Abstract
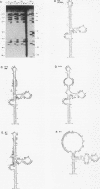
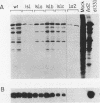
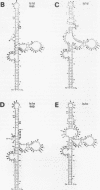
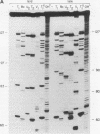
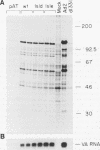
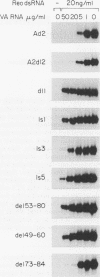
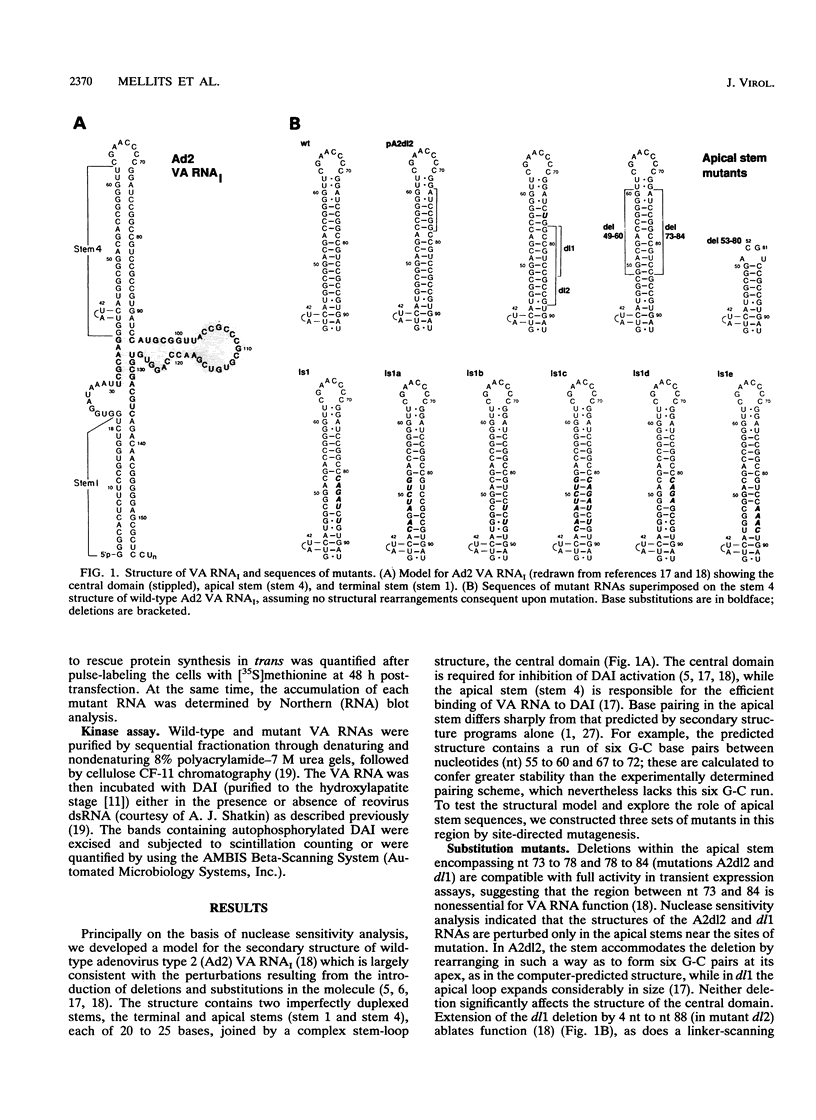
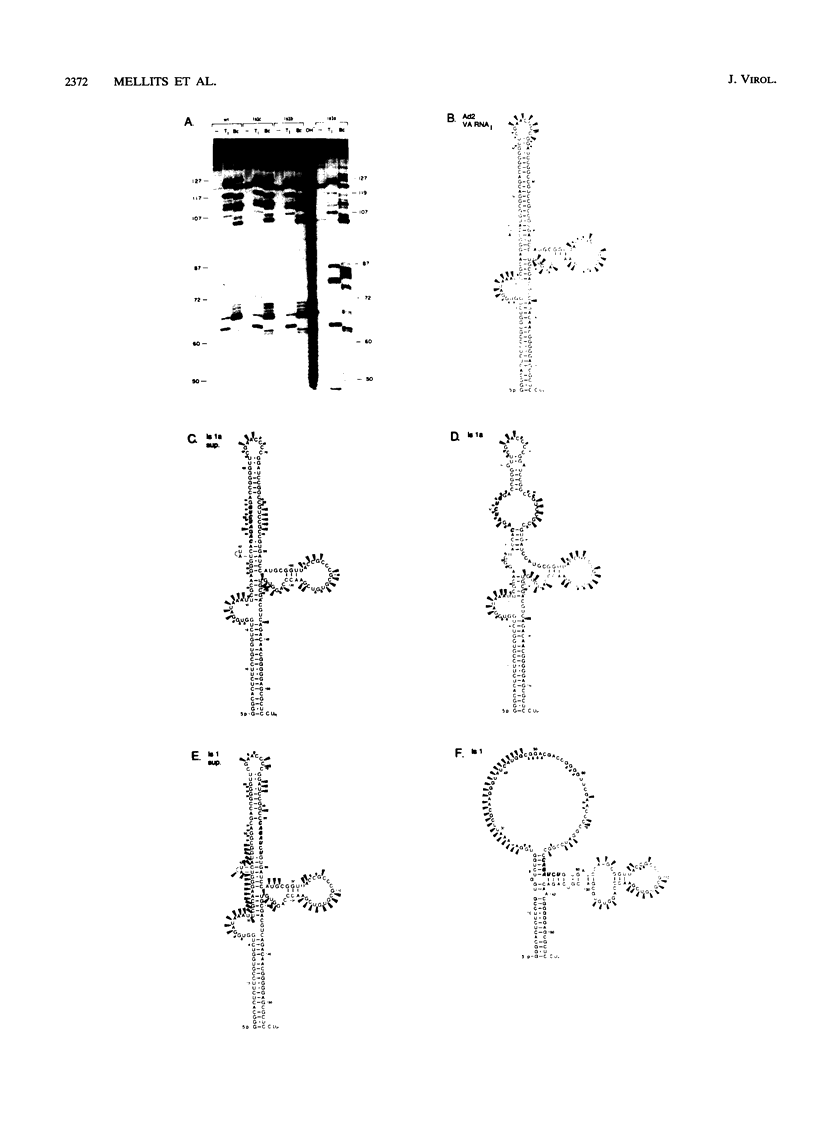
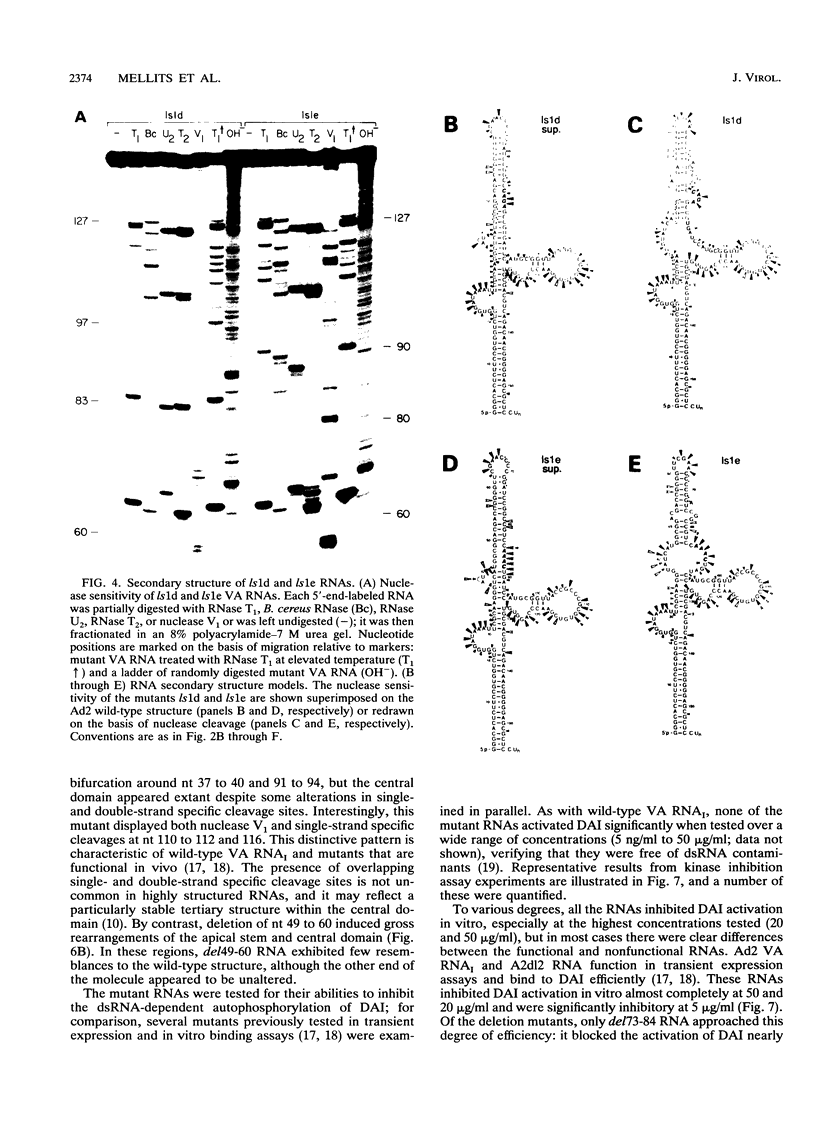
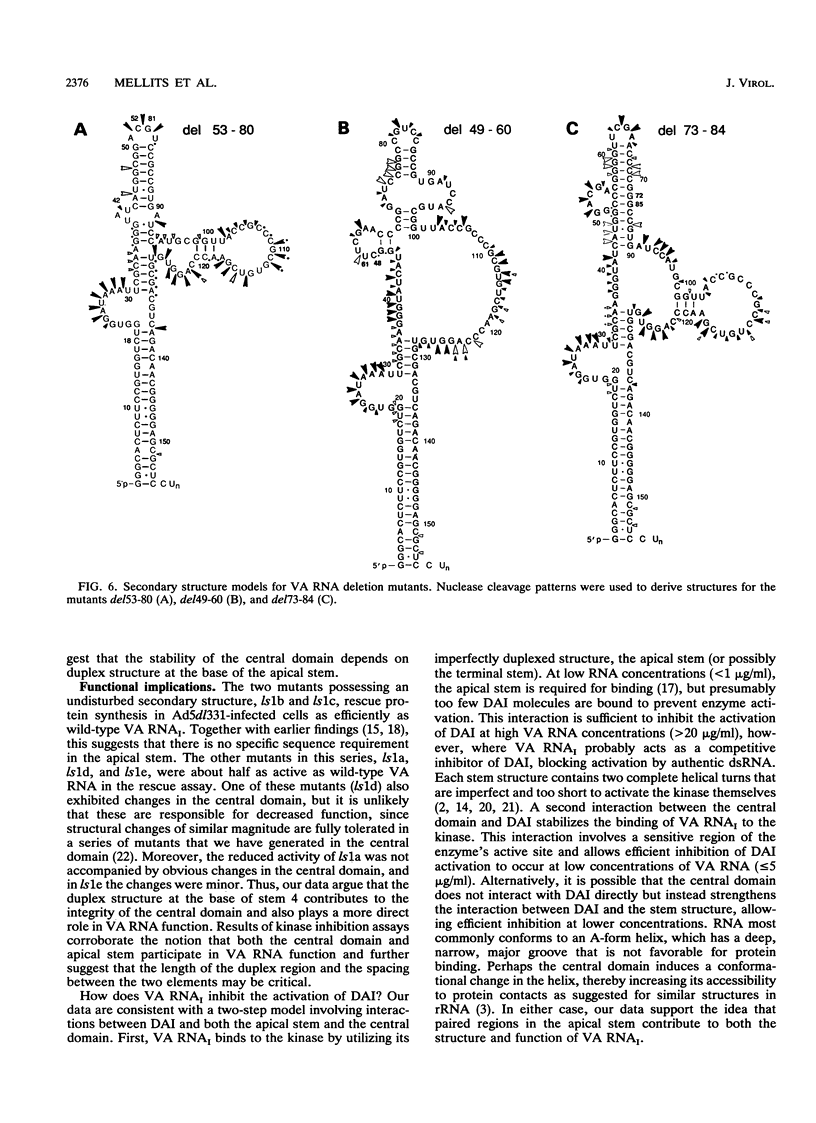
Adenovirus virus-associated (VA) RNAI maintains efficient protein synthesis during the late phase of infection by preventing the activation of the double-stranded-RNA-dependent protein kinase, DAI. A secondary structure model for VA RNAI predicts the existence of two stems joined by a complex stem-loop structure, the central domain. The structural consequences of mutations and compensating mutations introduced into the apical stem lend support to this model. In transient expression assays for VA RNA function, foreign sequences inserted into the apical stem were fully tolerated provided that the stem remained intact. Mutants in which the base of the apical stem was disrupted retained partial activity, but truncation of the apical stem abolished the ability of the RNA to block DAI activation in vitro, suggesting that the length and position of the stem are both important for VA RNA function. These results imply that VA RNAI activity depends on secondary structure at the top of the apical stem as well as in the central domain and are consistent with a two-step mechanism involving DAI interactions with both the apical stem and the central domain.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Mathews M. B., Andersson P., Vennström B., Pettersson U. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc Natl Acad Sci U S A. 1980 May;77(5):2424–2428. doi: 10.1073/pnas.77.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglioni C., Benvin S., Maroney P. A., Minks M. A., Nilsen T. W., West D. K. Interferon-induced enzymes: activation and role in te antiviral state. Ann N Y Acad Sci. 1980;350:497–509. doi: 10.1111/j.1749-6632.1980.tb20652.x. [DOI] [PubMed] [Google Scholar]
- Draper D. E. How do proteins recognize specific RNA sites? New clues from autogenously regulated ribosomal proteins. Trends Biochem Sci. 1989 Aug;14(8):335–338. doi: 10.1016/0968-0004(89)90167-9. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Furtado M. R., Subramanian S., Bhat R. A., Fowlkes D. M., Safer B., Thimmappaya B. Functional dissection of adenovirus VAI RNA. J Virol. 1989 Aug;63(8):3423–3434. doi: 10.1128/jvi.63.8.3423-3434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadge G. D., Swaminathan S., Katze M. G., Thimmapaya B. Binding of the adenovirus VAI RNA to the interferon-induced 68-kDa protein kinase correlates with function. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7140–7144. doi: 10.1073/pnas.88.16.7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle R., Weinmann R. Control region for adenovirus VA RNA transcription. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3378–3382. doi: 10.1073/pnas.78.6.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C. H., Mathews M. B. The adenovirus E1B 19-kilodalton protein stimulates gene expression by increasing DNA levels. Mol Cell Biol. 1989 Dec;9(12):5412–5423. doi: 10.1128/mcb.9.12.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean J. M., Draper D. E. Secondary structure of a 345-base RNA fragment covering the S8/S15 protein binding domain of Escherichia coli 16S ribosomal RNA. Biochemistry. 1985 Sep 10;24(19):5052–5061. doi: 10.1021/bi00340a015. [DOI] [PubMed] [Google Scholar]
- Kostura M., Mathews M. B. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989 Apr;9(4):1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M. B., Grodzicker T. Virus-associated RNAs of naturally occurring strains and variants of group C adenoviruses. J Virol. 1981 Jun;38(3):849–862. doi: 10.1128/jvi.38.3.849-862.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M. B., Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991 Nov;65(11):5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellits K. H., Kostura M., Mathews M. B. Interaction of adenovirus VA RNAl with the protein kinase DAI: nonequivalence of binding and function. Cell. 1990 Jun 1;61(5):843–852. doi: 10.1016/0092-8674(90)90194-j. [DOI] [PubMed] [Google Scholar]
- Mellits K. H., Mathews M. B. Effects of mutations in stem and loop regions on the structure and function of adenovirus VA RNAI. EMBO J. 1988 Sep;7(9):2849–2859. doi: 10.1002/j.1460-2075.1988.tb03141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellits K. H., Pe'ery T., Manche L., Robertson H. D., Mathews M. B. Removal of double-stranded contaminants from RNA transcripts: synthesis of adenovirus VA RNAI from a T7 vector. Nucleic Acids Res. 1990 Sep 25;18(18):5401–5406. doi: 10.1093/nar/18.18.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minks M. A., West D. K., Benvin S., Baglioni C. Structural requirements of double-stranded RNA for the activation of 2',5'-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J Biol Chem. 1979 Oct 25;254(20):10180–10183. [PubMed] [Google Scholar]
- Minks M. A., West D. K., Benvin S., Greene J. J., Ts'o P. O., Baglioni C. Activation of 2',5'-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells by 2'-O-methylated poly (inosinic acid) . poly(cytidylic acid), Correlations with interferon-inducing activity. J Biol Chem. 1980 Jul 10;255(13):6403–6407. [PubMed] [Google Scholar]
- Rohan R. M., Ketner G. A comprehensive collection of point mutations in the internal promoter of the adenoviral VAI gene. J Biol Chem. 1987 Jun 25;262(18):8500–8507. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya B., Weinberger C., Schneider R. J., Shenk T. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell. 1982 Dec;31(3 Pt 2):543–551. doi: 10.1016/0092-8674(82)90310-5. [DOI] [PubMed] [Google Scholar]
- Wu G. J., Railey J. F., Cannon R. E. Defining the functional domains in the control region of the adenovirus type 2 specific VARNA1 gene. J Mol Biol. 1987 Apr 5;194(3):423–442. doi: 10.1016/0022-2836(87)90672-3. [DOI] [PubMed] [Google Scholar]
- Zain S., Gingeras T. R., Bullock P., Wong G., Gelinas R. E. Determination and analysis of adenovirus-2 DNA sequences which may include signals for late messenger RNA processing. J Mol Biol. 1979 Dec 5;135(2):413–433. doi: 10.1016/0022-2836(79)90444-3. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]