Abstract
The neuronally expressed Gβ5 subunit is the most structurally divergent among heterotrimeric Gβ isoforms and unique in its ability to heterodimerize with the R7 subfamily of regulator of G protein signaling (RGS) proteins. The complex between Gβ5 and R7-type RGS proteins targets the cell nucleus by an unknown mechanism. Although the nuclear targeting of the Gβ5/R7-RGS complex is proposed to involve the binding of R7-binding protein (R7BP), this theory is challenged by the observations that endogenous R7BP is palmitoylated, co-localizes strongly with the plasma membrane, and has never been identified in the cytosol or nucleus of native neurons or untreated cultured cells. We show here mutant RGS7 lacking the N-terminal Disheveled, EGL-10, Pleckstrin homology (DEP) domain is expressed in transfected cells but, unlike wild type RGS7, is excluded from the cell nucleus. Because the DEP domain is essential for R7BP binding to RGS7, we studied the subcellular localization of Gβ5 in primary neurons and brain from mice deficient in R7BP. The level of endogenous nuclear Gβ5 and RGS7 in neurons and brains from R7BP KO mice is reduced by 50 to 70%. These results suggest that R7BP contributes significantly to the nuclear localization of endogenous Gβ5/R7-RGS complex in brain.
Keywords: heterotrimeric G protein, regulator of G protein signaling, R7BP, DEP domain, subcellular localization, palmitoylation
INTRODUCTION
The R7 subfamily of regulator of G protein signaling (RGS) proteins is differentially expressed in the nervous and visual systems where the timing of signaling events is critical for motor, sensory and autonomic function (Anderson et al. 2009b, Arshavsky et al. 2002). The R7-RGS subfamily in mammals consists of four principal gene products and their splice variants: RGS6, RGS7, RGS9 and RGS11. The members of the R7-RGS subfamily have a common protein domain structure (cf. Fig. 1A). The presence of multiple proteins domains in R7-RGS proteins allows specific protein-protein interactions and the organization of distinct multi-protein complexes to mediate essential R7-RGS functions in the nervous system and eye (Anderson et al. 2009b, Jayaraman et al. 2009). Like other members of the larger RGS protein family, R7-RGS proteins contain a core RGS homology domain that can act as a GTPase activating protein (GAP) for heterotrimeric G protein α subunits to rapidly terminate G protein signaling (Ross & Wilkie 2000). Another domain in R7-RGS proteins is the Gγ-like (GGL) domain (Snow et al. 1998) that provides an interface for tight and obligatory heterodimerization with G protein β5, structurally analogous to the interface between Gγ subunits and the four other Gβ isoforms (Cheever et al. 2008). In the absence of Gβ5 all R7-RGS proteins are unstable and proteolytically degraded (Chen et al. 2003).
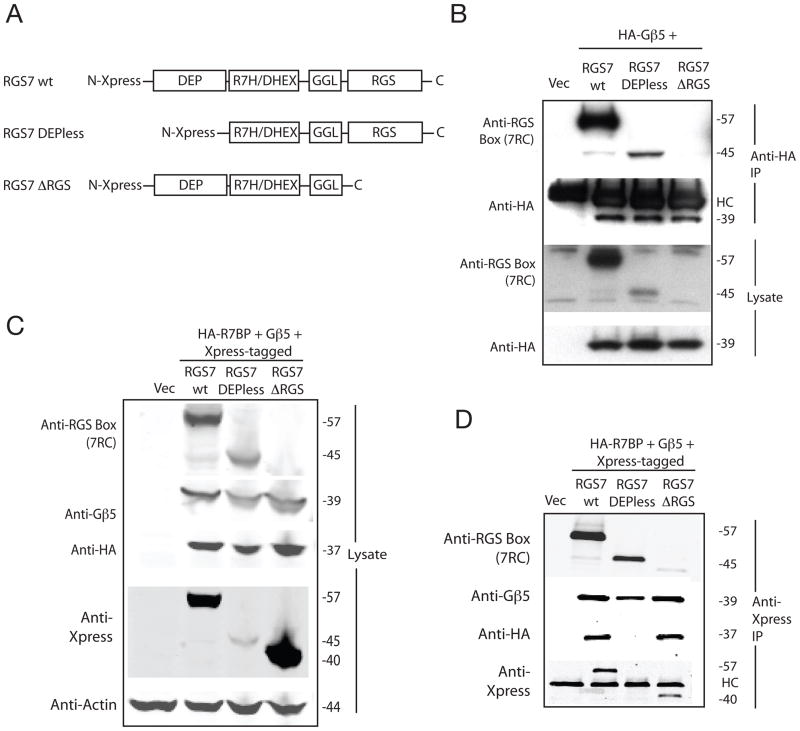
Figure 1. R7BP binds to wild type RGS7 but not Depless-RGS7.
(A) Schematic diagram showing the domain structures of wild-type RGS7 and the DEPless and ΔRGS RGS7 mutants used in the present study. DEP, Disheveled, EGL-10, Pleckstrin homology domain; R7H/DHEX, R7 homology domain also called DEP helical extension domain or interdomain; GGL, G-gamma-like domain; RGS, regulator of G protein signaling core homology domain (RGS box). For B, C, and D Total PC12 cell lysates were analyzed by immunoblotting for the expression of Gβ5, RGS7, RGS7 mutants and R7BP before (Lysate) and after immunoprecipitation (IP) using as shown. (B) Cells transfected with vector only, or HA- Gβ5 with RGS7 wt, Depless-RGS7 or ΔRGS-RGS7, and subsequent IP with anti-HA monoclonal antibody (C) and (D) Cells transfected with vector only, or HA-R7BP and Gβ5 with Xpress-tagged RGS7 wt, Depless-RGS7, or ΔRGS-RGS7, and subsequent IP with anti-Xpress monoclonal antibody. In B and D, HC indicates immunoglobulin heavy chain, of relative mobility ~50 kDa, present in the immunoprecipitate.
R7-RGS proteins contain two adjacent N-terminal protein domains that mediate protein-protein interactions critical for membrane anchoring: the Disheveled, EGL-10, Pleckstrin homology (DEP) domain and the DEP helical extension (DHEX) domain (Cheever et al. 2008) (also known as the R7 homology (R7H) domain (Anderson et al. 2007b)) together provide a binding surface for R9 anchoring protein (R9AP) (Hu & Wensel 2002) or R7 binding protein (R7BP) (Martemyanov et al. 2005, Drenan et al. 2005). All R7-RGS proteins can bind to R7BP but only RGS9 and RGS11 can bind R9AP (Martemyanov et al. 2005, Drenan et al. 2005, Cao et al. 2009, Song et al. 2007).
R9AP and R7BP have different mechanisms for their membrane attachment. While R9AP possesses a hydrophobic single-pass transmembrane helical domain near its C-terminus, R7BP is membrane anchored through the cooperation of hydrophobic interaction, via post-translational palmitoylation of one or two vicinal cysteines near the R7BP C-terminus, with electrostatic bonding involving a cluster of basic residues just upstream of the acylation sites (Drenan et al. 2005, Song et al. 2006). The membrane anchoring of R7-RGS/Gβ5 complexes via R7BP or R9AP is critical for termination by R7-RGS proteins of signals carried by G proteins from upstream GPCRs (Hu et al. 2003, Drenan et al. 2006).
There is conflicting evidence in the literature regarding the subcellular localization of endogenous R7-RGS/Gβ5 complexes in brain and neurons. Evidence was originally presented that endogenous RGS7/Gβ5 complexes localized to membrane and cytosolic fractions in rat and mouse brains (Witherow et al. 2000, Zhang et al. 2001). In addition endogenous RGS7/Gβ5 complexes were reported in the nuclear fraction of mouse brain and naive neuron-like PC12 cells (Zhang et al. 2001). This was in contrast to the bulk of Gβγ complexes, whose expression is restricted to the plasma membrane (though some Gβγ complexes are reported to target and/or function at intracellular membranes (Denker et al. 1996, Jamora et al. 1997)). Subsequent studies have corroborated the presence of endogenous Gβ5 and RGS7 in the cytosol and membranes of mouse brain using Gβ5 knockout (KO) mice as a negative control (Grabowska et al. 2008). A recent analysis however has suggested that the Gβ5 and RGS7 present in the nuclear fraction of mouse striatum may represent a contamination artefact (Mancuso et al. 2009). With respect to RGS9-2, studies in rat brain cortex and striatum employing immunohistofluorescence and subcellular fractionation concluded that RGS9-2 was localized primarily to the cytoplasm and nucleus (Bouhamdan et al. 2004). In contrast, subsequent investigations of RGS9-2 subcellular localization in mouse brain employing differential centrifugation, sucrose density gradient fractionation and electron microscopic analysis concluded that RGS9-2 is primarily localized to the plasma membrane (Song et al. 2006, Mancuso et al. 2009, Anderson et al. 2007a).
In the face of this disparate evidence, the current model proposes that R7BP is governs the distribution of the Gβ5/R7-RGS complex between the plasma membrane and the cell nucleus (Drenan et al. 2005, Song et al. 2006, Hepler 2005). This model is based on evidence from studies in cultured cells that mutational or chemical de-palmitoylation of R7BP promotes the nuclear targeting of a Gβ5/R7-RGS/R7BP complex mediated by a polybasic amino acid cluster in R7BP resembling a nuclear localization signal (NLS) (Drenan et al. 2005, Song et al. 2006, Drenan et al. 2006). But this model sidesteps the fact that in brain endogenous R7BP localizes strongly to the plasma membrane fraction (Song et al. 2006, Anderson et al. 2007a, Grabowska et al. 2008). Furthermore experiments with strong anti-R7BP antibodies could not detect R7BP in neuronal cell nuclei from multiple brain regions, nor identify R7BP associated with brain cytosolic Gβ5/RGS7, even though membrane-associated R7BP was readily demonstrable (Grabowska et al. 2008). Thus no evidence is currently available that an endogenous Gβ5/R7-RGS/R7BP complex traverses the cytoplasm or targets the cell nucleus.
To reexamine this issue we employed native primary neurons from R7BP KO mice and studied the properties of an R7BP binding-deficient RGS7 mutant in cultured cells. We report here that the nuclear targeting of the Gβ5/RGS7 signalling complex in transfected neuronal cell lines is blocked upon deletion of the RGS7 DEP domain that is essential for R7BP binding. Furthermore Gβ5/RGS7 nuclear targeting is greatly reduced in primary neurons and brains from R7BP-deficient mice. Plasma membrane localization of RGS7 was also partially reduced in certain brain subregions of the R7BP KO mice. These results demonstrate that the endogenous Gβ5/R7-RGS complex in brain depends on R7BP for proper nuclear and plasma membrane targeting.
EXPERIMENTAL PROCEDURES
Mammalian cDNA expression constructs
Complementary cDNAs encoding Gβ5 (in pCDNA3), HA epitope-tagged Gβ5 (in pCDNA3) and Xpress epitope-tagged bovine RGS7 (in pcDNA4/HisMax-C) were previously described (Rojkova et al. 2003). The Xpress epitope-tagged mutants DEPless RGS7 (in pcDNA4/HisMax-C) (comprising residues Pro-117 through Tyr-469 of wild-type bovine RGS7) and ΔRGS-RGS7 (in pcDNA4/HisMax-C) (comprising residues Ala-2 through Ser-319 of wild-type bovine RGS7) were prepared using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene). HA-tagged wild-type R7BP in pcDNA3 was prepared previously (Nini et al. 2007). The coding region of all epitope-tagged and mutant cDNAs was confirmed by DNA sequencing.
Mouse husbandry and genotyping
Generation of R7BP KO mice with heterozygous and homozygous deletion of the first 2 exons of R7bp in the germline was previously described (Anderson et al. 2007a). Mice were housed and treated in strict accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals. Mice were maintained in a pathogen-free facility, with 4–5 animals per cage in a temperature-controlled room with a 12-h light/dark cycle and access to food and water ad libitum. Age and sex matched littermate cohorts were used for all the experiments to account for potential strain differences. Mouse genotyping was performed by QPCR analysis of genomic DNA extracted from mouse tails using the DirectPCR (tail) solution (Viagen, Cat# 102-T) according to manufacturer’s instructions. The knock-out allele was identified employing the primer pair, forward (5′-CTGCAAGCCAGTAGTGCCAGTCCC-3′) and reverse (5′-GGAACTTCGCTAGACTAGTACGCGTG-3′). The wild type allele was identified employing the same forward primer as above with the reverse primer (5′-TGTTCTTAGTGTGATCGAGTGATATTGG-3′). PCR conditions used were 95°C for 10 min followed by 40 cycles of 95°C 30 sec, 57°C for 1 min and 72°C for 1 min. The specificity of the PCR products was evaluated by one cycle of dissociation curve analysis (95°C for 1 min, 55°C for 30 sec and 95°C for 30 sec).
Cell Culture
Rat pheochromocytoma PC12 cells (ATCC, Manassas, VA), human neuroblastoma SH-SY5Y cells (a kind gift from Dr. Jeffrey M. Brown, Department of Pharmacology, Uniformed Services University of the Health Sciences) and mouse Neuro-2a cells (a gift from Dr. Hong Lou, Section on Cellular Neurobiology, NICHD) were grown in 75 cm2 flasks in DMEM supplemented with 10% fetal bovine serum, 4mM L-glutamine and penicillin/streptomycin at 37° C and 5 % CO2. Empty vector or expression plasmids were transfected using Amaxa Nucleofector (solution V). The expression of the transfected proteins were verified by immunoblotting and/or immunofluorescence.
Mouse primary cortical neuron cultures were prepared as follows. Briefly, cortical tissues of previously genotyped 2 to 3 day-old mouse pups were placed into 1 ml of NeuroPrep medium (Gelantis, Cat. NM100100) containing NeuroPapain (Genlantis, Cat. NM100200, 1 mg/ml) in a 15 ml centrifuge tube. After incubation for 20 min at 30° C, the papain solution was replaced with 1 ml fresh Hibernate A-Ca medium (Neuromics, Cat. M36102). Neurons were dissociated by pipetting the papain-treated tissue up and down 20 times. The neuron suspension was filtered through a 40 μM nylon cell strainer (Thomas Sci., Cat. 4620E99) into a six-well plate and rinsed once with 1 ml of fresh Hibernate A-Ca medium. The neuron suspension (now 2 ml total) was transferred into a new 15 ml centrifuge tube and centrifuged at 170 × g for 3 min. The supernatant was removed and the cell pellet resuspended in 1 ml of Neurobasal™ -A (Invitrogen, Cat. 10888-022) medium containing B-27 Serum-Free Supplement (Invitrogen, Cat. 17504-044). After determination of the live cell count (using trypan blue and a hemocytometer), the cells were diluted using the same medium, spread onto plates freshly coated with Poly-D-lysine (Sigma P6407), and grown at 37° C and 5% CO2.
Immunofluorescence
PC12, SH-SY5Y and Neuro-2a cells were transfected as described above and plated in chambers of poly-D-lysine-coated chamber slides and incubated for 24h. Primary neuronal cells were isolated as described above and grown in chamber slides for 6 days. The culture medium was discarded and the cells were rinsed with PBS and fixed in 2% formaldehyde for 15 min. The slides were washed two times with PBS and incubated with PBS containing 8% FBS/azide (buffer A) for 5 min at room temperature. Then primary antibody was diluted in buffer A containing 0.2% saponin (buffer B) and incubated at room temperature for 1h. The slides were rinsed 3 times with buffer A and incubated with the secondary antibody (fluorescein Anti-rabbit or Anti-mouse IgG (H+L), Vector Laboratories, Burlingame, CA) diluted in buffer B and incubated for 1h at room temperature protected from light. The slides were washed 3 times with buffer A and then 2 times with PBS and removed chamber from the slides. Then added mounting media containing DAPI (Vectashield H-1200) and incubated for 5 minutes in order to provide nuclear counterstaining. Finally the slides were covered with a cover slip, sealed and kept at 4 degree till the time of confocal laser scanning microscopy.
Laser confocal scanning and intensity analysis
Images were acquired on a confocal microscope (Leica TCS-SP2 -Leica Microsystems GmbH, Mannheim, Germany) using a 60× oil immersion objective. Fluorescein was excited with an argon laser at 488nm and DAPI with an Argon laser (Enterprise model 651, Coherent Inc.) at 364nm. DIC images were acquired simultaneously using a transmitted light detector. Dyes were acquired in separate channels to minimize crosstalk. A stack of images were acquired using a step size of 0.7μm. Images were processed using Leica TCS-SP2 sofware, Imaris v6.2 (BItplane AG, Zurich Switzerland) and Adobe Photoshop CS3 (Adobe Systems).
For the quantification and analysis of nuclear Gβ5 or RGS7 in primary neurons, multiple images were acquired using 40 × oil immersion objective from different focal fields in the same slide, using the same laser power and gain. Intensity quantification was performed by rendering 3D volumes from the multi-channel Z-stacks using Imaris v6.2. To measure the fluorescein intensity within the nucleus of each cell, a 3D volume was created from the DAPI labeling and the mean intensity was calculated for the whole volume. From four sets of wild type and R7BP KO mice, 120 healthy neuronal cells from control and 144 healthy cells from R7BP KO mice were used for intensity analysis. Controls lacking primary antibody showed no labeling.
Immunoprecipitation
Immunoprecipitation with anti-HA or anti-Xpress antibody was performed according to the manufacturer’s instructions (Immunoprecipitation Starter Pack, cat. No. 309410, Amersham Biosciences). Briefly, after transfection and 24 h incubation, PC12 cells were lysed with buffer A (250 mM NaCl, 50 mM Tris, pH 8.0, 5 mM EDTA, 0.5 % NP-40 and 1 X complete protease inhibitor cocktail [cat. No. 1836170, Roche]). The lysate was centrifuged at 1500 × g for 10 min. The supernatant was incubated overnight with HA antibody at 4° C. The mixture was incubated with protein G-Sepharose previously equilibrated in buffer A for 2 hours at 4° C. The beads were then washed 3 times with buffer A by centrifugation at for 1 min, and the washes discarded. Proteins were eluted from the beads with SDS sample buffer and detected by immunoblotting.
Mouse brain nuclear-cytoplasmic fractionation
Nuclear complex Co-IP Kit (Active motif) was used for nuclear-cytoplasmic fractionation of mouse brain following the special protocol for fresh tissues with some modifications. Approximately 30 mg of fresh mouse brain cortex (3–4 week old mice) was homogenized in 10 volumes of cold hypotonic buffer containing protease inhibitors, detergent, phosphatase inhibitor and DTT using a manual glass tissue grinder (1ml) on ice (30 strokes). After incubation on ice for 15 minutes and centrifugation at 2000 rpm for 10 minutes, the supernatant (cytosol) was separated from the pellet. The pellet was resuspended in 500 μl of the homogenization buffer, incubated on ice for 15 minutes then, following the addition of 25 μl of detergent, vortexed for 10 seconds at a maximum speed and centrifuged at 13,000 rpm for 1 minute. This supernatant was added to the earlier one. The pellet (nuclear fraction) was re-suspended in 500 μl of homogenization buffer, centrifuged at 13,000 rpm for 1 minute, and the supernatant was discarded and this process was repeated for 2 more times. This washing process removed cytosolic proteins adhering to the tube as well as to the pellet. The nuclear pellet was dissolved in 225 μl of Laemmli’s 2X buffer and the cytosol was boiled with an equal volume of the Laemmli’s 2X buffer prior to western blot quantification analysis using the Odyssey infrared imaging system. For calculation of the relative cytosolic expression of Gβ5 or RGS7, the band intensities were divided by the tubulin band intensity form the same lane. Calculation of the nuclear expression of Gβ5, corrected for residual cytosolic contamination after washing (using tubulin as a marker for cytoplasmic proteins), was according the following formula: (raw nuclear Gβ5 – nuclear tubulin [cytoplasmic Gβ5/cytoplasmic tubulin])/nuclear histone H3. Calculation of the corrected nuclear expression of RGS7 followed a comparable formula.
Mouse brain plasma membrane-cytoplasmic fractionation
Striatum, hippocampus and approximately same amount of cortex were isolated from brains of C57Bl and R7BP knockout mice. Each sample was homogenized with a series of needles (18G, 23G, 25G) in 500 μl of PBS [1×PBS (Fisher Scientific), 150 mM NaCl, protease inhibitor cocktail (Roche), pH 7.4] and total protein concentration was measured using BSA kit (Pierce). Samples were diluted with PBS when needed to equalize concentrations and 1.5 mg (350 μl) of total protein was used for fractionation. Samples were centrifuged at 200 000 g for 20 min at 4° C to separate cytosol (supernatant) from membranes (pellet). Pellet was digested in 700 μl of Laemmli’s 2X buffer and sonicated to shear DNA. Equal amount of Laemmli’s 2X buffer was added to supernatant.
Immunoblotting, chemiluminescence and infrared imaging
Cell lysates were boiled with equal volume of Laemmli’s 2X gel loading buffer and the hot solution was loaded onto 4–20% Tris-Glycine SDS-PAGE gels (Invitrogen) to separate the proteins, followed by transfer of the proteins on to 0.45-micron nitrocellulose membrane. Membranes were blocked with TBS or PBS (pH 7.4) containing 0.1% Tween 20 and 5% nonfat dry milk (blocking buffer) and incubated overnight with primary antibodies in the same buffer. The membranes were then washed seven times for 5 minutes each with the above buffer without milk, followed by a 2-hour incubation in blocking buffer including appropriate horseradish peroxidase-conjugated secondary antibodies. Membranes were then washed as above, and the proteins detected by chemiluminescence on X-ray film using Super Signal West Dura Extended Duration Substrate (Pierce). For infrared imaging, IR-labeled secondary antibodies (dilution 1: 20,000) were used for detecting the protein signals in conjunction with the Odyssey infrared imaging system (LI-COR, Bioscience). Blocking and washing buffers used were same as described above, however the incubation time with the secondary antibody was half hour- one hour protected from light. For the quantification of the intensity of the protein bands membranes were dually probed, with the α-tubulin or histone signal used as loading controls.
Boiled fractions from mouse striatum, hippocampus and cortex were loaded onto in-house made 12.5% SDS-PAGE gels to separate proteins, followed by transfer of the proteins onto 0.45 μm Immobilon-FL PVDF membrane (Millipore). Membranes were blocked with Odyssey Blocking Buffer (LI-COR Bioscience) for 1 hour and incubated with primary antibody in the same buffer containing 0.1% Tween 20 for 1 hour. The membranes were then washed 3 times for 5 min each with 1 × PBS (pH 7.4) containing 0.1% Tween 20, followed by a 40 min incubation with IR-labeled secondary antibody (dilution 1:15,000 in Odyssey Blocking Buffer containing 0.1% Tween 20 and 0.01% SDS). Membranes then were washed as above followed by one 5 min wash with 1 × PBS. Protein signals were detected and densitometry performed using Odyssey infrared imaging system. Membranes were probed for RGS7 as well as for the plasma membrane markers Na+/K+ ATPase α1 and β2-subunits, which served as loading controls for the membrane fractions.
Antibodies
Antibodies used for immunoblots and immunoprecipitation were rabbit anti-Gβ5 polyclonal N-terminal antibody ATDG (Zhang & Simonds 2000), mouse anti-HA (Covance), mouse anti-α-tubulin (Calibiochem CP06), rabbit anti –Histone H3 (Abcam-Ab 8580), rabbit anti-RGS7 polyclonal antibody 7RC-1 (Rojkova et al. 2003), mouse anti-Na+/K+ ATPase α1 subunit monoclonal antibody (Abcam, ab7671) and rabbit anti-Na+/K+ ATPase β2 subunit (Sigma, A3979). Secondary antibodies utilized in immunoblots were bovine anti-mouse IgG-HRP-conjugated (Santa Cruz, sc-2380), sheep affinity-purified anti-rabbit IgG (H+L) HRP-conjugated (Binding Site, no. AP311), fluorescein anti-rabbit and anti-mouse IgG (H+L) from Vector Laboratories, nos. FI-1000 and FI-2000 respectively and IR secondary antibodies (anti-rabbit IR 800 and anti-mouse Red and Green) are from LI-COR Bioscience.
RESULTS
DEP domain of RGS7 essential for binding to R7BP
In order to better understand the mechanism of nuclear localization of the Gβ5/R7-RGS complex, RGS7 mutants were generated lacking either the N-terminal DEP domain (DEPless RGS7, lacking residues 1–116) or the C-terminal domain that includes the conserved RGS core (ΔRGS-RGS7, lacking residues 320–469) (Fig. 1A). Transfection of rat PC12 cells with the wild-type and mutant RGS7 constructs along with HA-tagged Gβ5 showed that the DEPless RGS7 mutant was expressed and bound Gβ5 as evidenced by its co-immunoprecipitation with anti-HA antibodies (Fig. 1B). The DEPless RGS7 mutant was expressed at a lower level than the wild-type presumably reflecting lower stability even in the presence of Gβ5 (Fig. 1B). Despite its expression in Gβ5 and HA-R7BP-cotranfected cells, the DEPless RGS7 mutant, unlike wild-type RGS7, failed to co-immunoprecipitate with R7BP (Fig. 1C, D). This confirms previous work demonstrating that the DEP domain of R7-RGS proteins is essential for binding to R7BP/R9AP (Hu et al. 2003, Martemyanov et al. 2003, Martemyanov et al. 2005).
The ΔRGS-RGS7, lacking residues 320–469, could not be detected with the anti-RGS7 antibody 7RC-1, which was generated against the conserved RGS domain of RGS7, residues 312–469 (Fig. 1B) (Rojkova et al. 2003). The expression of the ΔRGS-RGS7 mutant could be demonstrated however via its epitope tag with the anti-Xpress monoclonal antibody on lysates and anti-Xpress antibody immunoprecipitates from PC12 cells, experiments that also demonstrated its ability to bind Gβ5 and HA-R7BP (Fig. 1C, D).
DEP domain of RGS7 necessary for nuclear localization in PC12 cells
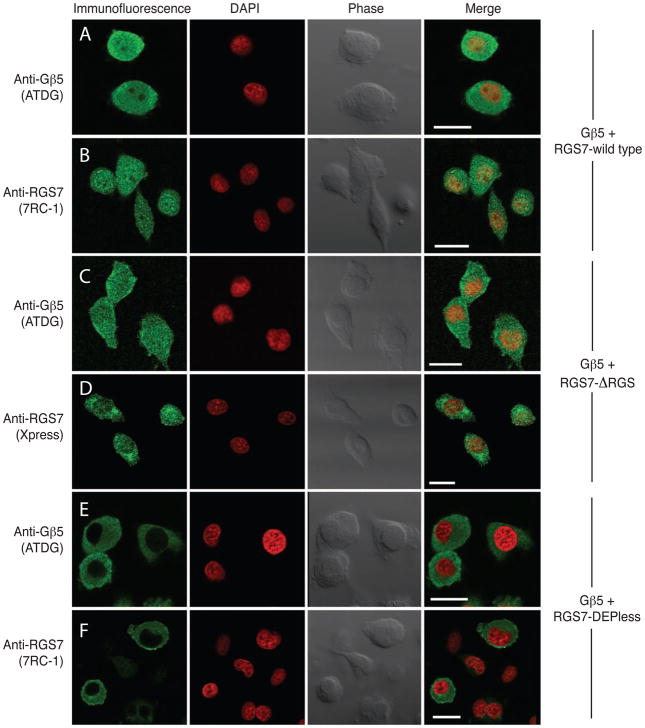
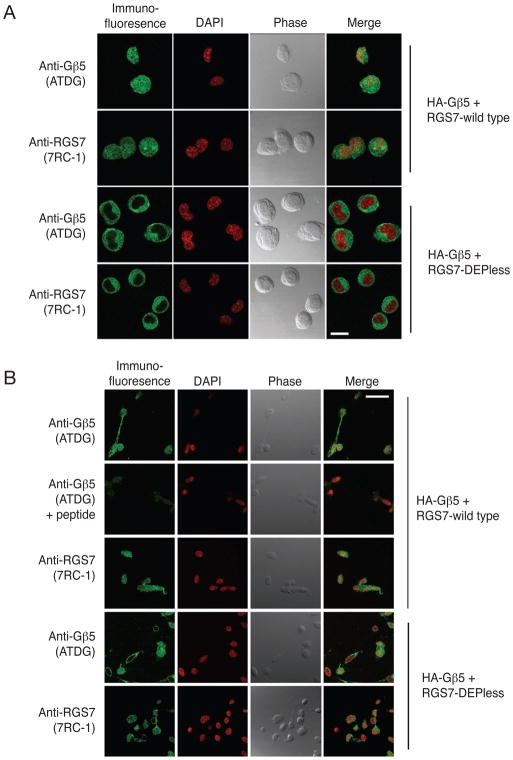
PC12 cells transfected with Gβ5 and either wild-type RGS7, ΔRGS-RGS7, or the DEPless RGS7 mutant were analyzed by immunofluorescence with anti- Gβ5 and anti-RGS7 or Xpress monoclonal antibodies employing laser confocal microscopy (Fig. 2). In cells transfected with Gβ5 and wild-type RGS7, anti- Gβ5 immunoreactivity, employing ATDG antibody directed against the N-terminal region of Gβ5, was evident throughout the cytoplasm and in the nucleus, the latter defined by DAPI counterstaining (Fig. 2, row A). No immunofluorescent signal was seen if the primary antibody was omitted or pre-absorbed with the cognate peptide (not shown). Like the Gβ5 signal, RGS7 immunoreactivity was evident in both the cytoplasm and nucleus (Fig. 2, row B). These results confirm previous immunofluorescence studies of the subcellular distribution of Gβ5/RGS7 expressed in PC12 cells (Zhang et al. 2001, Rojkova et al. 2003). In cells transfected with Gβ5 and the Xpress epitope-tagged ΔRGS-RGS7 mutant, both Gβ5 immunoreactivity and mutant RGS7 immunoreactivity (detected with the Xpress antibody) were evident in both the cytoplasm and nucleus in a pattern indistinguishable from that of Gβ5/wild type RGS7 (Fig. 2, rows C, D). In contrast, PC12 cells transfected with Gβ5 and the DEPless RGS7 mutant exhibited anti- Gβ5 and anti-RGS7 immunofluorescence in the cytoplasm but not in the nucleus (Fig. 2, rows E, F). Laser confocal microscopic analysis of Neuro-2a murine neuroblastoma cells transfected with Gβ5/RGS7 or Gβ5/DEPless RGS7 gave very similar results to those in PC12 cells (Fig. 3A). The same experiment in SH-SY5Y human neuroblastoma cells showed variable exclusion of anti- Gβ5 and anti-RGS7 immunofluorescence from the nuclei in Gβ5/DEPless RGS7-transfected cells (Fig. 3B). The reasons for the variability of Gβ5/DEPless RGS7 nuclear exclusion in SH-SY5Y cells compared to the uniformity of exclusion in PC12 and Neuro-2a cells is unclear, but analysis in the SH-SY5Y cells was hampered by the paucity of cytoplasm and highly variable cell morphology. Taken together these results suggest the absence of the RGS7 DEP domain may impair nuclear targeting of the Gβ5/RGS7 complex in multiple cellular contexts.
Figure 2. Gβ5/RGS7 and Gβ5/ΔRGS-RGS7, but not Gβ5/Depless-RGS7, colocalize to the nucleus of transfected PC12 cells.
PC12 cells transfected with the cDNA combinations HA- Gβ5/RGS7 (rows A,B), HA- Gβ5/ΔRGS-RGS7 (rows C, D), or HA- Gβ5/Depless-RGS7 (rows E, F) were immunostained and analyzed by confocal laser microscopy to determine the subcellular localization of the expressed proteins (green signal). DAPI was used to visualize the nuclei (red signal). Phase contrast and overlay (merge) of immunofluorescent signal with nuclear stain are also shown in all conditions. Gβ5 immunofluorescence ATDG polyclonal antibody (A, C, E). Immunofluorescence of RGS7 with 7RC-1 polyclonal antibody (B, F) or Xpress monoclonal antibody (D). Scale bar = 20 μm.
Figure 3. Gβ5/RGS7, but not Gβ5/Depless-RGS7, colocalize to the nucleus of transfected Neuro-2a cells and SH-SY5Y cells.
Neuro-2A (A) or SH-SY5Y (B) cells transfected with the cDNA combinations HA- Gβ5/RGS7 or HA- Gβ5/Depless-RGS7 were immunostained and analyzed by confocal laser microscopy to determine the subcellular localization of the expressed proteins (green signal). DAPI was used to visualize the nuclei (red signal). Phase contrast and overlay (merge) of immunofluorescent signal with nuclear stain are also shown in all conditions. Antibodies and conditions are as in the legend to Fig. 2. Scale bar = 20 μm.
In cells transfected with Gβ5 and wild-type RGS7, the nuclear localization of the ~95 kDa heterodimeric Gβ5/RGS7 complex is unlikely to result from diffusion from the cytosol since the size cutoff for passive entry through the nuclear pore complex is approximately 60 kDa (Allen et al. 2000). Furthermore no nuclear localization of the Gβ5/DEPless RGS7 complex was seen even though the mutant RGS7 is smaller than the wild-type resulting in a presumably more readily diffusible complex.
R7BP-dependent nuclear localization of Gβ5/RGS7 complex in primary neurons and brain
The requirement for the RGS7 DEP domain for Gβ5/RGS7 complex nuclear localization demonstrated above suggests a possible role for R7BP in the process. This is because of prior evidence identifying the R7-RGS DEP domain as a required element for R7BP/R9AP binding (Hu et al. 2003, Martemyanov et al. 2003, Martemyanov et al. 2005), and also because R7BP contains a C-terminal NLS that is sufficient to drive the nuclear localization of a Gβ5/RGS7/R7BP complex if R7BP is de-palmitoylated by chemical treatment or mutation (Song et al. 2006, Drenan et al. 2006). We therefore studied the nuclear localization of the endogenous Gβ5 complex in primary neurons and brains of wild-type and R7BP deficient mice.
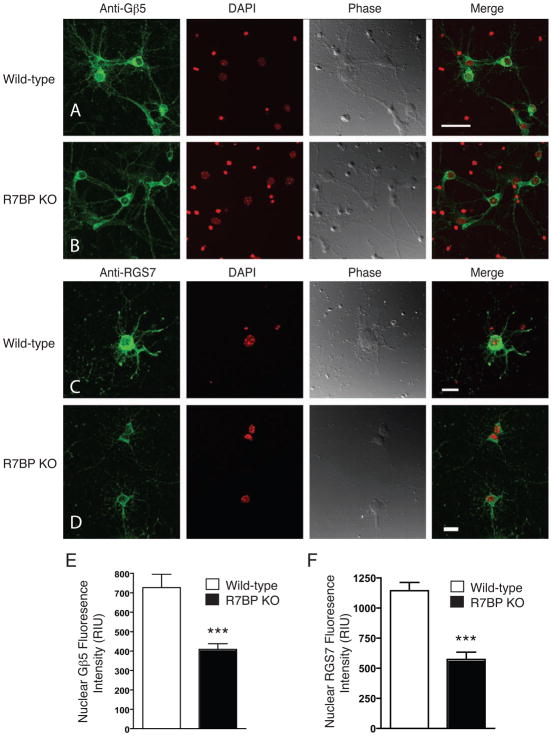
Primary cortical neurons from homozygous R7BP knockout (KO) mice or their wild-type littermates were analyzed for the nuclear localization of endogenous Gβ5/RGS7 by immunofluorescence with anti- Gβ5 (Fig. 4, rows A, B) and anti-RGS7 (Fig. 4, rows C, D) antibodies using laser confocal microscopy. The intensity of the Gβ5 and RGS7 signals in control and R7BP KO primary neurons was quantified by use of a 3-dimensional rendering of the nucleus (boundaries determined from DAPI labeling) from which the mean fluorescein intensity was calculated for the whole volume (Fig. 4E, F). The nuclear signal intensity of Gβ5 and RGS7 in R7BP KO primary neurons was approximately half that of the nuclear intensity in primary neurons from wild-type littermates (Fig. 4E, F). No immunofluorescent signal was seen if the primary antibody was omitted or pre-absorbed with the cognate peptide (not shown). Since no specific marker was used to delineate the plasma membrane of the primary neurons, and since the shapes of the neuronal soma and proximal processes were so variable, no quantitative estimate of the plasma membrane versus cytosolic distribution of Gβ5 and RGS7 was possible despite the acquisition of stacks of planar images by the laser confocal methodology. Careful comparison of the outer boundary of the cell determined from the phase contrast images with the Gβ5 and RGS7 immunofluorescence clearly indicated, however, the presence of both proteins at the cell surface in both wild-type and R7BP KO neurons.
Figure 4. Comparison of the nuclear localization of endogenous Gβ5 and RGS7 in primary cortical neuronal cells derived from wild type and R7BP homozygous KO mice.
Primary mouse cortical neurons isolated from wild-type mouse pups and littermates homozygous for germline R7BP KO were analyzed by immunostaining with antibody to endogenous Gβ5 (A, B) or RGS7 (C, D) (green signal) and confocal laser microscopy as described in Experimental Procedures. DAPI was used to visualize the nuclei (red signal), and phase contrast and overlay (merge) with nuclear stain are shown in all conditions. Neuronal DAPI stain is diffuse and fainter while glial cell nuclei give an intense and compact red DAPI signal. (A, C): Wild type mouse primary cortical neurons. (B, D): R7BP KO mouse primary cortical neurons. Scale bar = 20 μm. Quantification of the endogenous Gβ5 signal (E) or endogenous RGS7 (F) from the nuclei of wild type and R7BP KO mice primary neuronal cells. For (E) the quantification of 61 wild-type and 68 R7BP KO neurons were pooled; for (F) the analyses of 69 wild-type and 48 R7BP KO neurons were pooled. ***p< 0.0001, Student’s t test. The confocal images used (A–D) are representative of those used for nuclear Gβ5 or RGS7 quantification (E, F) as described in Experimental Procedures.
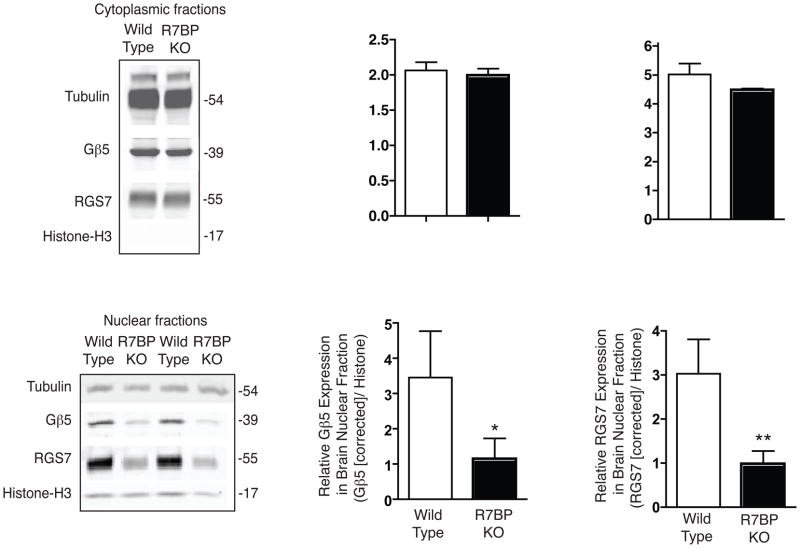
The subcellular localization of endogenous Gβ5/RGS7 complexes in the cerebral cortex of wild type and R7BP KO mice was studied by fractionation followed by quantitative immunoblotting (Fig. 5). Immunoblots of purified cytosolic fraction from mouse brain cortex readily demonstrated the presence of Gβ5 Gβ5 and RGS7 (Fig. 5A), consistent with previous reports (Witherow et al. 2000, Zhang et al. 2001, Grabowska et al. 2008). No immunoreactivity of histone H3, a marker of the nuclear fraction, was evident in the isolated cytoplasmic fractions (Fig. 5A). Quantification of the expression of cytosolic Gβ5 and RGS7 relative to tubulin by infrared imaging of immunoblots showed no difference between wild type and R7BP KO mice (Fig. 5B, C). Immunoblots of the purified brain nuclear fractions demonstrated endogenous Gβ5 and RGS7 in both mouse strains (Fig. 5D) (Zhang et al. 2001). Quantification of the expression of nuclear Gβ5 and RGS7 relative to histone H3 by infrared imaging, using the raw band intensity values, estimated the level of both Gβ5 and RGS7 in R7BP KO mice to be about 40% that of wild type controls (not shown). After correction for a small amount of cytosolic protein contamination (evident as tubulin immunoreactivity in the nuclear fraction), the level of both Gβ5 and RGS7 in R7BP KO mice in the nuclear fraction was found to be about 30% that of the wild type (Fig. 5E, F).
Figure 5. Subcellular fractionation and quantification of endogenous cytoplasmic and nuclear Gβ5 and RGS7 from brains of wild-type and R7BP KO mice.
Cytosolic and nuclear fractions were prepared from mouse brain cortex from wild-type or homozygous R7BP KO female mouse pups and analyzed by immunoblotting with antibodies to endogenous Gβ5 and RGS7 as indicated. Protein band detection and quantification employed infrared-dye tagged secondary antibodies and an infrared imaging system as described in Experimental Procedures. (A) Immunoblot showing the Gβ5 and RGS7 expression in cytosolic fraction, with tubulin shown as a loading control, and histone H3 shown to indicate the absence of nuclear contamination. (B) and (C) Quantification of protein band intensity showing the relative expression of Gβ5 and RGS7 in the cytosolic fraction (D) Immunoblot showing the Gβ5 and RGS7 expression in nuclear fraction along with histone H3 as loading control, with tubulin shown to indicate extent of contamination with cytosolic proteins. (E) and (F) Quantification of protein band intensity showing the relative expression of Gβ5 and RGS7 in the nuclear fraction, after correction for cytoplasmic contamination as described in Experimental Procedures. *, P< 0.05, **, P<0.02, Student’s paired t-test.
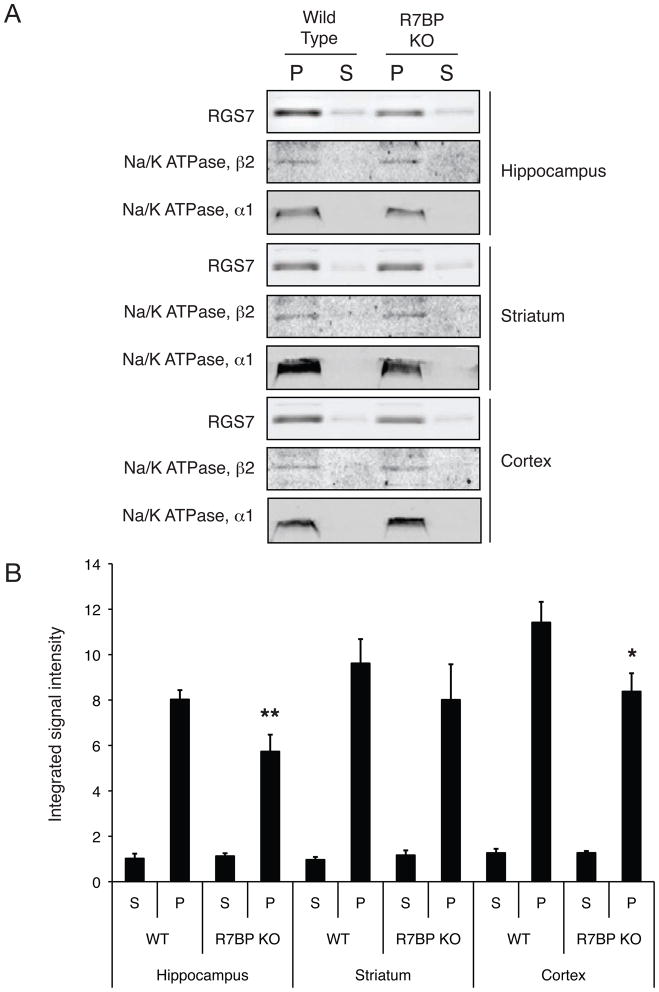
R7BP may govern the distribution of the Gβ5/R7-RGS complex between the plasma membrane and the cell nucleus (Drenan et al. 2005, Song et al. 2006, Drenan et al. 2006). Since there was a significant loss of nuclear localization of the Gβ5/RGS7 complex in brains from R7BP KO mice but no change in the cytosolic expression, the extent of plasma membrane localization of RGS7 was estimated in different subregions of brains from R7BP KO mice and their wild-type littermates by quantitative immunoblotting with anti-RGS7 antibody and compared to the distribution of two plasma membrane markers (Fig. 6A). While there was no difference between wild-type and R7BP KO mice in the RGS7 expression in plasma membranes from striatum, both the hippocampal and cortical plasma membrane expression of RGS7 was reduced by some 25% with no differences observed in the abundance of the plasma membrane markers in the corresponding particulate fractions (Fig. 6B).
Figure 6. Subcellular fractionation and quantification of endogenous cytoplasmic and membrane RGS7 from different brain regions of wild-type and R7P KO mice.
Cytosolic and membrane fractions were prepared from striatum, hippocampus and brain cortex of wild type or homozygous R7BP KO mice and analyzed by immunoblotting with antibodies to endogenous RGS7 as indicated. Protein band detection and quantification was performed using infrared-dye tagged secondary antibodies and infrared imaging system as described in Experimental Procedures. (A) Immunoblots showing RGS7 expression in cytosolic (S) and membrane (P) fractions in hippocampus, striatum and cortex, with Na+/K+ ATPase β2 subunit and α1 subunits as loading controls. (B) Quantification of protein band intensity. **, p< 0.01, wild-type vs. R7BP KO hippocampal membranes; *, p < 0.05, wild-type vs. R7BP KO cortical membranes, Student’s paired t-test.
DISCUSSION
While the experiments presented here focused primarily on the nuclear localization of the Gβ5/R7-RGS complex, it was perhaps surprising that the plasma membrane targeting in brain of RGS7 was so minimally affected in R7BP KO mice (Fig. 6). In the striatum, where the majority of R7BP is complexed with RGS9-2 (Anderson et al. 2009a), the lack of R7BP KO effect on RGS7 membrane localization suggests the existence there of R7BP-independent membrane targeting mechanism(s). Such mechanism(s) also seem to be active in the hippocampus and cortex since the majority of RGS7 remains membrane associated in the R7BP KO mice. It is interesting to note that a recent study of RGS9-2 localization in brain also found evidence for R7BP-independent membrane targeting, based on the differential detergent solubility of R7BP and RGS9-2 from the striatal particulate fraction (Mancuso et al. 2009), although studies of brains from R7BP KO mice documented almost complete mislocalization of RGS9-2 away from the plasma membrane (Anderson et al. 2007a).
Consideration of the nuclear localization of the Gb5/R7-RGS suggests a mechanism of regulation or action of this complex outside the canonical model of heterotrimeric G protein signaling (Zhang et al. 2001, Rojkova et al. 2003). Studies of other RGS proteins may offer instructive parallels for consideration. In the case of RGS10, its phosphorylation-dependent subcellular redistribution appears to dampen its GAP function at the plasma membrane by sequestration in the nucleus (Burgon et al. 2001). Recently RGS13 was found to be a nuclear repressor of CREB-dependent transcription by a mechanism involving complex formation with phosphorylated CREB and CBP/p300 (Xie et al. 2008). Like RGS13 and RGS10, none of the identified components of a potential Gβ5/R7-RGS/R7BP complex contains a basic or zinc finger protein domain comparable to those found in direct DNA-binding transcription factors.
The mechanism of nuclear localization by the Gβ5/R7-RGS complex is not clear. Our results suggest that the DEP domain of RGS7 is important in this process. The adjacent DEP and R7H/DHEX domains of R7-RGS proteins together comprise the binding site for R7BP/R9AP (Anderson et al. 2007b), and as expected the DEPless RGS7 mutant employed here failed to bind R7BP. Our demonstration of significantly reduced Gβ5/RGS7 nuclear localization in mice lacking R7BP supports a model that loss of R7BP interaction contributes to the lack of nuclear targeting by the DEPless RGS7 mutant.
With respect to the nuclear localization of the endogenous Gβ5/R7-RGS complex, a polybasic cluster of amino acids resembling a NLS has been identified in the C-terminus of R7BP that can drive the nuclear localization of a Gβ5/R7-RGS/R7BP complex in cultured cells when R7BP palmitoylation is blocked by 2-bromopalmitate treatment or mutation of the putative C-terminal Cys-252/Cys-253 palmitoylation site (Drenan et al. 2005, Song et al. 2006, Drenan et al. 2006). But whether endogenous neuronal R7BP is ever resident in the cell nucleus (or in the cytoplasm through which it must necessarily traverse, for any hypothetical trafficking between plasma membrane and nucleus) is an open question. Using strong anti-R7BP antibodies Grabowska and co-workers failed to detect R7BP in neuronal cell nuclei from multiple brain regions, and could not identify R7BP associated with brain cytosolic Gβ5/RGS7, even though membrane-associated R7BP was readily demonstrable (Grabowska et al. 2008). Our results here tend to exclude a model in which de-palmitoylated R7BP is an obligatory partner for the nuclear localization of Gβ5 and RGS7, since residual nuclear localization of Gβ5/RGS7 was clearly evident in neurons and brains of R7BP KO mice. The residual nuclear localization of Gβ5/RGS7 in the brains and neurons of R7BP KO, at some 30 to 50% of control levels, points to the existence of R7BP-independent mechanisms for Gβ5/RGS7 nuclear localization, though it is possible utilization of such pathway(s) is exaggerated in the absence of R7BP.
Since knockout of R7BP does not affect the overall stability or expression level of Gβ5 or RGS7 (Anderson et al. 2007b, Anderson et al. 2007a), the significant reduction in nuclear Gβ5/RGS7 complex seen in the R7BP KO mice may reflect an indirect effect on complex nuclear localization resulting from loss of R7BP function/expression at the plasma membrane. Indeed modest but significant reductions were seen in hippocampal and cortical RGS7 plasma membrane targeting. It is conceivable therefore that interaction of R7BP with Gβ5/R7-RGS at the plasma membrane, where R7BP facilitates the regulatory effect of the Gβ5/R7-RGS complex on GPCR-to-effector signalling (Drenan et al. 2006), could somehow “mark” the complex, perhaps by loss or gain of an unknown posttranslational modification, and thereby promote subsequent nuclear localization. Alternatively the functional interaction of R7BP with Gβ5/R7-RGS and Gβ5/R7-RGS/R7BP with Gα subunits or other proteins at the plasma membrane might normally result in the increased expression or activity of one or more components of the nuclear import machinery. Whether such a mechanism exists in neurons, to provide feedback to the nucleus regarding the intensity of GPCR signaling at the postsynaptic density for example, remains to be elucidated in future work.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (W.F.S.) and grants NIH DA021743 and NIH DA026405 (K.A.M.).
Abbreviations used
- RGS
regulator of G protein signaling protein
- R7BP
R7 binding protein
- R9AP
R9 anchoring protein
- DEP domain
Disheveled, EGL-10, Pleckstrin homology domain
- R7H domain
R7 homology domain
- DHEX domain
DEP helical extension domain
- GGL domain
Gγ-like domain
- GAP
GTPase activating protein
- PBS
phosphate-buffered saline
- KO
knockout
Footnotes
None of the authors has a financial or any other conflict of interest.
References
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113 (Pt 10):1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Martemyanov KA. Changes in striatal signaling induce remodeling of RGS complexes containing Gbeta5 and R7BP subunits. Mol Cell Biol. 2009a;29:3033–3044. doi: 10.1128/MCB.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Semenov A, et al. Expression and localization of RGS9-2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J Neurosci. 2007a;27:14117–14127. doi: 10.1523/JNEUROSCI.3884-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Posokhova E, Martemyanov KA. The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys. 2009b;54:33–46. doi: 10.1007/s12013-009-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Semenov A, Song JH, Martemyanov KA. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9-2. J Biol Chem. 2007b;282:4772–4781. doi: 10.1074/jbc.M610518200. [DOI] [PubMed] [Google Scholar]
- Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- Bouhamdan M, Michelhaugh SK, Calin-Jageman I, Ahern-Djamali S, Bannon MJ. Brain-specific RGS9–2 is localized to the nucleus via its unique proline-rich domain. Biochim Biophys Acta. 2004;1691:141–150. doi: 10.1016/j.bbamcr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Burgon PG, Lee WL, Nixon AB, Peralta EG, Casey PJ. Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10) J Biol Chem. 2001;276:32828–32834. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- Cao Y, Masuho I, Okawa H, et al. Retina-specific GTPase accelerator RGS11/G beta 5S/R9AP is a constitutive heterotrimer selectively targeted to mGluR6 in ON-bipolar neurons. J Neurosci. 2009;29:9301–9313. doi: 10.1523/JNEUROSCI.1367-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nature structural & molecular biology. 2008;15:155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker SP, McCaffery JM, Palade GE, Insel PA, Farquhar MG. Differential distribution of alpha subunits and beta gamma subunits of heterotrimeric G proteins on Golgi membranes of the exocrine pancreas. J Cell Biol. 1996;133:1027–1040. doi: 10.1083/jcb.133.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan RM, Doupnik CA, Boyle MP, Muglia LJ, Huettner JE, Linder ME, Blumer KJ. Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J Cell Biol. 2005;169:623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan RM, Doupnik CA, Jayaraman M, Buchwalter AL, Kaltenbronn KM, Huettner JE, Linder ME, Blumer KJ. R7BP augments the function of RGS7/Gbeta;5 complexes by a plasma membrane-targeting mechanism. J Biol Chem. 2006;281:28222–28231. doi: 10.1074/jbc.M604428200. [DOI] [PubMed] [Google Scholar]
- Grabowska D, Jayaraman M, Kaltenbronn KM, Sandiford SL, Wang Q, Jenkins S, Slepak VZ, Smith Y, Blumer KJ. Postnatal induction and localization of R7BP, a membrane-anchoring protein for regulator of G protein signaling 7 family-G®5 complexes in brain. Neuroscience. 2008;151:969–982. doi: 10.1016/j.neuroscience.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler JR. R7BP: a surprising new link between G proteins, RGS proteins, and nuclear signaling in the brain. Sci STKE 2005. 2005:pe38. doi: 10.1126/stke.2942005pe38. [DOI] [PubMed] [Google Scholar]
- Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci U S A. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Zhang Z, Wensel TG. Activation of RGS9-1GTPase acceleration by its membrane anchor, R9AP. J Biol Chem. 2003;278:14550–14554. doi: 10.1074/jbc.M212046200. [DOI] [PubMed] [Google Scholar]
- Jamora C, Takizawa PA, Zaarour RF, Denesvre C, Faulkner DJ, Malhotra V. Regulation of Golgi structure through heterotrimeric G proteins. Cell. 1997;91:617–626. doi: 10.1016/s0092-8674(00)80449-3. [DOI] [PubMed] [Google Scholar]
- Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol Sci. 2009;30:17–24. doi: 10.1016/j.tips.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso JJ, Qian Y, Long C, Wu GY, Wensel TG. Distribution of RGS9-2 in neurons of the mouse striatum. J Neurochem. 2009 doi: 10.1111/j.1471-4159.2009.06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martemyanov KA, Lishko PV, Calero N, et al. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J Neurosci. 2003;23:10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- Nini L, Waheed AA, Panicker LM, Czapiga M, Zhang JH, Simonds WF. R7-binding protein targets the G protein beta 5/R7-regulator of G protein signaling complex to lipid rafts in neuronal cells and brain. BMC biochemistry. 2007;8:18. doi: 10.1186/1471-2091-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojkova AM, Woodard GE, Huang TC, Combs CA, Zhang JH, Simonds WF. Gγ subunit-selective G protein β5 mutant defines regulators of G protein signaling protein binding requirement for nuclear localization. J Biol Chem. 2003;278:12507–12512. doi: 10.1074/jbc.M207302200. [DOI] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, et al. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gβ5 subunits. Proc Nat Acad Sci USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Song H, Wensel TG, Sokolov M, Martemyanov KA. Localization and differential interaction of R7 RGS proteins with their membrane anchors R7BP and R9AP in neurons of vertebrate retina. Mol Cell Neurosci. 2007;35:311–319. doi: 10.1016/j.mcn.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Song JH, Waataja JJ, Martemyanov KA. Subcellular targeting of RGS9-2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J Biol Chem. 2006;281:15361–15369. doi: 10.1074/jbc.M600749200. [DOI] [PubMed] [Google Scholar]
- Witherow DS, Wang Q, Levay K, Cabrera JL, Chen J, Willars GB, Slepak VZ. Complexes of the G protein subunit Gβ5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J Biol Chem. 2000;275:24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- Xie Z, Geiger TR, Johnson EN, Nyborg JK, Druey KM. RGS13 acts as a nuclear repressor of CREB. Mol Cell. 2008;31:660–670. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Barr VA, Mo Y, Rojkova AM, Liu S, Simonds WF. Nuclear localization of Gβ5 and regulator of G protein signalling 7 in neurons and brain. J Biol Chem. 2001;276:10284–10289. doi: 10.1074/jbc.M009247200. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Simonds WF. Copurification of brain G-protein β5 with RGS6 and RGS7. J Neurosci. 2000;20:RC59, 1–5. doi: 10.1523/JNEUROSCI.20-03-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]