overall, the effects of increased methylation counteract the conformational and functional effects of ligand occupancy, re-establishing the conformational and signaling state of the chemoreceptor before ligand occupancy.
If the model of a two-state, piston-sliding equilibrium is valid, then artificial means of moving the signaling helix should have predictable consequences on kinase activation and propensity for adaptational modification. Such experiments have been done by introducing or moving charged or aromatic residues near the periplasmic and cytoplasmic membrane–water interfaces of the signaling helix with the aim of creating electrostatic or hydrophobic forces that would slide the helix along its long axis perpendicular to the membrane [51–53]. In support of the piston mechanism, substitutions expected to drive the signaling helix towards the cytoplasm shifted the receptor to the off-state of lower kinase activation and higher methylation rate, whereas substitutions driving movement towards the periplasm shifted the receptor to the on-state, displaying higher kinase activation and lower methylation rate [51–53]. These results indicate that native residues near the membrane–water interface in the signaling helix have an important role in modulating receptor on–off bias by stabilizing that helix in a proper transmembrane register. Analogous approaches should be useful in determining whether piston movements mediate signaling in other receptors.
Signaling in the signal conversion module
In chemoreceptors, the HAMP signal conversion module interconverts (i) attractant-induced helix sliding in the transmembrane sensing module and (ii) a different conformational change that alters the subunit interface of the kinase control module. The recently determined structure of an isolated, archaeal HAMP domain [43] appears well-suited to mediate conformational conversions (Box 3, Figure I), although the protein containing this HAMP domain lacks a kinase control module. Piston movements of HAMP helix AS-1, which abuts the transmembrane signaling helix, could easily perturb the packing interface of the HAMP four-helix bundle to shift its conformation. Altered HAMP packing might in turn shift the AS-2 helices, thereby altering the subunit interface of the adjoining kinase control module. Conversely, a shift in the stability of the subunit interface of that module, induced by a methylation change, would trigger corresponding conformational changes in the HAMP domain. Whatever the details of conformational signaling in the signal conversion module, the movements must not involve large magnitude helical displacements because attractant signals can be transmitted through modules constrained by disulfides across the AS-2/AS-2′ or adjacent subunit interface [44,54].
Figure I.
The chemoreceptor dimer. A ribbon diagram and a schematic show the 3D organization of a chemoreceptor dimer from E. coli. Modules are indicated on the left, roles or identities of module segments in the middle and notable features on the right. The model is based on the shape of an intact, membrane-embedded receptor revealed by electron microscopy [41], the X-ray structure of a periplasmic fragment [67], the X-ray structure of a cytoplasmic fragment [25], patterns of disulfide formation between introduced cysteines [49,57,68] and a model of the signal conversion module based on an NMR structure of a homologous HAMP domain [43].
Signaling in the kinase control module
The mechanism of conformational signaling in the kinase control module is less clear than in the trans-membrane sensing module, in part because the kinase control module is significantly more dynamic [25,55]. However, considerable evidence indicates that changes at the subunit–subunit interface within the kinase control module are important in signaling [56–58]. This contrasts with the transmembrane sensing domain, in which signaling movements occur between the two helices in a single subunit and the subunit interface is static [49]. In the otherwise dynamic kinase control module, stabilization of inter-subunit packing by disulfide bonds or amino acid replacements can lock the receptor to a kinase-on state, implying that attractant-generated, kinase-inhibiting signals shift or destabilize the subunit interface via mechanical forces [54,57]. In addition, the adaptation region is highly anionic and is regulated by an electrostatic mechanism involving the adaptation site glutamates and several other anionic side-chains lining the subunit–subunit interface [58]. Covalent neutralization of an interfacial anion by methyl esterification or amidation stabilizes the interface and activates kinase, whereas simultaneous neutralization of all anions locks the receptor in the kinase-activating state [58] Conformational signaling in the protein interaction region, where the kinase interacts, remains poorly understood. However, like the adaptation region, its subunit interface seems to be crucial for signal transmission because many ‘lock-on’ mutations occur at interfacial locations [54,59].
Macroscopic transitions in the trimer-of-dimers
Two types of inter-dimer motions have been proposed to occur during on–off switching within the trimer-of-dimers. One is rotational movement of the periplasmic segment of each dimer about its long axis, detected by modest changes in rates of inter-dimer disulfide formation [60]. The other is tilting of dimers relative to the central trimer axis, detected by changes in dimer-to-dimer FRET [47,48]. In principle, such macroscopic transitions could be driven by changes in supercoiling, or supercoiling-dependent flexibility of the four-helix bundle of the kinase control module [46,58]. By triggering changes in inter-dimer distances or contacts, macroscopic transitions could have an important role in the strong positive signaling cooperativity between dimers, within or between trimers.
Concluding remarks and future perspectives
The high-performance features of bacterial chemoreceptors emerge from several levels of macromolecular organization. The core receptor structural unit, the homodimer, performs many essential functions, but multiple dimers, probably at least three, are required to enable receptors to control kinase activity. Trimers-of-dimers provide the first level of higher-order organization; the full spectrum of high-performance operation involves interactions of tens of dimers. We do not yet understand the mechanisms by which tens, much less thousands, of receptors localize in patches or the means by which those receptors communicate with one another. Receptor patches are probably built by interactions among trimers, but we do not know how this occurs. Interestingly, signaling within a single chemoreceptor dimer involves distinct, bidirectional conformational changes in its three modules. These intradimer changes might trigger changes in the trimer, but these effects are not yet fully defined.
To make additional progress we need deeper molecular insights at each organizational level. For the individual receptor dimer, we need a better understanding of signaling-related conformational changes, particularly in the signal conversion and kinase control modules where the structural and dynamic details of on–off switching remain mysterious. Until high-resolution structures are available for these modules in their on- and off-states, in vivo and in vitro mutational, biochemical and spectroscopic approaches should provide important clues.
For trimers-of-dimers, we need to understand how conformational changes in individual receptor dimers influence structural relationships between members of trimer-based signaling teams. Do trimers expand and contract in response to chemotactic stimuli? Do all of their component dimers behave in the same way? In vivo and in vitro cross-linking and spectroscopic studies could provide answers to these questions. We also need to define the minimal receptor signaling unit: how many receptor, CheA, and CheW molecules compose a functional signaling team? Where are crucial interaction determinants located on each component? Studies of receptors in Nanodiscs offer considerable promise for answering these questions.
Finally, we need to understand the architecture of receptor arrays: what causes receptors to form patches? How are component signaling units interconnected? Answers should come through light, atomic force, and electron microscopy, probably in combination with innovative methods for tagging signaling components. The pursuit of these issues will keep the study of bacterial chemoreceptors at the forefront of molecular research in biological signaling.
Dedication
We dedicate this article to the memory of Daniel E. Kosh-land, Jr, a central figure in the field of biochemistry who studied bacterial chemotaxis for thirty years. Dr Koshland passed away July 23, 2007.
Figure I.
The chemoreceptor signaling pathway in E. coli. Components and reactions in red promote counter clockwise (CCW) flagellar rotation; those in green promote clockwise (CW) flagellar rotation. Components in gray represent inactive forms. Solid lines represent enzymatic reactions; broken lines indicate binding interactions. CheA-derived phosphoryl groups are shown as blue spheres. Receptor modification sites are shown as white (unmethylated) and black (methylated) circles.
Figure I.
Diagnostic cross-linking sites for the trimer-of-dimer organization. (a) Backbone traces of receptor signaling tips in a mixed trimer-of-dimers containing one Tsr molecule (yellow) and two Tar molecules (light blue). Residues chosen for cysteine reporter replacements: V384 (white, Tar), V398 (black, Tsr), S366 (blue, Tar and Tsr). (b) View down the trimer axis looking towards the tip. Note the trigonal arrangement of residues at position 366 in the inward-facing subunits of the dimers. (c) View of the trimer tip looking towards the membrane. Note the proximity of Tar residue 384 and Tsr residue 398 at a dimer–dimer interface in the trimer.
Acknowledgements
Because of space constraints it was not possible to discuss or cite all research papers relevant to the subject. We thank S. Subramaniam for original versions of the components of Figure 1 and W.C. Lai for assistance with the manuscript. Work in the authors’ laboratories was supported by NIH grants GM29963 (G.L.H.), GM40731 (J.J.F.) and GM19559 (J.S.P.).
References
- 1.Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 2.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson JS, et al. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Baker MD, et al. Systems biology of bacterial chemotaxis. Curr. Opin. Microbiol. 2006;9:187–192. doi: 10.1016/j.mib.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 7.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr. Opin. Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Segall JE, et al. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 12.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 13.Bornhorst JA, Falke JJ. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: development of a heterogeneous two-state model. J. Mol. Biol. 2003;326:1597–1614. doi: 10.1016/s0022-2836(03)00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai R-Z, et al. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol. Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 16.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endres RG, et al. Chemotaxis receptor complexes: from signaling to assembly. PLoS Comput. Biol. 2007;3:e150. doi: 10.1371/journal.pcbi.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 19.Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P, et al. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiem S, et al. Positioning of chemosensory clusters in E. coli and its relation to cell division. EMBO J. 2007;26:1615–1623. doi: 10.1038/sj.emboj.7601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kentner D, et al. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 23.Lybarger SR, et al. Clustering requires modified methyl-accepting sites in low-abundance but not high-abundance chemoreceptors of Escherichia coli. Mol. Microbiol. 2005;56:1078–1086. doi: 10.1111/j.1365-2958.2005.04593.x. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim KK, et al. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 26.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ames P, et al. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ames P, Parkinson JS. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9292–9297. doi: 10.1073/pnas.0602135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asinas AE, Weis RM. Competitive and cooperative interactions in receptor signaling complexes. J. Biol. Chem. 2006;281:30512–30523. doi: 10.1074/jbc.M606267200. [DOI] [PubMed] [Google Scholar]
- 31.Levit MN, et al. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem. 2002;277:36748–36754. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu TS, et al. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 33.Gestwicki JE, et al. Evolutionary conservation of methyl-accepting chemotaxis protein location in bacteria and archaea. J. Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S-Y, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 35.Miller AS, et al. CheA Kinase of bacterial chemotaxis: chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francis NR, et al. Three-dimensional structure and organization of a receptor/signaling complex. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17480–17485. doi: 10.1073/pnas.0407826101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolanin PM, et al. Self-assembly of receptor/signaling complexes in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14313–14318. doi: 10.1073/pnas.0606350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boldog T, et al. Using nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 39.Nath A, et al. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 40.Boldog T, et al. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weis RM, et al. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 2003;185:3636–3643. doi: 10.1128/JB.185.12.3636-3643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 43.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 44.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams SB, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 1999;33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 46.Coleman MD, et al. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaknin A, Berg HC. Osmotic stress mechanically perturbs chemoreceptors in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2006;103:592–596. doi: 10.1073/pnas.0510047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai W-C, et al. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol. Microbiol. 2006;61:1081–1090. doi: 10.1111/j.1365-2958.2006.05296.x. [DOI] [PubMed] [Google Scholar]
- 51.Miller AS, Falke JJ. Side chains at the membrane-water interface modulate the signaling state of a transmembrane receptor. Biochemistry. 2004;43:1763–1770. doi: 10.1021/bi0360206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Draheim RR, et al. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 53.Draheim RR, et al. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry. 2005;44:1268–1277. doi: 10.1021/bi048969d. [DOI] [PubMed] [Google Scholar]
- 54.Bass RB, Falke JJ. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Structure. 1999;7:829–840. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seeley SK, et al. The cytoplasmic fragment of the aspartate receptor displays globally dynamic behavior. Biochemistry. 1996;35:5199–5206. doi: 10.1021/bi9524979. [DOI] [PubMed] [Google Scholar]
- 56.Bass RB, et al. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surface: cysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winston SE, et al. Evidence that the adaptation region of the aspartate receptor is a dynamic four-helix bundle: cysteine and disulfide scanning studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: Electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bass RB, Falke JJ. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J. Biol. Chem. 1998;273:25006–25014. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irieda H, et al. Control of chemotactic signal gain via modulation of a pre-formed receptor array. J. Biol. Chem. 2006;281:23880–23886. doi: 10.1074/jbc.M600018200. [DOI] [PubMed] [Google Scholar]
- 61.Mello BA, Tu Y. Effects of adaptation in maintaining high sensitivity over a wide range of backgrounds for Escherichia coli chemotaxis. Biophys. J. 2007;92:2329–2337. doi: 10.1529/biophysj.106.097808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studdert CA, Parkinson JS. In vivo crosslinking methods for analyzing the assembly and architecture of chemoreceptor arrays. Methods Enzymol. 2007;423:414–431. doi: 10.1016/S0076-6879(07)23019-8. [DOI] [PubMed] [Google Scholar]
- 63.Milligan DL, Koshland DE., Jr Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 64.Boldog T, Hazelbauer GL. Accessibility of introduced cysteines in chemoreceptor transmembrane helices reveals boundaries interior to bracketing charged residues. Protein Sci. 2004;13:1466–1475. doi: 10.1110/ps.04648604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ulrich LE, Zhulin IB. Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics. 2005;21(Suppl 3):iii45–iii48. doi: 10.1093/bioinformatics/bti1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai W-C, et al. Diagnostic cross-linking of paired cysteine pairs demonstrates homologous structures for two chemoreceptor domains with low sequence identity. Protein Sci. 2006;15:94–101. doi: 10.1110/ps.051802806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milburn MV, et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 68.Peach ML, et al. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 2002;11:912–923. doi: 10.1110/ps.4640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J, et al. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 70.Barnakov A, et al. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]