Abstract
Background
Biochemical diagnostics of ethanol intake would improve alcohol abuse treatment and have applications in clinical trial and public safety settings. Self-reporting of alcohol use has clinical utility, but lacks the desired reliability. Previously proposed single-analyte biochemical tests of alcohol intake suffer from low sensitivity and specificity or examine only acute drinking and have therefore seen limited clinical use.
Methods
To address this unmet need, plasma protein biomarker discovery and validation were performed using an alcohol self-administering non-human primate model system to develop a diagnostic that accurately classifies subjects into non-drinking, non-abusive drinking, and abusive drinking categories.
Results
A 17 plasma protein panel was determined that correctly classifies abusive drinking with 100% sensitivity and also differentiates any level of drinking from alcohol abstinence with 88% accuracy.
Conclusions
The biomarker panel reflects changes in multiple organ systems and suggests robust changes in the plasma proteome with drinking that may serve as a sensitive and specific diagnostic test. The specific plasma proteins altered with alcohol self-administration may represent indicators of alcohol-induced stress on a variety of organ systems.
Keywords: Alcoholism, Alcohol Abuse, Proteomics, Biomarker, Diagnostic, Non-human primate
Introduction
Alcohol abuse and alcoholism exact a tremendous cost on society. In economic terms, over $170,000,000 is lost each year to the effects of excessive drinking (1). This economic burden is in addition to well-documented emotional and personal costs. Consumption of alcohol in the U.S. is unevenly distributed, with 64% of the adult population actively drinking alcohol but 20% of the drinking population consuming approximately 80% of all alcohol sold. Unfortunately, the clinical treatment community lacks reliable biomarkers to monitor at-risk populations such as recovering alcoholics, pregnant women, and critical members of the community (e.g., public transportation employees, active duty military, and healthcare providers) (2). Self-reported alcohol use can be extremely informative but, in some scenarios (e.g., social stigma associated with alcohol abuse and the potential legal/social ramifications of frank alcoholism), can be inconsistent (3;4). Indeed, there is strong motivation for some subjects to deny drinking (5). This has led to the search for objective biochemical markers of alcohol abuse. Several biochemical markers of alcoholism have been proposed (6;7), but there are limitations to the accuracy and sensitivity of these assays (8–10). To overcome the limitations of these unitary analyte diagnostics, we performed a screen of 90 known plasma proteins and used machine learning algorithms to develop a panel of biomarkers with high sensitivity and specificity.
Non-human primates serve as a valuable model system for studies of the effects of alcohol. Macaque neuroanatomy, neurochemistry, and physiology are similar to humans and are subject to similar disease processes (11). Monkeys display alcohol absorption and metabolism pharmacokinetics that are similar to human beings (12) and display commonalities with changes in human plasma proteins (13), while simultaneously offering a well controlled experimental model. This study used a non-human primate model of chronic ethanol exposure where animals self-administer large quantities of ethanol continuously over 12–18 months (14–17).
Methods and Materials
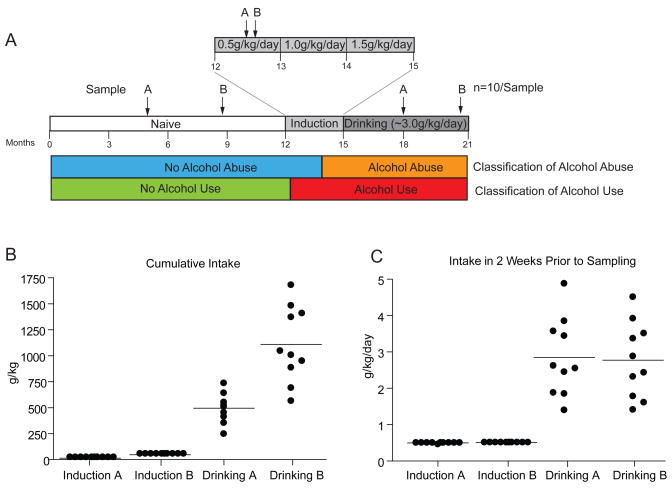
Ten male cynomolgus monkeys (macaca fascicularis), were part of a 21-month experimental time line (Figure 1A). For the first year (Naïve samples), monkeys were acclimatized to the study environment and operant instrumentation. Monkeys were induced to consume liquids under a schedule of food pellet deliveries (i.e., schedule-induced polydipsia (18)) as described previously (15). Following one month of 0.5 g/kg/day ethanol (two drink equivalents, Induction samples), the animals were escalated to drink 1.0 g/kg/day for 30 consecutive days, and finally, 1.5 g/kg/day for 30 consecutive days. Following the 90-day induction period of alcohol consumption, animals were given unlimited access (22 hours per day) to either ethanol or water for the next six months (Drinking samples) (15;17). Two independent samples (A and B) were collected on different days from each state in the experimental time line (Figure 1A).
Figure 1.
(A) Time course of non-human primate alcohol self-administration and plasma sampling points. Two independent samples (A and B) were collected from each condition (Naïve, Induction, and Drinking) for biomarker discovery and classification analysis testing. (B) The cumulative self-administered intake of the monkeys at Induction and Drinking time points. (C) In the two weeks prior to plasma sampling time points, the average daily intake was the same for A and B samples in the Induction or Drinking states. Data are presented with mean lines and dots for each animal and time point (n=10 for each sample).
Plasma protein profiling was performed at Rules-Based Medicine, Inc. (Austin, Texas) using standard Luminex technology. Differential abundance of individual plasma proteins was determined using a conservative approach with a One-Way repeated measures ANOVA and Bonferroni multiple testing correction (p<0.05). To identify the most consistent plasma protein changes, only those differences significant by a Student Newman-Keuls pair-wise post-hoc test (p<0.05) between two time points and for both the A and B samples were considered. Support Vector Machine (SVM) classification analysis was performed using GeneSpring 7.3 (Agilent) with a polynomial kernel function and no scaling factor (Figure S1). Database searching for tissue origin of proteins was performed using Ingenuity Pathway Analysis software (Ingenuity, Redwood City CA). Additional methodological details see presented in the Supplementary Methods.
Results
Ethanol Self-Administration
The cumulative intake for these animals at the collection time points is presented in Figure 1B. With chronic and compulsive drinking, there was naturally a higher total level of total consumption. During the 2 weeks prior to sample collection for the A and B Drinking samples, however, the level of alcohol consumption was not significantly different (Figure 1C). There was a tightly controlled 0.5 g/kg/day (2 drink equivalents per day) consumption during the initial month of ethanol induction which resulted in blood ethanol levels between 20–40 mg/dL depending on the drinking typography (15). The two Drinking time points (following 3 and 6 months of 22/hr/day access) had essentially the same amount of ethanol intake over the two weeks preceding sample collection when given unlimited assess (ca. 3 g/kg/day; approximately 12 drink equivalents per day).
Biomarker Analysis
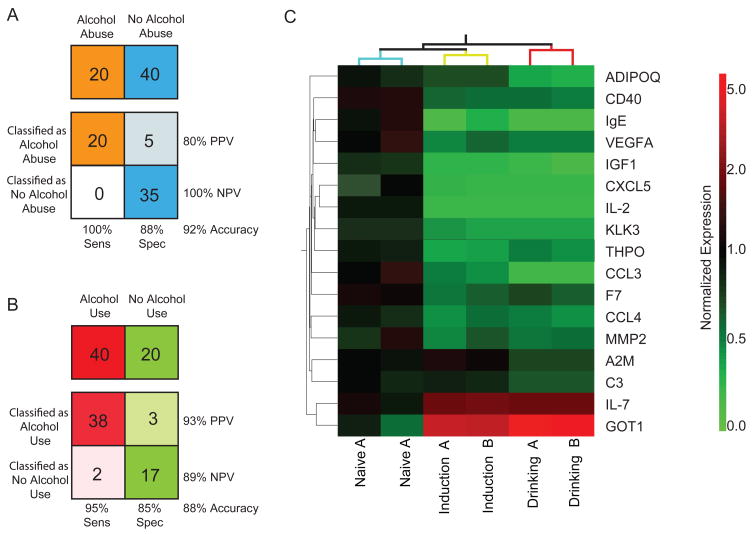
Initial biomarker discovery was conducted by multiplex Luminex analysis of 90 known plasma cytokines, growth factors, and other proteins (Table S1). Two independently collected samples (A and B) were analyzed for each of the three drinking states (Naïve, Induction, Drinking) (Figure 1A) to identify the most consistently altered proteins. Sixty-one of the 90 proteins were present at detectable levels. Correlation of ethanol consumption with the level of protein concentration changes did not reveal significant associations (data not shown). Proteins to be included in the biomarker panel were selected based on statistically-significant differences in protein abundance between both A and B samples in at least two of the three pair-wise drinking state comparisons (Figure S2). The 17 proteins that met this criterion were tested for their classification accuracy using a Support Vector Machine (SVM) algorithm. Using a cross-validation approach, alcohol abuse was correctly classified with 92% accuracy (Drinking vs. Naïve and Induction) (Figure 2A) using a three protein biomarker set [adiponectin (ADIPOQ), alpha-2-macroglobulin (A2M), complement component 3 (C3)]. Alcohol use (Induction and Drinking samples) was correctly classified with 88% accuracy from non-alcohol using samples (Naïve) (Figure 2B) using a 14 protein biomarker panel [(CD40, chemokine ligand 5 (CXCL5), Factor VII (F7), IgE, IGF1, interleukin 2 (IL2), interleukin 7 (IL7), chemokine (C-C motif) ligand 3 (CCL3), chemokine (C-C motif) ligand 4 (CCL4), matrix metallopeptidease 2 (MMP2), kallikrein-related peptidase 3 (KLK3), glutamic-oxaloacetic transaminase 1 (GOT1), thrombopoietin (THPO), vascular endothelial growth factor A (VEGFA)]. The combined biomarker panel is presented in Figure 2C. While additional data sets will need to be collected before truly independent Training Set/Test Set analysis can be conducted, using A samples as a Training Set and B samples as a Test Set, the accuracy of the alcohol abuse and alcohol use classifications were 87% and 97% respectively (Figure S3). Reversal of the training and test sets returned similar results and use of an alternate algorithm (K-means nearest neighbor) resulted in similar accuracy (data not shown).
Figure 2.
(A) Using a three protein panel and Support Vector Machine (SVM) classification algorithm, alcohol abuse samples (Drinking) were correctly classified from non-alcohol abusing samples (Naïve and Induction) with 92% accuracy and 100% sensitivity. (B) Using a fourteen protein panel and SVM classification algorithm, alcohol using samples (Induction and Drinking) were correctly classified from non-alcohol using samples (Naïve) with 88% accuracy and 95% sensitivity. (C) Heatmap representation of the seventeen protein biomarker panel and clustering of the sample groups. Mean expression levels, normalized to a mean naïve level of 1, for each drinking state and time point were clustered by condition. Increased abundance, compared to mean Naive, is presented in red and reduced abundance in green. Independent samples from each drinking state clustered together.
Discussion
The combined set of 17 differentially-regulated plasma proteins could potentially be used as a diagnostic to differentiate subjects into non-drinking, non-abusive drinking, and abusive drinking categories. This panel includes both inductions and reductions in protein levels (Figure 2C). Notably, several of these proteins have been previously reported to be responsive to alcohol intake. We have previously described ApoAI induction in non-human primates (13) and this has been observed in humans as well (19). GOT1 is well known to be induced in humans with alcohol consumption and reduced IGF1 plasma levels have been observed in rodent studies (20). In general, this panoply of circulating proteins may represent indicators of alcohol-induced stress. That is, specific proteins that are altered may reflect well-known clinical sequela of alcohol consumption such as cardiovascular disease (F7, THPO), liver disease (ADIPOQ, GOT1), central and peripheral nervous system damage (IGF1), osteoporosis (MMP2), and immune deficiency (A2M, C3, CCL3, CCL4, CD40, CXCL5, IgE, IL-2, IL-7). Subsets of proteins from the biomarker panel may also have application as indicators of tissue damage to specific organs. It is important to note that many proteins comprising the diagnostic panel do not originate solely or at all from the liver (Table S2). A common problem for liver-enzyme based diagnostics to alcohol abuse is that they are not specific for alcohol abuse, but rather indicate general liver dysfunction. By using a panel of proteins from different tissue sources, a potential test is less likely to return false-positive for other disease states.
Future studies will require examination of the biomarker panel’s persistence with cessation of ethanol self-administration. This is an important step in the development of a clinical diagnostic, as the biomarker panel should return to a normal state with alcohol abstinence. It remains possible that the observed changes may not normalize with alcohol abstinence, which would indicate long-lasting organ damage or epigenetic changes. Additionally, these findings must be replicated in a clinical setting to examine: 1) the classification ability of the biomarker panel in the heterogeneous (e.g., diet, genetics, co-morbid drug use) human population; 2) the duration of alcohol abuse required to initiate the biomarker signature; and 3) the persistence of the these plasma protein changes with cessation of drinking. While this study pursued a directed biomarker discovery approach, additional studies using proteome-wide methods may identify additional proteins for development as biomarkers.
This study demonstrates that a panel of 17 plasma proteins can be used to accurately differentiate non-human primates into non-drinking, non-abusive drinking, and abusive drinking categories. This model system provides the closest animal model to human alcoholism with similar physiology and durations of drinking. Quantitative analysis of plasma samples by the Luminex measurement technology is well-suited for direct translation into the clinical setting without additional technical optimization. This biomarker panel will now require testing in human subjects to establish its clinical utility.
Supplementary Material
Acknowledgments
This study was supported by grants from the US National Institutes of Health (AA016613 to K.E.V., and AA11997, AA13510 and AA13641 to K.A.G).
Footnotes
Financial Disclosures
Drs. Freeman, Vrana, and Grant, and Mr. Gonzales report a patent application on diagnostics of alcohol intake in preparation. Ms. Salzberg reports no biomedical financial interests or potential conflicts of interest.
References
- 1.Harwood H. Updating Estimates of the Economic Costs of Alcohol Abuse in the United States: Estimates, Update Methods, and Data. Rockville, MD: National Institute on Alcohol Abuse and Alcoholism; 2000. [Google Scholar]
- 2.Hannuksela ML, Liisanantti MK, Nissinen AE, Savolainen MJ. Biochemical markers of alcoholism. Clin Chem Lab Med. 2007;45:953–961. doi: 10.1515/CCLM.2007.190. [DOI] [PubMed] [Google Scholar]
- 3.Phillips M. Sweat-patch testing detects inaccurate self-reports of alcohol consumption. Alcohol Clin Exp Res. 1984;8:51–53. doi: 10.1111/j.1530-0277.1984.tb05032.x. [DOI] [PubMed] [Google Scholar]
- 4.Toneatto T, Sobell LC, Sobell MB. Predictors of alcohol abusers’ inconsistent self-reports of their drinking and life events. Alcohol Clin Exp Res. 1992;16:542–546. doi: 10.1111/j.1530-0277.1992.tb01414.x. [DOI] [PubMed] [Google Scholar]
- 5.Pernanen K. Validity of survey data on alcohol use. In: Gibbons, et al., editors. Research Advances in Alcohol and Drug Problems. New York: 1974. pp. 355–374. [Google Scholar]
- 6.Haber H, Jahn H, Ehrenreich H, Melzig MF. Assay of salsolinol in peripheral blood mononuclear cells of alcoholics and healthy subjects by gas chromatography-mass spectrometry. Addict Biol. 2002;7:403–407. doi: 10.1080/1355621021000005991. [DOI] [PubMed] [Google Scholar]
- 7.Helander A. Biological markers in alcoholism. J Neural Transm Suppl. 2003:15–32. doi: 10.1007/978-3-7091-0541-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Conigrave KM, Degenhardt LJ, Whitfield JB, Saunders JB, Helander A, Tabakoff B. CDT, GGT, and AST as markers of alcohol use: the WHO/ISBRA collaborative project. Alcohol Clin Exp Res. 2002;26:332–339. [PubMed] [Google Scholar]
- 9.Anton RF, Lieber C, Tabakoff B. Carbohydrate-deficient transferrin and gamma-glutamyltransferase for the detection and monitoring of alcohol use: results from a multisite study. Alcohol Clin Exp Res. 2002;26:1215–1222. doi: 10.1097/01.ALC.0000023986.42254.F5. [DOI] [PubMed] [Google Scholar]
- 10.Alte D, Ludemann J, Piek M, Adam C, Rose HJ, John U. Distribution and dose response of laboratory markers to alcohol consumption in a general population: results of the study of health in Pomerania (SHIP) J Stud Alcohol. 2003;64:75–82. doi: 10.15288/jsa.2003.64.75. [DOI] [PubMed] [Google Scholar]
- 11.Grant KA, Bennett AJ. Advances in nonhuman primate alcohol abuse and alcoholism research. Pharmacol Ther. 2003;100:235–255. doi: 10.1016/j.pharmthera.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Green KL, Szeliga KT, Bowen CA, Kautz MA, Azarov AV, Grant KA. Comparison of ethanol metabolism in male and female cynomolgus macaques (Macaca fascicularis) Alcohol Clin Exp Res. 1999;23:611–616. [PubMed] [Google Scholar]
- 13.Freeman WM, Gooch RS, Lull ME, Worst TJ, Walker SJ, Xu AS, et al. Apo-aii is an elevated biomarker of chronic non-human primate ethanol self-administration. Alcohol Alcohol. 2006;41:300–305. doi: 10.1093/alcalc/agl021. [DOI] [PubMed] [Google Scholar]
- 14.Floyd DW, Friedman DP, Daunais JB, Pierre PJ, Grant KA, McCool BA. Long-term Ethanol Self-administration by Cynomolgus Macaques Alters the Pharmacology and Expression of GABA(A) Receptors Expressed in Basolateral Amygdala. J Pharmacol Exp Ther. 2004 doi: 10.1124/jpet.104.072025. [DOI] [PubMed] [Google Scholar]
- 15.Grant KA, Leng X, Green HL, Szeliga KT, Rogers LS, Gonzales SW. Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcohol Clin Exp Res. 2008;32:1824–1838. doi: 10.1111/j.1530-0277.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivester P, Roberts LJ, Young T, Stafforini D, Vivian J, Lees C, et al. Ethanol self-administration and alterations in the livers of the cynomolgus monkey, Macaca fascicularis. Alcohol Clin Exp Res. 2007;31:144–155. doi: 10.1111/j.1530-0277.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 17.Vivian JA, Green HL, Young JE, Majerksy LS, Thomas BW, Shively CA, et al. Induction and maintenance of ethanol self-administration in cynomolgus monkeys (Macaca fascicularis): long-term characterization of sex and individual differences. Alcohol Clin Exp Res. 2001;25:1087–1097. [PubMed] [Google Scholar]
- 18.Falk D. Production of polydipsia in normal rats by an intermittent food schedule. Science. 1961;133:195–196. doi: 10.1126/science.133.3447.195. [DOI] [PubMed] [Google Scholar]
- 19.Gottrand F, Beghin L, Duhal N, Lacroix B, Bonte JP, Fruchart JC, et al. Moderate red wine consumption in healthy volunteers reduced plasma clearance of apolipoprotein AII. Eur J Clin Invest. 1999;29:387–394. doi: 10.1046/j.1365-2362.1999.00484.x. [DOI] [PubMed] [Google Scholar]
- 20.Sonntag WE, Boyd RL. Diminished insulin-like growth factor-1 levels after chronic ethanol: relationship to pulsatile growth hormone release. Alcohol Clin Exp Res. 1989;13:3–7. doi: 10.1111/j.1530-0277.1989.tb00274.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.