Abstract
Aim
Since inflammation is an important contributor to atherosclerosis, gene variants mediating inflammation are of interest. We investigated gene variants in acute phase serum amyloid-A (SAA), a sensitive indicator of inflammatory activity, and their associations with cardiovascular disease (CVD) and HDL cholesterol. Interaction of the SAA genes with genetic variants of their regulators, IL-1, IL-6 and TNF-α in influencing CVD was also explored.
Methods
SNPs characterizing common variation in the SAA1 and SAA2 genes were genotyped in European-(EA) and African-American (AA) participants (n=3969 and n=719) of the Cardiovascular Health Study. Using linear and Cox proportional hazards regression, we assessed associations of SNPs with baseline carotid artery intima-media thickness (cIMT) and risk of incident myocardial infarction, ischemic stroke, total CVD events or mortality during ~14 years of follow-up.
Results
No associations between SAA SNPs and outcomes were observed in EA, with the exception of total CVD events; each rs4638289 minor allele was associated with an increased risk in obese individuals, HR=1.2 (95%CI: 0.98-1.4; p=0.086) and decreased risk among non-obese, HR=0.9 (95%CI: 0.8-0.99; p=0.026). In AA, we observed modest associations between SAA SNPs and cIMT, potentially modified by HDL. SAA SNPs were also associated with lower HDL in EA and AA. Suggestive gene-gene interaction findings for cIMT in AA and CVD mortality in EA were not significant in subsequent model selection tests.
Conclusion
Associations of SAA SNPs with cIMT, HDL and total CVD events were identified, unadjusted for multiple testing. These findings should be regarded as hypothesis-generating until confirmed by other studies.
Keywords: acute phase reactants, carotid IMT, myocardial infarction, ischemic stroke, genetic variants
Background
Similar to C-reactive protein, serum amyloid-A (SAA) is a sensitive marker of the acute inflammatory state and its plasma concentrations markedly increase in response to infection, trauma or stress.[1] As the precursor to amyloid protein-A, the small apolipoprotein SAA is required for the amyloidosis disease process, but also has reported functions in tissue factor expression in endothelial cells,[2] lipid metabolism and transport, chemotaxis and regulation of the inflammatory process.[1] During the acute phase response, SAA proteins associate with high density lipoprotein cholesterol (HDL), displacing its apolipoprotein A-I/II components.[3] Sustained high expression of SAA may contribute to atherogenesis through its interference with the cholesterol transport and anti-oxidant functions of HDL.[1, 4] Indeed, elevated SAA level is associated with increased progression of atherosclerosis[5, 6] and an increased risk of cardiovascular disease (CVD) events,[7, 8] though it is unclear whether it acts as a mediator or a marker of atherosclerosis.[9]
The heritability of SAA has been estimated as 49-67%[10] and two genes on chromosome 11 (SAA1 and SAA2) encode for the acute phase SAA proteins SAA1 and SAA2, respectively.[11] SAA synthesis is regulated by interleukin-6 (IL-6), interleukin-1 (IL-1) family, and tumor necrosis factor-α (TNF-α cytokines and primarily occurs in the liver, but extra-hepatic expression in atherosclerotic lesions as well as in epithelial cells of normal tissues has been reported.[1] Because cytokine regulation of SAA and other acute phase reactants determines the magnitude and duration of the immune system response, genetic variation in cytokines and acute phase reactants may be related to the development of age-related inflammatory diseases such as CVD,[12], [13] yet few studies have described the relation between genetic variants in SAA1/2 and CVD.
In this exploratory study, we investigated the association between common genetic variants in SAA1/2 and sub-clinical atherosclerosis as well as incident CVD events (broadly-defined), myocardial infarction (MI), ischemic stroke (IS), and CVD mortality in participants of the Cardiovascular Health Study. Because SAA has the propensity to affect HDL concentration and composition, we also examined whether the relationship between SAA genetic variants and CVD is modified by HDL concentration. Using the data reduction method, logic regression[14] we assessed whether genetic variants in cytokines involved in acute phase SAA regulation may interact with SAA1/2 genetic variants to influence risk of CVD.
Methods
Study Population
The Cardiovascular Health Study (CHS) is a prospective population-based cohort study of men and women aged 65 and older at recruitment.[15] Participants were randomly selected from Medicare eligibility files at four different sites in the U.S. between 1989-90 (primarily European-Americans) and 1992-3 (primarily African-Americans) for a total n=5888. In these analyses, we excluded participants who did not consent to DNA testing, for whom DNA was not available or whose self-reported race was not White (EA) or Black (AA). Additionally, individuals with a history of stroke or MI (n=698) at baseline were excluded, for totals of 3969 EA and 719 AA participants.
Data collection
Demographic, lifestyle, medical history and physical examination information were collected at baseline. Lipids (triglycerides, total, HDL, and LDL cholesterol) were measured in fasting blood samples; blood collection procedures and laboratory methods have been reported previously.[16] BMI was calculated as weight in kg divided by the square of height in meters (kg/m2) and obesity was defined as body mass index >30 kg/m2. Blood pressure (BP) was determined from the average of two seated measurements; hypertension was defined as systolic BP>=140mmHg or diastolic BP>=90mmHg or having history of hypertension with concurrent use of anti-hypertensive medication. C-reactive protein (CRP) was measured on stored EDTA plasma from the baseline examination using a high-sensitivity enzyme-linked immunosorbent assay (coefficient of variation, 6.2%).[17] Maximum carotid artery intima-media thickness (cIMT) in mm was determined at the baseline examination using high-resolution B-mode ultrasonography.[18] cIMT measures were calculated for the common and internal carotid arteries by averaging maximum wall thicknesses obtained from scans of the near and far walls on the left and right sides. Non-normally distributed measurements were ln-transformed.
Participants were followed for events through June 2005. Censoring date was defined by death, loss to follow-up, study drop out or event date. Incident, non-procedure-related (not occurring during surgery or re-vascularization) MI, ischemic stroke and CVD events and CVD mortality were included in the current analyses. MI was defined using standard CHS criteria: history of chest pain, cardiac enzyme levels, and characteristic changes on serial electrocardiograms. Stroke was validated based on criteria that included onset of symptoms, duration of deficits, and findings on computed tomography or magnetic resonance imaging. Strokes were further classified by subtype: ischemic, hemorrhagic, or unknown. CVD events included incident MI, angina, stroke, transient ischemic attack, claudication, and atherosclerotic causes of death. CVD mortality included fatal MI, stroke and coronary heart disease. Participants with multiple incident events were censored on the date of the first event. All events were adjudicated by CHS committee as reported previously.[19]
Genotyping
Genomic sequence variation data were available for the IL6, TNF, and 2 IL-1 family genes, IL1B (coding for IL-1β protein) and IL1RN (coding for IL-1 receptor antagonist (IL-1RA) protein) through the SeattleSNPs Program for Genomics Applications (PGA).[20] Using the LDselect algorithm of Carlson et al.[21] we selected a set of tagSNPs capturing common race-specific patterns of genetic variation within each gene, using minor allele frequency (MAF) ≥5% and linkage disequilibrium patterns with r2>0.64 as a cutoff. For genes not re-sequenced by the PGA (SAA1/SAA2), we used data available from the International HapMap project to select race-specific tagSNPs across the two genes including potential regulatory areas in the 5′ regions of both genes. IL1RN and the majority of SAA2 SNPs were genotyped using the Illumina GoldenGate platform by the Center for Inherited Disease Research, Johns Hopkins University (Baltimore, Maryland, USA). SAA1/2, IL1B and TNF SNPs were typed at the Laboratory for Clinical Biochemistry Research, University of Vermont with the ABI TaqMan platform under standard conditions (TaqMan® SNP Genotyping Assays Protocol, Rev. B, Part #4332856b, Applied Biosystems, Foster City, CA).
Race-specific minor allele frequencies of SNPs were calculated and consistency of observed genotype frequencies with Hardy-Weinberg equilibrium (HWE) was assessed by performing Fischer’s exact chi-squared test. Of the 12 SAA1/2 SNPs genotyped, two SAA1 SNPs were not in HWE in EA and one was not in HWE for AA, with p<0.05. Genotyping errors were found for rs2045272 which was in not in HWE in either racial group. Re-genotyping efforts failed and it was dropped from further analyses. No evidence of genotyping error was found for the other SNP out of HWE in EA, rs4638289 and it was retained in analyses. SNPs with MAF < 5% in our study population were not included in the analyses of main effects leaving a total of 8 SAA1/2 tagSNPs in EA and 10 in AA.
Using pair-wise linkage disequilibrium statistics (r2), we tested for intra-gene correlation of SNPs and for inter-gene correlation between SNPs of IL1RN and IL1B which are located approximately 300kb apart on chromosome 2. In EA, correlations among SAA1/2 SNPs were generally low, with r2= 0.0-0.18 with the exception of two pairs of SNPs: r2=0.24 for rs7113375 and rs7130337; and r2=0.33 for rs2460824 and rs2468844. In AA, correlations among SAA1/2 SNPs ranged from 0.0-0.17, with the exception of two pairs of SNPs: r2 =0.25 for rs7130337 and rs10832911; and r2 =0.56 for rs7130337 and rs7933280. These modestly correlated pairs were retained in analyses. In EA, a few highly correlated (r2>0.6) SNPs within IL1RN and IL6 were found. The correlated SNP with the lowest minor allele frequency was dropped from analyses; in EA we dropped IL6 rs1800795 which was highly correlated with rs1554606 (r2~0.9) and for IL1RN, three SNPs were dropped: rs397211, rs431726 and rs315952. In AA, only IL6-rs1524107 was dropped. For gene-gene interaction analyses, SNPs with MAF >0.02 were included for the following totals: IL6 (n=5), IL1B (n=2), TNF (n=2), IL1RN (n=9) and SAA1/2 (n=10) in EA. In AA, the following SNPs were included: IL6 (n=8), IL1B (n=5), TNF (n=2), IL1RN (n=17) and SAA1/2 (n=11). [Table 1]
Table 1.
Summary of IL6, IL1B, IL1RN and TNF Genotyping Results
| MAF§ | |||||
|---|---|---|---|---|---|
| Gene | SNP | PGA ID | EA | AA | Context |
| IL6 | rs1800796 | 1111 | 0.049 | 0.083 | chr7 (p15.3):intergenic |
| rs1800795 | 1510 | 0.410 | 0.091 | intergenic | |
| rs2069830 | 2002 | 0.076 | coding, non-synonymous SNP | ||
| rs1474347 | 2989 | 0.177 | intron | ||
| rs2069837 | 2892 | 0.083 | 0.117 | intron; HWD in EA, p=0.001; excess homozygotes | |
| rs1524107 | 3084 | 0.067 | intron | ||
| rs1554606 | 3572 | 0.436 | 0.339 | intron | |
| rs2069849 | 6021 | 0.026 | 0.133 | coding, synonymous SNP | |
| rs1818879 | 7592 | 0.313 | 0.196 | intergenic (downstream) | |
| IL1B | rs1143625 | 0302 | 0.258 | 0.124 | chr2 (q14.2): intergenic (upstream) |
| rs1143629 | 2143 | 0.427 | intron | ||
| rs1143630 | 4006 | 0.225 | intron | ||
| rs1143634 | 5277 | 0.237 | 0.124 | coding synonymous SNP | |
| rs2853550 | 8546 | 0.305 | intergenic (downstream) | ||
| IL1RN | rs4251955 | 0422 | 0.031 | chr2 (q13): intergenic | |
| rs4251961 | 1018 | 0.375 | 0.191 | intergenic | |
| rs4251962 | 1193 | 0.024 | intergenic | ||
| rs315919 | 2765 | 0.396 | 0.488 | intron | |
| rs2254511 | 3374 | 0.002 | 0.111 | intron | |
| rs3213448 | 5848 | 0.129 | 0.227 | intron | |
| rs315934 | 10257 | 0.202 | 0.103 | intron | |
| rs4252010 | 13027 | 0.044 | intron | ||
| rs2232354 | 13888 | 0.203 | 0.042 | intron | |
| rs432014 | 15132 | 0.273 | 0.172 | intron | |
| rs380092 | 15453 | 0.315 | 0.644 | intron | |
| rs431726 | 15559 | 0.260 | 0.137 | intron | |
| rs315955 | 15983 | 0.002 | 0.093 | in HWD in AA, p=0.022 | |
| rs315952 | 16857 | 0.282 | 0.412 | coding synonymous SNP | |
| rs4252023 | 16926 | 0.043 | coding synonymous SNP | ||
| rs4252041 | 17163 | 0.043 | 0.007 | UTR | |
| rs397211 | 18694 | 0.302 | 0.150 | intergenic (downstream); in HWD in AA, p=0.023 | |
| rs315949 | 19327 | 0.413 | 0.361 | intergenic (downstream) | |
| TNF | rs1800628 | 4101 | 0.099 | 0.040 | chr6(p21.33): intergenic (downstream) |
| rs361525 | 0352 | 0.052 | 0.043 | intergenic (upstream) | |
Bolded MAF indicates SNPs included in logic regression analyses; others were not included due to correlation or very low MAF.
Statistical Analysis
All analyses were race-specific to minimize potential confounding attributable to population stratification, and were minimally adjusted for age, sex and recruitment site. Furthermore, AA models were adjusted for % African Ancestry (%AA), a continuous variable that quantifies the proportion of an individual’s genome that is of African origin using ancestry informative markers as previously described.[22] We investigated effect modification by baseline HDL concentration and obesity status for all outcomes, testing the significance of SNP*HDL level or SNP*obesity interaction terms. Model adjustment for current smoking, LDL cholesterol, HDL cholesterol, obesity and BMI was also investigated.
For the continuous outcomes of common cIMT, ln(internal cIMT) and HDL, we assessed associations between the measures and SAA1/2 SNPs using linear regression and a minimally-adjusted additive genetic model, assuming a constant effect size for each additional copy of the minor allele so that the regression coefficient (β) represents the adjusted allele-specific difference in estimated mean IMT in mm or HDL in mg/dL. Associations between SAA1/2 genotypes and risk of incident events were assessed using Cox proportional hazards models. Due to the exploratory nature of all analyses, results were not adjusted for multiple testing.
To evaluate the role of gene-gene interactions and their association with sub-clinical and clinical CVD, logic regression[14] was used. SAA1/2, IL6, TNF, IL1B and IL1RN SNPs were coded as binary predictor variables (for both dominant and recessive genetic models). Briefly, logic regression searches for a variable or Boolean expression (termed ‘tree’) that best predicts outcome (i.e. minimizes the scoring function); this process is repeated until additional variables do not improve scores. It is adaptable to different forms of regression-based methods including linear regression and Cox proportional hazards methods. To simplify its interpretation and for discovery of combinations of predictors affecting relatively larger groups of individuals, we limited final model size a priori, by setting the maximum tree size to 2 and the number of leaves (i.e. variables) to 8, as suggested by Kooperberg et al.[14]
Since logic regression is an adaptive, exploratory method, appropriate model selection is necessary to prevent ‘over-fitting’ of the data. The software includes cross-validation and permutation tests which we used for model selection and for interpretation of our results. In a screening stage, called the signal test, we compared scores from a null model (with no SNPs) to scores of the best model identified by the logic regression annealing algorithm during the course of permutations (n=200). For linear regression models, the score represents the residual sum of squares and the Cox proportional hazards model is scored using partial likelihood. Provided the signal test indicated that predictors have some discriminatory power (we chose the threshold of p<0.20), we pursued additional model selection tests.
Potential models were selected using 10-fold cross-validation in which the data are divided and used to build models (training) and the remaining data are used to test the model (testing). Model scores are averaged and models with the lowest (best) scores selected. Additionally, we employed permutation tests conditioning on successive models of increasing size; a successive model was only chosen if it fit the data better (had a smaller score) than the prior smaller model. For this test, we used a liberal cutoff of p<0.20 for significance, but describe the best models (with the lowest p-values). Gene-gene interaction models were minimally adjusted for age, sex and recruitment site and in AA, %AA, since acquired factors are unlikely to confound the gene-outcome association. However, in all models we did investigate effect modification by obesity status, a strong correlate of inflammation. R software and the logic regression package (v. 1.4.7) on the Linux platform were used for logic regression analyses. STATA/SE 10.0 software was used for all other analyses.
Results
Population Characteristics
In both EA and AA populations, the mean age at baseline was 73 years and the majority of participants were female and hypertensive. While CVD risk factors such as obesity, BMI and ln(CRP) were higher in AA than EA, blood lipids were lower, with the exception of HDL which was slightly higher in AA than in EA.[Table 2]
Table 2.
Study Population Baseline Characteristics
| Baseline Characteristic† | European-Americans | African-Americans |
|---|---|---|
| Number (%)§ | 3969 (80.6) | 719 (77.8) |
| Age (years) | 72.6 ± 5.5 | 72.8 ± 5.7 |
| Female sex (%) | 2359 (59.4) | 463 (64.4) |
| Current smokers (%) | 432 (10.9) | 116 (16.2) |
| Obese (%) | 706 (17.8) | 234 (32.6) |
| Hypertension (%) | 2168 (54.8) | 530 (73.8) |
| Body mass index (kg/m2) | 26.4 ± 4.5 | 28.6 ± 5.6 |
| Total cholesterol (mg/dl) | 212.3 ± 38.9 | 209.7 ± 38.4 |
| LDL cholesterol (mg/dl) | 130.1 ± 35.6 | 128.9 ± 35.3 |
| HDL cholesterol (mg/dl) | 54.5 ± 15.8 | 58.3 ± 15.4 |
| Triglycerides (mg/dl) | 141.7 ± 75.6 | 114.6 ± 57.0 |
| ln(CRP) (mg/dl) | 0.9 ± 1.0 | 1.2 ± 1.1 |
Data are presented as number (%) or mean ± standard deviation.
As a percentage of the initially recruited CHS cohort.
Association between SAA1/2 SNPs and cIMT
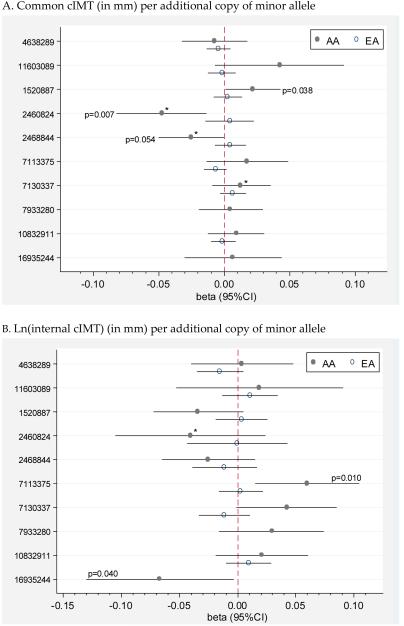
Maximum mean common cIMT was 0.99mm ± 0.20 in EA and 1.11mm ± 0.23 in AA. Ln-transformed maximum mean internal cIMT values in EA were slightly larger than in AA, 0.29mm ± 0.44 vs. 0.25mm ± 0.38. In EA, no SAA1/2 SNPs were significantly associated with common or ln(internal cIMT), all p>0.05.[Figure 1] In AA, SAA2 SNPs were significantly associated with lower common cIMT, namely rs2460824 (β= −0.05, 95%CI: −0.08, 0.01; p=0.007) and rs2468844 (β=−0.03, 95%CI:−0.05, 0; p=0.054) and lower ln(internal cIMT), rs16935244 (β=−0.07 95%CI: −0.13, −0.003; p=0.040). Additionally, in AA, two SAA2 SNPs were associated with higher common cIMT, rs1520887 (β =0.02, 95%CI: 0.001, 0.04; p=0.038) and higher ln(internal IMT), rs7113375 (β =0.06, 95%CI: 0.02, 0.11; p=0.010). There was no evidence of effect modification by obesity or BMI on the association between SAA1/2 SNPs and cIMT in EA or AA; all interaction p-values >0.05. Similarly, we did not find evidence of interaction between SAA1/2 SNPs and HDL levels on cIMT in EA (all interaction term p>0.05). In AA, three SAA2 SNP–HDL level interaction terms were significant in common cIMT models involving the SNPs: rs2468844, p=0.043; rs2460824, p=0.012 and rs7130337, p=0.029. Significant interaction between rs2460824 and HDL level, p=0.034 was also found in AA ln(internal cIMT) models. These significant interactions imply that the relationship between SAA1/2 SNPs and IMT may vary depending on HDL level or equivalently that the association between IMT and HDL may vary by SAA2 genotype. Upon closer inspection, it appears that the significance of interaction terms for rs2460824 and rs2468844 may be related to the larger sample sizes of the common homozygous groups compared to the size of the other group.[Table 3] Results for rs7130337 are perhaps more interesting in that for each additional minor allele, higher HDL is more strongly associated with lower cIMT, which may suggest a beneficial effect for this minor allele. However, as shown, effect sizes and confidence intervals are very similar, regardless of genotype. Model adjustment for smoking, LDL-cholesterol, HDL-cholesterol, BMI or obesity did not appreciably change results in EA or AA.
Figure 1. Association between SAA1/2 SNPs and cIMT in EA† and AA§.
A. Common cIMT (in mm) per additional copy of minor allele
B. Ln(internal cIMT) (in mm) per additional copy of minor allele
†Models adjusted for age, sex and clinic; §Models adjusted for age, sex, clinic and %AA *HDL-SNP interaction term significant.
Table 3.
Association between IMT and HDL by SAA2 genotype in AA
| Outcome | SNP | # of Minor Alleles |
N | IMT in mm per 10 mg/dL HDL β (95%CI) |
p-value |
|---|---|---|---|---|---|
| common cIMT | rs2460824 | 0 | 519 | −0.03 (−0.04, −0.02) | <0.001 |
| 1 | 123 | 0.003 (−0.02, 0.03) | 0.831 | ||
| 2 | 8 | −0.07 (−1.66, 1.52) | 0.683 | ||
|
| |||||
| rs2468844 | 0 | 306 | −0.04 (−0.05, −0.02) | <0.001 | |
| 1 | 270 | −0.01 (−0.03, 0.04) | 0.129 | ||
| 2 | 74 | −0.02 (−0.06, 0.02) | 0.309 | ||
|
| |||||
| rs7130337 | 0 | 135 | −0.01 (−0.03, 0.01) | 0.466 | |
| 1 | 352 | −0.02 (−0.04, −0.01) | 0.004 | ||
| 2 | 196 | −0.04 (−0.06, −0.02) | 0.001 | ||
|
| |||||
| ln(internal cIMT) |
rs2460824 | 0 | 517 | −0.01 (−0.04, 0.01) | 0.216 |
| 1 | 122 | 0.03 (−0.01, 0.07) | 0.104 | ||
| 2 | 8 | 0.07 (−1.29, 1.42) | 0.654 | ||
SNPs for which the SNP*HDL interaction term was significant for the IMT outcome in AA (see Figure 1) are shown above. The significant interaction term (SNP*HDL) implies that the association between SAA genotype and IMT may differ by HDL level or similarly, the association between HDL and IMT may differ according to SAA genotype. This table shows the difference in mean IMT in mm per 10 mg/dL HDL by SAA genotype, using a separate model for each genotype. The three significant models (p-values shown in bold) suggest that IMT and HDL are negatively correlated as might be expected—for each 10 mg/dL increase in HDL level, IMT is slightly smaller.
Association between SAA1/2 SNPs and risk of CVD events
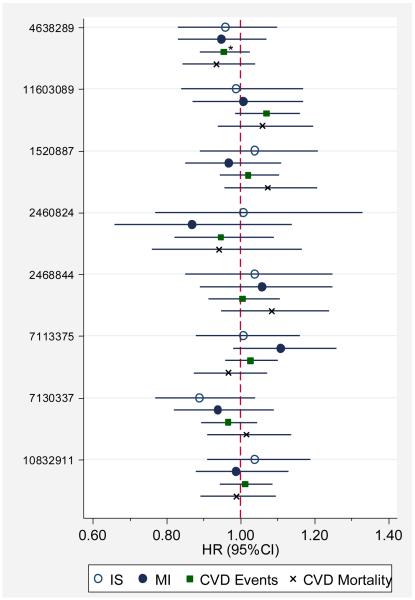
In EA, incidence rates of MI, ischemic stroke, CVD events, and CVD mortality were 12.2, 10.2, 44.6, and 17.2 events per 1000 person-years (py), respectively, during an approximate median follow-up of 13.6 years. In our analyses of effect modification of the relationship between SAA1/2 SNPs and events by HDL concentration or obesity status we found evidence of an interaction for obesity (p=0.011) so that among non-obese, each additional minor allele of rs4638289 was associated with a modestly reduced hazard of CVD events: HR=0.9 (95%CI: 0.8-0.99; p=0.026) and among obese, each additional allele was associated with an increased risk, HR=1.2 (95%CI: 0.98-1.4; p= 0.086). No other significant interactions or associations between common SAA1/2 SNPs and risk of events were observed.[Figure 2]
Figure 2. Association between SAA1/2 SNPs and Risk of CVD Events in European-Americans.
*Significant evidence (p=0.011) of interaction by obesity status; among obese, HR=1.2 (95%CI: 0.98-1.4; p=0.086) and among not obese, HR=0.9 (95%CI: 0.8-0.99; p=0.026)
In AA, 85 incident MIs (12.0 events per 1000 py) and 84 incident ischemic strokes (11.9 events per 1000 py) were observed; median follow-up time for both events was 12.1 years. Given the small number of events in AA, we did not model hazard ratios for SNP-event outcomes since power to detect an HR of 1.5 was low, estimated at approximately 44%.
Association between SAA1/2 SNPs and baseline HDL cholesterol
In cross-sectional analyses, we investigated whether genetic variation in SAA1/2 is associated with baseline HDL-cholesterol levels. In EA, SAA1 rs11603089 and SAA2 rs10832911 were associated with slightly lower HDL levels, β=−0.9 mg/dL (95%CI: −1.7, −0.1; p=0.028) and β =−0.7 mg/dL (95%CI: −1.4, −0.03; p=0.042), respectively.[Table 4] Similarly SAA1 rs1520887 was associated with slightly lower levels of HDL in AA, β =−1.7 mg/dL (95%CI: −3.2, −0.2; p=0.030).
Table 4.
Association of SAA1/2 SNPs with HDL-cholesterol
| European-Americans | African Americans | ||||
|---|---|---|---|---|---|
| Gene | SNP | MAF† | β in mg/dL (95%CI) | MAF† | β in mg/dL (95%CI) |
| SAA1 | rs4638289 | 0.330 | 0.6 (−0.1, 1.3) p=0.083 |
0.334 | −1.0 (−2.6, 0.6) p=0.215 |
| rs11603089 | 0.202 |
−0.9 (−1.7, −0.1)
p=0.028 |
0.085 | 1.0 (−2.0, 3.9) p=0.513 |
|
| SAA2 | rs1520887 | 0.242 | −0.5 (−1.2, 0.3) p=0.224 |
0.477 |
−1.7 (−3.2, −0.2)
p=0.030 |
| rs2460824 | 0.058 | 0.2 (−1.2, 1.6) p=0.780 |
0.117 | 0.1 (−2.5, 2.7) p=0.947 |
|
| rs2468844 | 0.139 | 0.02 (−0.9, 1.0) p=0.967 |
0.327 | −0.8 (−2.4, 0.8) p=0.338 |
|
| rs7113375 | 0.449 | 0.6 (−0.1, 1.3) p=0.080 |
0.239 | 0.1 (−1.9, 2.0) p=0.935 |
|
| rs7130337 | 0.245 | −0.2 (−0.9,0.5) p=0.603 |
0.535 | 0.1 (−1.5, 1.6) p=0.931 |
|
| rs7933280 | 0.033 | MAF<0.05 | 0.399 | −0.5 (−2.2, 1.3) p=0.612 |
|
| rs10832911 | 0.342 |
−0.7 (−1.4, −0.03)
p=0.042 |
0.500 | 0.04 (−1.5, 1.5) p=0.963 |
|
| rs16935244 | <0.01 | MAF<0.05 | 0.114 | 1.0 (−1.6, 3.6) p=0.443 |
|
MAF= minor allele frequency
Interaction between SAA1/2 and IL6, IL1, IL1RN and TNF SNPs on risk of CVD
Using a subset of EA with complete genotype data for all non-correlated SNPs (n=28), we had a total of n=3698 participants. Demographics for this subgroup were similar to the full cohort, participants had a mean age=73 ± 5.5 yrs. and were 57% female. In logic regression models, we first performed a signal test, which tests the null hypothesis that none of the predictors (SNPs) are associated with the outcome of interest. A significant finding, which we defined as p<0.20, suggests evidence of an association between the outcome and at least one SNP or combination of SNPs; subsequent model selection tests may be warranted. In the signal tests, we did not find evidence of association between SNPs and the outcomes of IMT, ischemic stroke, MI or total CVD events, signal test p-values>0.20.[Table 5] HDL and BMI adjusted results did not differ appreciably from unadjusted results. However, we did find suggestive evidence of an association between predictors and CVD mortality, signal test p-value=0.017, and pursued cross-validation and conditional permutation tests to identify the minimal set of variables that best modeled CVD mortality. Conditional permutation tests did not identify any significant models, all p≥0.30, but the three best models included SAA2-rs7933280, associated with increased risk of CVD mortality. The first model (p=0.300) included only a main effect term for the variable and the second model (p=0.300) included its main effect term as well as an interaction between IL1RN-rs315934 and IL1B-rs1143634. The third best model (p=0.400) included a main effect term for SAA2-rs1520887 and an interaction between SAA2-rs7933280 and IL1RN-rs4251961.
Table 5.
Logic Regression Signal Test Results for Within Gene and Gene-Gene Interactions between SAA1/2, IL1RN, IL1B, IL6, and TNF SNPs in Relation to Outcomes
| Race | Outcome | ‘p-value’ |
|---|---|---|
| AA | Common cIMT | 0.045 |
| ln(Internal cIMT) | 0.455 | |
| EA | Common cIMT | 0.375 |
| ln(Internal cIMT) | 0.315 | |
| Ischemic stroke | 0.560 | |
| Myocardial infarction | 0.285 | |
| CVD events | 0.545 | |
| CVD mortality | 0.017 |
‘P-value’ is a pseudo p-value representing the proportion of times model scores from randomized data are better than scores from the best model using the actual dataset, n=200 permutations. p <0.20 provides modest evidence against the Hϕ: no association between any predictors and the outcome. All models were adjusted for age, sex and site; AA models were also adjusted for %AA.
In AA, using a subset (n=630) of the population with complete data for all non-correlated SNPs (n=43), we found evidence of an association between SNPs and common cIMT, but not ln(internal cIMT), signal test p-values=0.045 and 0.455, respectively.[Table 5] Using model selection tools, we found the following three best models for common cIMT: 1) cIMT= −0.0506 * ≥ 1 minor allele of IL1RN-rs4251961; p=0.15, 2) cIMT=0.124 * (≥ 1 SAA2-rs7113375 minor allele and 2 IL1RN-rs315919 minor alleles); p=0.35, and 3) −0.124 * ((no minor alleles of SAA2-rs7113375) or (<2 IL1RN-rs315919 minor alleles));p=0.40. In the first model, having at least one copy of the IL1RN-rs4251961 minor allele was associated with lower common cIMT; this association was significant at p<0.20. The second and third models, which were not significant, involve the same intronic SNPs in SAA2 and IL1RN, namely SAA2-rs7113375 in a dominant genetic model and IL1RN-rs315919 in a recessive genetic model. These two models are logically equivalent according to DeMorgan’s Theorem.[23]
Discussion
We investigated the association of genetic variants in acute phase SAA genes and the risk of CVD. Only one of the common SNPs that we studied is known to have functional consequences, minor alleles of rs2468844 cause the substitution of the larger and more basic arginine amino acid residue for histidine at position 89 of the SAA2 protein.[24] Whether this substitution affects the stability or binding of the protein is not known, but interestingly the association of this SNP with cIMT appeared to be modified by HDL in our population. The remaining SNPs studied reflect common variation across the entire two gene locus including potential regulatory regions which may be important for initiation of the acute phase response. We did not find evidence that the SNPs are strongly associated with clinical events, though we did find modest associations with HDL and in AA, with cIMT. However, given the number of tests performed in this analysis, these novel findings would not be statistically significant after adjustment for multiple testing and should be regarded as hypothesis-generating until confirmed.
Using vaccination as an acute stimulus, we previously found more robust IL-6, CRP and SAA responses in men with severe carotid artery disease than in men without; though only the SAA response was significantly different between the two groups.[25] It remains unclear whether these differences are a result of severe disease or contributed to its development. However, genetic variation in the inflammatory response may influence its regulation and susceptibility to atherosclerosis,[26] which could ultimately predispose individuals to develop severe disease or conversely, to undergo healthy aging.[12] In this study, we investigate cIMT, a quantitative sub-clinical marker of atherosclerosis that has been positively associated with increased risk of cardiovascular events,[27, 28] though studies of its relation with IL-6,[29, 30] SAA,[29] or CRP[30, 31] levels report conflicting or weak associations. cIMT has been positively associated with the IL-6 -174G>C polymorphism[32] (rs1800795) which, in CHS EA, is highly correlated (r2~0.9) with and tags the same common haplotype as rs1554606 which was included in the current study.
We identified two SAA2 SNPs associated with increased cIMT in AA; similar non-significant trends for these SNPs were also seen in EA. Additionally, we identified SAA2 SNPs associated with decreased cIMT in AA only. One of these SNPs was not investigated in EA due to its low MAF; the other SNPs included the 3′ intergenic SNP rs2460824 and the non-synonymous variant rs2468844, both of which were in low LD in the AA population. These significant findings, limited to AA, should be interpreted cautiously given the smaller sample size, but differences in allele frequencies, interaction with other genes or environmental exposures, or variability/precision of the IMT measurement may account for our race-specific results. Our findings also suggest that genetic variants in SAA1/2 may be weakly associated with lower levels of HDL cholesterol in both EA and AA, though the race-specific variants differ. One possible explanation for this association may be explained by a recent study[33] indicating that SAA is capable of generating HDL using cellular lipid, a function that is mediated by ATP-binding cassette transporter proteins, ABCA1 or ABCA7. Furthermore, during the inflammatory state, HDL composition is altered as its apoA-I and apoA-II components are displaced by SAA in the form of predominately SAA1 protein (>70%) and to a lesser extent, SAA2 protein.[34, 35] Though it is possible that they have different binding affinities for HDL[36], SAA1 and SAA2 are 93% identical in their primary structures and little is known about their functional differences.[37] Since apoA-I plays a central role in reverse cholesterol transport and thus may be protective against atherosclerosis, its displacement by SAA is viewed as potentially pro-atherosclerotic. An additional lipid-related function of SAA is that it binds cholesterol and promotes its cellular uptake; SAA-HDL particles have a higher affinity for macrophages and lower affinity for hepatocytes than HDL,[4] suggesting that they may be more likely to be incorporated into plaques. Once attached to HDL, SAA also acts as a chemo-attractant of immune cells,[4] thus contributing to local inflammation and potentially promoting atherosclerotic plaque destablization.[38] We speculated that variation in SAA1/2 could lead to altered binding affinity of the SAA proteins for HDL and hypothesized that the association between SAA1/2 genetic variants and CVD may be modified by HDL. In AA only, we found modest evidence of interaction between SAA SNPs and HDL on the outcome of cIMT; these SNPs were not the same SNPs found to be associated with HDL level. Furthermore, the clinical significance of such effect modification may not be meaningful since differences in the slopes describing the relationship between HDL and cIMT by SAA genotype were very small.
A recent analysis of the Atherosclerosis Risk in Communities (ARIC) cohort demonstrates that aggregating information from multiple SNPs into a risk score can be used to improve prediction of incident coronary heart disease.[39] Similarly, we hypothesized that a consideration of SNP combinations may be useful in identifying risk variants that might not be found in single SNP analyses. This approach may better reflect the underlying biological complexity of inflammation and atherosclerosis than traditional tests of individual SNPs and multiplicative interaction terms. For example, Hagihara et al. found that either IL-6 and IL-1β, or IL-6 and TNF-α, but not IL-1β and TNF-α resulted in the synergistic induction of SAA1 and SAA2 genes in vitro.[40] Such a relationship is easily screened using the Boolean expressions in logic regression, but may be overlooked using a traditional approach in linear regression model building. Although we did not find robust evidence of gene-gene interaction among the SAA genes and major genes modulating its expression and CVD, we did identify a combination of two SNPs (one SAA2 and one IL1RN) potentially associated with IMT in AA which may deserve further investigation. One of these, IL1RN-rs4251961 has been previously reported to be associated with the systemic inflammation phenotype in this cohort and other cohorts; specifically, it was associated with higher CRP and IL-6 levels and with reduced cellular IL-1 receptor antagonist (IL-1RA) production ex vivo in response to an inflammatory stimulus.[41] These findings are consistent with its biological role as a pro-inflammatory protein.[42]
Several limitations of our study deserve mention. We can not rule out chance as a possible explanation for our findings, though our logic regression analyses are perhaps more robust to type 1 error, given our use of permutation and cross-validation tests in model selection. Additionally, our investigation was limited to tagSNPs representing common variation in the genes; we did not specifically investigate rare variants or target structural variants such as insertions or deletions which may also be associated with outcomes. We do however, report findings from one rare SAA2 variant rs7933280 included in our logic regression analyses only, which was associated with CVD mortality in our EA population. It is unknown whether SAA levels are associated with prevalent CVD or predictive of future events in this older population, since plasma measurements were not available. Similarly, associations between SAA levels and regulatory SNPs could also not be assessed; though a promoter polymorphism in the TNF-α gene associated with increased levels of SAA has been previously reported.[43] Furthermore, the impact of genetic variation on CVD risk as well as the underlying pathophysiological mechanisms of MI or stroke may vary in older populations relative to younger ones[44, 45] so our results may not be applicable in younger populations.
We hypothesized that the acute phase response may contribute to CVD, just as chronic systemic inflammation is a likely contributor. To our knowledge this is the first study to investigate common allelic variants in the SAA1 and SAA2 genes and their association with CVD. Our analysis of the acute phase response ‘pathway’ involving key regulators of SAA expression and its association with CVD in a human population is also novel. The CHS cohort, well characterized with respect to cardiovascular endpoints, allowed us to investigate a number of cardiovascular outcomes ranging from sub-clinical disease to the more severe non-fatal and fatal clinical events. Furthermore, we were able to reduce disease heterogeneity by restricting incident events to relevant subcategories; for example, we investigated ischemic stroke as an outcome since atherosclerosis is a potential underlying cause.
Inflammation is an important contributor to atherosclerosis and gene variants mediating inflammation are of interest. Regulation of levels of acute phase proteins in addition to their structure and function may be important in disease risk; our findings in this older population weakly support this hypothesis and may warrant further research. Overall, we expect that data regarding SAA, a key acute phase reactant may contribute to future research on the regulation of the inflammatory response and its role in CVD.
Acknowledgements
The research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and grant number U01 HL080295 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. CLC received support from the NHLBI grant, T32 HL007902. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the National Institutes of Health.
References
- (1).Upragarin N, Landman WJM, Gaastra W, Gruys E. Extrahepatic production of acute phase serum amyloid A. Histol Histopathol. 2005;20:1295–307. doi: 10.14670/HH-20.1295. [DOI] [PubMed] [Google Scholar]
- (2).Zhao Y, Zhou S, Heng C-K. Impact of serum amyloid A on tissue factor and tissue factor pathway inhibitor expression and activity in endothelial cells. Arterioscler Thromb Vasc Biol. 2007;27:1645–50. doi: 10.1161/ATVBAHA.106.137455. [DOI] [PubMed] [Google Scholar]
- (3).Jahangiri A, de Beer MC, Noffsinger V, Tannock LR, Ramaiah C, Webb NR, van der Westhuyzen DR, de Beer FC. HDL remodeling during the acute phase response. Arterioscler Thromb Vasc Biol. 2008 doi: 10.1161/ATVBAHA.108.178681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–23. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- (5).Schillinger M, Exner M, Mlekusch W, Sabeti S, Amighi J, Nikowitsch R, Timmel E, Kickinger B, Minar C, Pones M, Lalouschek W, Rumpold H, Maurer G, Wagner O, Minar E. Inflammation and Carotid Artery Risk for Atherosclerosis study (ICARAS) Circulation. 2005;111:2203–9. doi: 10.1161/01.CIR.0000163569.97918.C0. [DOI] [PubMed] [Google Scholar]
- (6).Fyfe AI, Rothenberg LS, DeBeer FC, Cantor RM, Rotter JI, Lusis AJ. Association between serum amyloid A proteins and coronary artery disease: Evidence from two distinct arteriosclerotic processes. Circulation. 1997;96:2914–9. doi: 10.1161/01.cir.96.9.2914. [DOI] [PubMed] [Google Scholar]
- (7).Johnson BD, Kip KE, Marroquin OC, Ridker PM, Kelsey SF, Shaw LJ, Pepine CJ, Sharaf B, Merz CN Bairey, Sopko G, Olson MB, Reis SE. Serum amyloid A as a predictor of coronary artery disease and cardiovascular outcome in women: The National Heart, Lung, and Blood Institute-sponsored Women’s Ischemia Syndrome Evaluation (WISE) Circulation. 2004;109:726–32. doi: 10.1161/01.CIR.0000115516.54550.B1. [DOI] [PubMed] [Google Scholar]
- (8).Danesh J, Whincup P, Walker M, Lennon L, Thomson A, Appleby P, Gallimore JR, Pepys MB. Low grade inflammation and coronary heart disease: Prospective study and updated meta-analyses. BMJ. 2000;321:199–204. doi: 10.1136/bmj.321.7255.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chait A, Han C, Oram J, Heinecke J. Thematic review series: The immune system and atherogenesis. Lipoprotein-associated inflammatory proteins: Markers or mediators of cardiovascular disease. J Lipid Res. 2005;46:389–403. doi: 10.1194/jlr.R400017-JLR200. [DOI] [PubMed] [Google Scholar]
- (10).MacGregor AJ, Gallimore JR, Spector TD, Pepys MB. Genetic effects on baseline values of C-reactive protein and serum amyloid A protein: A comparison of monozygotic and dizygotic twins. Clin Chem. 2004;50:130–4. doi: 10.1373/clinchem.2003.028258. [DOI] [PubMed] [Google Scholar]
- (11).Whitehead AS, de Beer MC, Steel DM, Rits M, Lelias JM, Lane WS, de Beer FC. Identification of novel members of the serum amyloid A protein superfamily as constitutive apolipoproteins of high density lipoprotein. J Biol Chem. 1992;267:3862–7. [PubMed] [Google Scholar]
- (12).Candore G, Vasto S, Colonna-Romano G, Lio D, Caruso M, Rea IM, Caruso C. Atherosclerosis. In: Vandenbroeck K, editor. Cytokine Gene Polymorphisms in Multifactorial Diseases. CRC Press, Taylor & Francis Group; Boca Raton, FL: 2006. [Google Scholar]
- (13).Baba S, Masago SA, Takahashi T, Kasama T, Sugimura H, Tsugane S, Tsutsui Y, Shirasawa H. A novel allelic variant of serum amyloid A, SAA 1 gamma: Genomic evidence, evolution, frequency, and implication as a risk factor for reactive systemic a-amyloidosis. Hum Mol Genet. 1995;4:1083–7. doi: 10.1093/hmg/4.6.1083. [DOI] [PubMed] [Google Scholar]
- (14).Kooperberg C, Ruczinski I, LeBlanc ML, Hsu L. Sequence analysis using logic regression. Genet Epidemiol. 2001;21(S1):S626–31. doi: 10.1002/gepi.2001.21.s1.s626. [DOI] [PubMed] [Google Scholar]
- (15).Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TM, Mittelmark MB, Newman A, O’Leary DH, Psaty B, Rautaharju P, Tracy RT, Weiler PG. The Cardiovascular Health Study: Design and rationale. Ann Epidemiol. 1991;1:263–76. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- (16).Cushman M, Cornell ES, Howard PR, Bovill EG, Tracy RP. Laboratory methods and quality assurance in the Cardiovascular Health Study. Clin Chem. 1995;41:264–70. [PubMed] [Google Scholar]
- (17).Macy E, Hayes T, Tracy R. Variability in the measurement of C-reactive protein in healthy subjects: Implication for reference interval and epidemiological applications. Clin Chem. 1997;43:52–8. [PubMed] [Google Scholar]
- (18).O’Leary DH, Polak JF, Wolfson SK, Jr., Bond MG, Bommer W, Sheth S, Psaty BM, Sharrett AR, Manolio TA. Use of sonography to evaluate carotid atherosclerosis in the elderly. The Cardiovascular Health Study. CHS Collaborative Research Group. Stroke. 1991;22:1155–63. doi: 10.1161/01.str.22.9.1155. [DOI] [PubMed] [Google Scholar]
- (19).Ives DG, Fitzpatrick AL, Bild DE, Psaty BM, Kuller LH, Crowley PM, Cruise RG, Theroux S. Surveillance and ascertainment of cardiovascular events. The Cardiovascular Health Study. Ann Epidemiol. 1995;5:278–85. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- (20).SeattleSNPs . NHLBI program for genomic applications. Seattle SNPs; Seattle, WA: [Accessed September 2006]. URL: http://pga.gs.washington.edu/ [Google Scholar]
- (21).Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM, Kwok PY. Population structure, admixture, and aging-related phenotypes in African American adults: The Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–77. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Whitesitt JE. Boolean algebra and its applications. Dover Publications; Mineola, N.Y.: 1995. [Google Scholar]
- (24).Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Carty CL, Heagerty P, Nakayama K, McClung EC, Lewis J, Lum D, Boespflug E, McCloud-Gehring C, Soleimani BR, Ranchalis J, Bacus TJ, Furlong CE, Jarvik GP. Inflammatory response after influenza vaccination in men with and without carotid artery disease. Arterioscler Thromb Vasc Biol. 2006;26:2738–44. doi: 10.1161/01.ATV.0000248534.30057.b5. [DOI] [PubMed] [Google Scholar]
- (26).Elkind MS, Cole JW. Do common infections cause stroke? Semin Neurol. 2006;26:88–99. doi: 10.1055/s-2006-933312. [DOI] [PubMed] [Google Scholar]
- (27).Chambless LE, Folsom AR, Clegg LX, Sharrett AR, Shahar E, Nieto FJ, Rosamond WD, Evans G. Carotid wall thickness is predictive of incident clinical stroke: The Atherosclerosis Risk in Communities (ARIC) study. Am J Epidemiol. 2000;151:478–87. doi: 10.1093/oxfordjournals.aje.a010233. [DOI] [PubMed] [Google Scholar]
- (28).Chambless LE, Heiss G, Folsom AR, Rosamond W, Szklo M, Sharrett AR, Clegg LX. Association of coronary heart disease incidence with carotid arterial wall thickness and major risk factors: The Atherosclerosis Risk in Communities (ARIC) study, 1987-1993. Am J Epidemiol. 1997;146:483–94. doi: 10.1093/oxfordjournals.aje.a009302. [DOI] [PubMed] [Google Scholar]
- (29).Wohlin M, Helmersson J, Sundström J, Ärnlöv J, Vessby B, Larsson A, Andrén B, Lind L, Basu S. Both cyclooxygenase- and cytokine-mediated inflammation are associated with carotid intima-media thickness. Cytokine. 2007;38:130–6. doi: 10.1016/j.cyto.2007.05.014. [DOI] [PubMed] [Google Scholar]
- (30).Walston JD, Fallin MD, Cushman M, Lange L, Psaty B, Jenny N, Browner W, Tracy R, Durda P, Reiner A. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the cardiovascular health study. Hum Genet. 2007;122:485–94. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- (31).Lange LA, Carlson CS, Hindorff LA, Lange EM, Walston J, Durda JP, Cushman M, Bis JC, Zeng D, Lin D, Kuller LH, Nickerson DA, Psaty BM, Tracy RP, Reiner AP. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. Jama. 2006;296:2703–11. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- (32).Rauramaa R, Väisänen SB, Luong L, Schmidt-Trücksäss A, Penttilä IM, Bouchard C, Töyry J, Humphries SE. Stromelysin-1 and interleukin-6 gene promoter polymorphisms are determinants of asymptomatic carotid artery atherosclerosis. Arterioscler Thromb Vasc Biol. 2000;20:2657–62. doi: 10.1161/01.atv.20.12.2657. [DOI] [PubMed] [Google Scholar]
- (33).Abe-Dohmae S, Kato KH, Kumon Y, Hu W, Ishigami H, Iwamoto N, Okazaki M, Wu CA, Tsujita M, Ueda K, Yokoyama S. Serum amyloid A generates high density lipoprotein with cellular lipid in an ABCA1- or ABCA7-dependent manner. J Lipid Res. 2006;47:1542–50. doi: 10.1194/jlr.M600145-JLR200. [DOI] [PubMed] [Google Scholar]
- (34).Kluve-Beckerman B, Dwulet FE, Benson MD. Human serum amyloid a. Three hepatic mRNAs and the corresponding proteins in one person. J Clin Invest. 1988;82:1670–5. doi: 10.1172/JCI113779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Strachan AF, de Beer FC, van der Westhuyzen DR, Coetzee GA. Identification of three isoform patterns of human serum amyloid A protein. Biochem J. 1988;250:203–7. doi: 10.1042/bj2500203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang L, Colon W. The interaction between apolipoprotein serum amyloid A and high-density lipoprotein. Biochem Biophys Res Commun. 2004;317:157–61. doi: 10.1016/j.bbrc.2004.03.027. [DOI] [PubMed] [Google Scholar]
- (37).Xu Y, Yamada T, Satoh T, Okuda Y. Measurement of serum amyloid A1 (SAA 1), a major isotype of acute phase saa. Clin Chem Lab Med. 2006;44:59–63. doi: 10.1515/CCLM.2006.012. [DOI] [PubMed] [Google Scholar]
- (38).Johnson BA, Fram EK, Dayer BP, Dean BL, Keller PJ, Jacobowitz R. Evaluation of shared-view acquisition using repeated echoes (SHARE): A dual-echo fast spin-echo mr technique. AJNR Am J Neuroradiol. 1994;15:667–73. [PMC free article] [PubMed] [Google Scholar]
- (39).Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, Pankow JS, Devlin JJ, Willerson JT, Boerwinkle E. Prediction of coronary heart disease risk using a genetic risk score: The atherosclerosis risk in communities study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- (40).Hagihara K, Nishikawa T, Isobe T, Song J, Sugamata Y, Yoshizaki K. Il-6 plays a critical role in the synergistic induction of human serum amyloid a (SAA) gene when stimulated with proinflammatory cytokines as analyzed with an SAA isoform real-time quantitative rt-PCR assay system. Biochemical and Biophysical Research Communications. 2004;314:363–9. doi: 10.1016/j.bbrc.2003.12.096. [DOI] [PubMed] [Google Scholar]
- (41).Reiner AP, Wurfel MM, Lange LA, Carlson CS, Nord AS, Carty CL, Rieder MJ, Desmarais C, Jenny NS, Iribarren C, Walston JD, Williams OD, Nickerson DA, Jarvik GP. Polymorphisms of the il1-receptor antagonist gene (IL1RN) are associated with multiple markers of systemic inflammation. Arterioscler Thromb Vasc Biol. 2008;28:1407–12. doi: 10.1161/ATVBAHA.108.167437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Arend WP. The balance between IL-1 and IL-1RA in disease. Cytokine & Growth Factor Reviews. 2002;13:323–40. doi: 10.1016/s1359-6101(02)00020-5. [DOI] [PubMed] [Google Scholar]
- (43).Gonzalez S, Rodrigo L, Martinez-Borra J, Lopez-Vazquez A, Fuentes D, Nino P, Cadahia V, Saro C, Dieguez MA, Lopez-Larrea C. TNF-alpha-308a promoter polymorphism is associated with enhanced tnf-alpha production and inflammatory activity in crohn’s patients with fistulizing disease. The American Journal of Gastroenterology. 2003;98:1101–6. doi: 10.1111/j.1572-0241.2003.07416.x. [DOI] [PubMed] [Google Scholar]
- (44).Choudhury L, Marsh JD. Myocardial infarction in young patients. Am J Med. 1999;107:254–61. doi: 10.1016/s0002-9343(99)00218-1. [DOI] [PubMed] [Google Scholar]
- (45).Carr FJ, McBride MW, Carswell HV, Graham D, Strahorn P, Clark JS, Charchar FJ, Dominiczak AF. Genetic aspects of stroke: Human and experimental studies. J Cereb Blood Flow Metab. 2002;22:767–73. doi: 10.1097/00004647-200207000-00001. [DOI] [PubMed] [Google Scholar]