Abstract
Environmental fluctuations lead to a rapid adjustment of the physiology of Escherichia coli, necessitating changes on every level of the underlying cellular and molecular network. Thus far, the majority of global analyses of E. coli stress responses have been limited to just one level, gene expression. Here, we incorporate the metabolite composition together with gene expression data to provide a more comprehensive insight on system level stress adjustments by describing detailed time-resolved E. coli response to five different perturbations (cold, heat, oxidative stress, lactose diauxie, and stationary phase). The metabolite response is more specific as compared with the general response observed on the transcript level and is reflected by much higher specificity during the early stress adaptation phase and when comparing the stationary phase response to other perturbations. Despite these differences, the response on both levels still follows the same dynamics and general strategy of energy conservation as reflected by rapid decrease of central carbon metabolism intermediates coinciding with downregulation of genes related to cell growth. Application of co-clustering and canonical correlation analysis on combined metabolite and transcript data identified a number of significant condition-dependent associations between metabolites and transcripts. The results confirm and extend existing models about co-regulation between gene expression and metabolites demonstrating the power of integrated systems oriented analysis.
Keywords: Escherichia coli, metabolomic, response to stress, time course, transcriptomic
Introduction
The response of biological systems to environmental perturbations is characterized by a fast and appropriate adjusting of physiology on every level of the cellular and molecular network. Stress response, as reflected on the level of gene expression, displays some conserved features largely independent of the organism.
Gene expression stress responses are transient, leading to new steady state levels similar to the unstressed cells even in the presence of a persistent stress (Lopez-Maury et al, 2008). Stress response is usually represented by a combination of both specific responses, aimed at minimizing deleterious effects (e.g. catalase during oxidative stress), or repairing damage (e.g. protein chaperones under temperature stress) and general responses which, in part, comprise the downregulation of genes related to translation and ribosome biogenesis (Hengge-Aronis, 2000). This in turn is reflected by growth cessation or reduction observed under essentially all stress conditions and is an important strategy to adjust cellular physiology to the new condition.
Escherichia coli has been intensively investigated in relation to stress responses (Zheng et al, 2001; Chang et al, 2002; Patten et al, 2004; Phadtare and Inouye, 2004; Gadgil et al, 2005; Durfee et al, 2008). Major components of the general and specific response regulate key cellular processes ensuring global control upon perturbation. σs (RpoS) is a central regulator during the response to many stress conditions. σs controls expression of >140 genes involved in metabolism, protein processing, stress adaptation, transport, and transcriptional regulation (Weber et al, 2005). Another important global regulator is (p)ppGpp, involved in the stringent response, one of the mechanisms bacteria use to tune metabolism to available resources. The stringent response is observed when depleting the system of amino acids, and during carbon starvation (Irr, 1972).
The majority of global analyses of the E. coli response to environmental changes have been limited to just one level of information processing, transcription. Although this may be explained by both the central importance of gene expression and the availability of mature techniques, which permit the study of transcriptional changes on a genome-wide level, it is also true that similar approaches on different molecular levels are largely missing. Specifically, comprehensive analyses of changes on the level of metabolites are very rare (Brauer et al, 2006). This is particularly true for the integrated and parallel analysis of the systems response on two levels of genome information processing such as the transcriptome and the metabolome (Bradley et al, 2009).
To better understand system response to perturbation, we designed a time-resolved experiment to compare and integrate metabolic and transcript changes of E. coli using four stress conditions including non-lethal temperature shifts, oxidative stress, and carbon starvation relative to cultures grown under optimal conditions. The resulting data set allowed us to identify parallel and distinct response patterns, represented by conserved patterns on both the metabolic and gene expression levels, across all stress conditions, which indicates a systematic adjustment to suboptimal growth conditions through the impediment of energy demanding growth-related processes. In addition to this conserved component, each response displayed a large amount of stress specificity, thus allowing the clear discrimination of the various stresses through clustering of the metabolomic or transcriptomic data. Performing a time-resolved analysis of the response, however, showed a higher degree of stress specificity for the metabolomic response when compared with the transcriptomic response during the early time points after stress application. As well, metabolic profiles of cultures entering stationary phase are, in contrary to transcript changes, highly dissimilar to metabolic responses to all other tested perturbations.
Clustering and canonical correlation approaches were followed to identify coordinated changes on the transcriptome and the metabolite level, which revealed previously known specific pathway regulations (such as Kleefeld et al, 2009) as well as potential new ones that will require biological validation through further experimentation.
Results and discussion
Experiment design
An established metabolic profiling platform was used to characterize the metabolic responses of an E. coli to four different environmental perturbations, comprising oxidative stress, glucose-lactose diauxic shift, heat and cold treatments, and using an unperturbed culture as a control. Each experimental condition was independently repeated three times and in each of these three biological repetitions, three technical replicas were made, thereby yielding a total of >550 samples. Metabolic profiles containing 188 metabolites (95 could be positively identified, 58 could be chemically classified, and 35 of unknown structure) from E. coli cultures before, during, and after acclimation to the four perturbations plus controls were obtained.
In parallel to gas chromatography mass spectrometry (GC-MS) measurements, microarray-based transcript profiling was carried out for samples from time points 10–50 min post-perturbation plus two control time points before each perturbation for all conditions except the oxidative stress experiment in which all samples (12 time points) were used for transcript profiling covering the entire growth curve, including the stationary phase. Again, three biological replicates were analyzed for each time point, but in contrast to the metabolic profiling no technical repetitions were performed.
The overall measurement reproducibility was determined for all independently performed biological experiments. Relative standard deviation (RSD) of technical and biological replicates was calculated, and showed high reproducibility (Supplementary Figure 1). Median RSD of metabolic measurements for all biological replicates lay within the range of 19.5 (cold) to 27.1% (oxidative stress).
The experiments were designed to both compare and contrast the growth phases within any single applied condition, and also of similar (parallel) time points from the different perturbations on both the metabolic and transcript level. However, of greatest interest was the dynamic response of the system to each of the different conditions applied. Therefore, each experiment was sampled with at least 11 non-linear time points with the highest sampling resolution during the adaptation phase of the culture immediately after perturbation. The five experimental conditions resulted in three distinct growth curves. Exponentially growing cells confronted with oxidative stress and glucose-lactose shift arrested growth for ∼40 min and then resumed logarithmic growth (40–210 min after stress) until reaching stationary phase at about 210 min after stress. After both heat and cold stress application, E. coli stopped growing for approximately 40–50 min and then slowly recovered growth (50–260 min) although at a much slower rate. Within the time frame of the experiment (260 min after stress application), heat and cold stressed cultures did not reach stationary phase. Unperturbed control cultures reached stationary phase about 210 min after having reached optical density (OD) 0.6 (the time point of stress application for the treated cultures). Further details about the growth and sampling time points can be found in Supplementary Figure 2.
Growth phase has a predominant influence on metabolic profiles
Here, we describe the significant metabolic changes (P⩽0.05, ratio ⩾2) relative to time points before perturbation, illustrating the influence growth phase has on the metabolic composition. We first analyzed the changes of the metabolite composition across all time points of all conditions. As cultures were harvested before perturbation in mid-logarithmic growth, a comparison of the metabolic response from each condition to the average of metabolites taken before perturbation is possible (Figure 1).
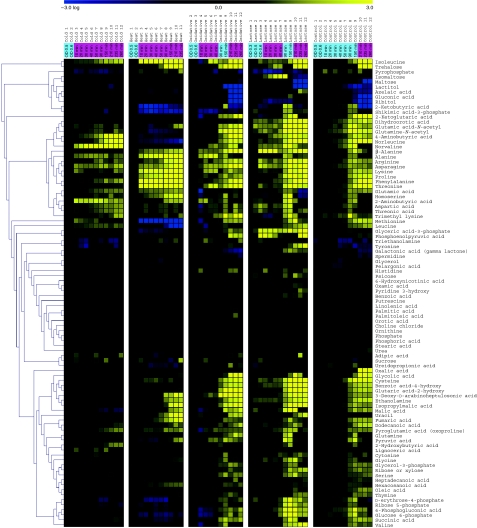
Figure 1.
Metabolite changes during growth and as a result of stress treatment. Median for metabolite levels based on three independent biological repetitions of each stress condition plus control growth relative to time points before perturbation are shown in columns. Hierarchical clustering was performed on those 95 with known annotations. Within each condition, time points are ordered chronologically, the numbering refers to the numbering given in Supplementary Figure 2. Sampling time is shown in a panel at the top of the figure indicating the time after application of the different stresses in minutes. The color of the panel indicates the growth phase: blue-exponential growth, magenta-growth reduction or cessation. Time points before stress application are indicated by their optical density. Source data is available for this figure at www.nature.com/msb.
Figure 1 shows the metabolic profiles of all identified metabolites for all four stress conditions and the control relative to time points before perturbation. One of the most striking features of the heat map is the strong influence of the growth phase on metabolite levels.
During both temperature experiments (cold and heat stress), the temperature was maintained at the altered level after the initial shock treatment. In consequence, no resumption of exponential growth was observed (Supplementary Figure 2). In this sense, the applied cold and heat stresses are ‘permanent,’ which is largely reflected in the metabolic readout. After application of cold or heat, metabolic levels stay fixed or gradually recover after the initial perturbations immediately after stress. This is in contrast to the more transient changes seen after hydrogen peroxide treatment and carbon source shift, which both restore exponential growth after 40 min post-perturbation. A detailed description of the metabolite changes is given in the Supplementary information.
The conserved metabolic response pattern is in agreement with the energy conservation program
The requirement to conserve energy is an important feature of all stress responses, and this necessity has been associated with many stress response mechanisms including the stringent response (Durfee et al, 2008), and the general stress response (Weber et al, 2005). The implementation of the latter has been shown through gene expression studies to reduce energy expenditure through the repression of genes involved in growth, cell division, and protein synthesis (Weber et al, 2005). The repression of transcripts involved in aerobic metabolism has also been seen in response to oxidative stress (Chang et al, 2002) and carbon starvation (Nystrom, 2004). It has been shown that the stringent response involves the downregulation of transcripts involved in transcription and translation (Barker et al, 2001).
In light of these transcriptome-based observations, we decided to see whether the general decrease of central metabolism is also reflected on the metabolite level across the different stress conditions. As induction of the general stress response takes place directly after perturbation, we concentrated on the changes specifically during the first 40 min after application of the stress, the time where cells had not yet resumed growth (Supplementary Figure 2). Metabolic profiles of all identified metabolites are presented in Figure 1, whereas all significant changes are shown in Supplementary Table 1.
Consistent decrease in the levels of metabolites related to glycolysis, the pentose phosphate pathway (p.p.p.), and the TCA cycle is one of the most pronounced effects of the stress application (Supplementary Figure 3). Those include rapid decrease of glucose-6-phosphate (glc-6-P), glyceric acid-3-phosphate (3PGA), pyruvic acid followed by decrease of succinic acid, erythrose-4-phosphate (E-4-P), and ribose-5-phosphate (ribose-5-P) within 40 min, and 6-phosphogluconic acid 90 min after heat-stress application. After oxidative stress application within 20 min, glc-6-P, 3PGA, malic acid, and 2-ketoglutaric acid decreased. Levels of 2-ketoglutaric acid decreased also 10 min after glucose-lactose shift. At 90 min after cold stress, levels of malic acid and ribose-5-P significantly decreased. Noteworthy is the decrease in levels of ribose-5-P, which is precursor of the nucleotide biosynthesis. The decrease in nucleotide biosynthesis is strongly reflected also on transcript level (see below), being one of the most pronounced responses common to different stress conditions (Gasch et al, 2000).
The only glycolytic intermediate that accumulates during the adaptation phase is phosphoenolpyruvic acid (PEP), which transiently increases 10 min after the glucose-lactose shift. As PEP serves as phosphate donor for the phosphotransferase system responsible for glucose import, swift accumulation of PEP was recently proposed to be a direct effect of decreased glucose import caused by low-glucose concentration in the medium (Brauer et al, 2006).
Another general effect of stress application is the accumulation of various amino acids (Supplementary Figure 3). During the adaptation phase, levels of alanine, asparagine, lysine, isoleucine, methionine, leucine, aspartic acid, glutamic acid, phenylalanie, and homoserine significantly increase under cold; isoleucine, threonine, phenylalanine, lysine, alanine, asparagine, glutamic acid, and homoserine under heat; asparagine in lactose shift; and alanine and asparagine in oxidative stress experiment.
The increase in amino-acid levels could be, at least in part, a result of increased protein degradation (Mandelstam, 1963). Degradation of proteins can be caused by the need to eliminate abnormal proteins formed as a result of stress, or can be interpreted as a means to increase the availability of amino acids required for the synthesis of new proteins important for survival under the new, less favorable condition (Willetts, 1967). It has been shown that protein degradation is influenced by the increase of ppGpp levels during amino acid and carbon starvation, and this degradation was suggested to be dependent on the action of Lon and Clp proteases (Kuroda et al, 2001). Proteins that are preferentially degraded by proteases are free ribosomal proteins, tagged with a polyphosphate chain, which stimulates proteolytic attack (Kuroda et al, 2001). In line with those findings, we observed a massive increase in the levels of various amino acids on the entry to the stationary phase of growth starting from 210 min after oxidative stress, lactose shift, and in parallel time points in the control cultures (Supplementary Table 1).
Although many amino acids accumulate, some do show a decrease. Methionine levels significantly decrease after both heat and oxidative stress (Supplementary Table 1), which is in agreement with methionine synthase (MetE) being very sensitive to oxidation. The addition of methionine to the growth medium leads to increased survival of E. coli during heat stress and a shortened growth lag during oxidative stress (Hondorp and Matthews, 2004). As oxidized MetE is inactive, the resulting methionine limitation might affect protein translation (Gold, 1988). In line with these findings, we observe an increase in methionine levels on growth resumption in the oxidative stress experiment (Figure 1; Supplementary Figure 3).
Taken together, the changes observed on metabolic level, specifically the decrease in most measured metabolites of the TCA cycle and the glycolysis pathway (cf. also Supplementary Figure 3 for a better presentation of these metabolites), are in agreement with the general energy conservation strategy previously reported for the transcriptomic response.
Major changes at the metabolic and transcript level coincide with growth transitions
As discussed in the Introduction, both specific Figure 1 and general responses (Gasch et al, 2000; Weber et al, 2005) were observed. To further probe conserved and non-conserved responses, we analyzed time points displaying the highest number of changes. To this end, the number of metabolites and transcripts that significantly differ (change) between two neighboring time points (the time point of interest and the directly preceding one) within each condition were calculated.
When performing this analysis for all conditions, a highly conserved pattern emerged for both transcripts and metabolites (Figure 2A and B). Thus, on both levels, the largest number of changes is observed within the first time point after stress application with the largest number of changes on the transcriptome level displayed by the heat-stress conditions. As to the metabolite pattern, the diauxic shift displays the largest number of changes followed by cold stress, oxidative stress, and heat stress. It is important to note that no significant changes were observed for the control cultures during this growth period (mid-log growth phase), indicating that exponential growth phase is represented on both levels by few if any changes on the level of transcripts and metabolites, which is in agreement with transcript level observations (Chang et al, 2002).
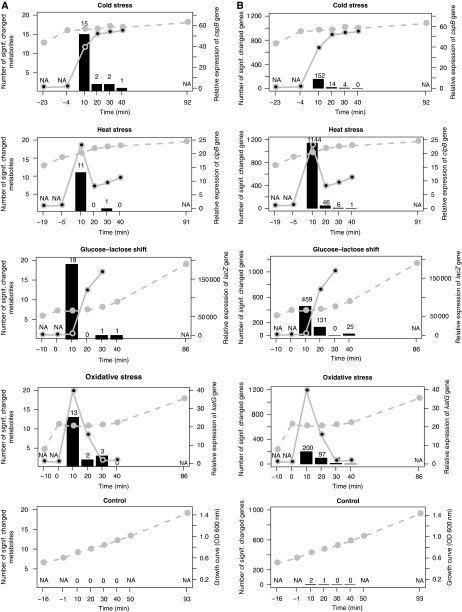
Figure 2.
Different perturbations result in similar dynamics of responses on metabolite and transcript level. Number of changing metabolites (A) or transcripts (B) between neighboring time points is shown as a histogram for time points 10–90 min after stress (for significance threshold see Materials and methods section). The actual numbers of changes are shown above each bar. The dotted gray line indicates the growth curve, whereas the solid gray line represents the expression of genes indicative for each condition: oxidative stress-katG; heat stress-clpB; glucose-lactose shift-lacZ; cold stress-cspB).
Overrepresentation analysis of functional categories (based on gene ontology—GO) of genes, which change at 10 min past stress application reveals a conserved pattern across all conditions. Genes associated with amino acid, amine, nucleotide, and ribonucleotide biosynthetic processes and ATP synthesis, proton transport were downregulated (Supplementary Figure 4). These findings are in agreement with comparable experiments performed for both yeast and E. coli (Gasch et al, 2000; Chang et al, 2002; Durfee et al, 2008). Interestingly, we observed downregulation of genes assigned to ‘flagella motility’ GO term across all conditions. As flagella motility requires a steep proton gradient between the periplasmatic space and the cytoplasm, decreased cell motion could indicate energy deficiency. Other biological processes that depend on proton gradient are ATP synthesis and transmembrane transport. However, in contrast to genes involved in ATP synthesis, which decrease after all perturbations, genes encoding general transport increase during glucose-lactose shift and oxidative stress. This could indicate that transport of external carbon sources is favored over chemotaxis (Lemuth et al, 2008).
The coincidence of the response on both levels can indicate that the changes on the metabolic level are not transcriptionally dependent. Global proteomics analyses indicated that protein levels, post-translational modifications, and stability are directly affected by different perturbations (for review see Kultz, 2005). As enzyme abundance and activity have predominant influence on biochemical reactions, the possibility that metabolic changes are caused by enzymes, directly influenced by environmental conditions, cannot be excluded. This possibility could be tested by application of transcription inhibitors (e.g. Rifampicin) and analyzing the kinetics of metabolic response. It would be interesting to further extend this concept by applying protein synthesis or protein post-translational modifications inhibitors.
Stress response displays higher specificity on the metabolite as compared with the transcript level with respect to the individual stress applied
As described above, the general response pattern on both metabolite and transcript level is similar with respect to its kinetics within 40 min post-perturbation. To see whether this pattern is due to similar or rather dissimilar responses, we determined which metabolites and transcripts change significantly (for significance thresholds see Materials and methods section) during the different stress treatments in comparison to the relative time points from control. Subsequently, we asked whether the observed changes display a significant overlap between different conditions by applying Fisher exact test. This analysis enables us to compare the specificity (as defined in the Materials and methods section) of E. coli response to perturbation on the metabolome with the transcriptome. Figure 3 displays these results for all pairwise comparisons of experimental conditions in a binary form: 1 encodes a significant overlap or dependence of the response of two conditions, whereas a 0 entry corresponds to no significant overlap, that is an independent response. The absolute numbers of changing genes and metabolites are shown in Supplementary Figure 5).
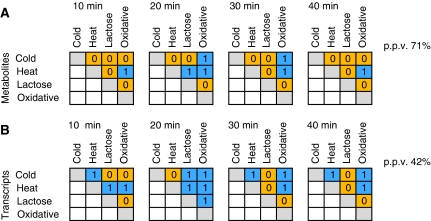
Figure 3.
Metabolic profiles are more stress specific as compared with the changes at gene expression level. Similarities between responses to different conditions on metabolic (A) and transcript (B) level relative to control condition. Parallel time points post-perturbation (t1: 10 min; t2: 20 min; t3: 30 min; t4: 40 min) from different experiments were compared against corresponding time points from control. The significance of the overlaps between compared conditions was calculated based on Fisher exact test. The significant overlaps (P⩽0.05) are marked by 1 (blue), whereas no significant overlaps are marked with 0 (orange). The number of significant overlaps between conditions was compared on both levels and is shown in percentage to the total number of possible comparisons. The actual number of metabolites and transcripts that overlap between compared conditions is given in Supplementary Figure 5.
With respect to the metabolites as shown in Figure 3A for the first post-perturbation time point (10 min), stress specificity is high with only one of the six possible comparisons displaying significant similarity (heat and oxidative stress). At later time points (20 and 30 min post-perturbation), three out of six conditions show overlap, whereas after 40 min, only heat and oxidative stress still overlap. We summarize these findings by the positive predictive value (PPV) of the metabolic response of 71%.
We next analyzed the overlap on the transcriptome level. However, as the number of metabolites analyzed is less than the number of transcripts, a direct comparison between both data sets would be biased. Moreover, this could lead to a higher level of conservation on the transcript level due to the inclusion of many general transcriptional responses (as exemplified by ESR in yeast, Gasch et al, 2000) not paralleled by any metabolite data. Therefore, the transcriptome analysis included only those 288 genes, which are directly linked to metabolic enzymes (based on EcoCyc), by considering genes where either the substrate or the product was contained in the metabolite data set (Supplementary Table 2). In contrast to the metabolite data, more pairwise comparisons of different conditions show dependence in the transcriptome response (Figure 3B). Our results show a significant overlap for three comparisons within 10 min, and five pairwise comparisons 20 min after stress (Figure 3B). The number of dependent responses decreases with increasing time; specifically, the response of the diauxic shift experiment loses similarity to other responses correspondingly to the metabolic response (Figure 3A). The highest similarity was found for the response toward heat and oxidative stress at both levels. This corroborates the link between responses to heat and oxidative stress observed in earlier studies (Farr and Kogoma, 1991) and is in further agreement with the results of the HCA presented in Supplementary Figure 7.
Taken together, the response on metabolic level is obviously more specific as the PPV on the metabolites is 71% in contrast to 42% on the transcript level. Our observation that the metabolic response displays a higher level of specificity as compared with the transcriptomics response cannot be explained in a straightforward way. One interpretation is that metabolism has both the capacity to react faster and the need to react more specifically compared with the more mid-term adjustment based on reprogramming of the transcription–translation machinery. A fast delivery of metabolites needed to protect the system could be crucial for the initial survival of the system before more massive changes brought about by changes on the gene expression program come into play. One example of such mechanism is osmotic stress response in Synechocystis, where concentration of compatible solute is regulated on the post-transcriptional level of protein activity triggered directly by the stress and paralleled by a more time-consuming induction of gene expression (Hagemann, 1996).
In contrast to the highly conserved transcriptional response pattern, the metabolite response is different for growth arrest induced by stress and by reaching stationary phase
E. coli responds to stress by ceasing or reducing growth. It has been shown previously that changes on the transcript level, as a result of stress-induced growth arrest, significantly overlap with changes observed when cells cease to grow due to entering stationary phase (Chang et al, 2002; Weber et al, 2005). In light of the observation that the stress-induced changes on the metabolite level in the initial response phase display a higher stress specificity compared with the transcript level, we were interested to determine the degree of similarity of the changes on the metabolite level observed in response to the two different growth cessation conditions.
To this end, we compared time points from the stress adaptation phase and time points taken 210 min after stress application (for details see Materials and methods section). At this time point, the lactose shift, oxidative stress, and the control experiment had entered the stationary phase (Supplementary Figure 2). Both temperature stress experiments were excluded from this comparison as, due to the maintained temperature stress, these cultures do not resume exponential growth and therefore do not run out of nutrients and enter stationary phase.
When comparing only the metabolic profiles for the three stationary phase samples, a high degree of similarity is seen (Figure 4A) in agreement with the results of the HCA shown in Supplementary Figure 7, suggesting an underlying common cause. Among the metabolites that change consistently in all stationary phase conditions PEP, isoleucine, and phenylalanine all increased, whereas homoserine consistently decreased. A decrease in homoserine levels and an increase in PEP has previously been shown under carbon and nitrogen starvation (Brauer et al, 2006). The assumption of carbon starvation as the common underlying source is further supported by transcriptome data revealing an upregulation of carbon starvation-induced genes (csiD, csiE, cstA).
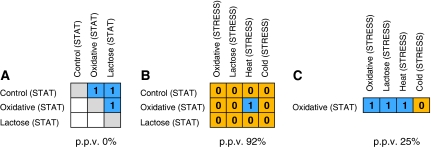
Figure 4.
Metabolite profiles of stationary phase culture differ from metabolic profile of stress-arrested cultures. Changes in metabolites during stationary phase are similar between different cultures (A) but different from metabolites changing as a result of stress (B), whereas transcripts changing during stationary phase or in response to stress are very similar (C). The significance of the overlaps between conditions was calculated based on the Fisher exact test. Significant overlaps (P⩽0.05) are marked by 1 (blue), whereas insignificant overlaps are marked with 0 (orange). The number of significant overlaps between different conditions is higher for transcript responses (PPV=25%) as compared with metabolic responses (92%). The actual number of metabolites and transcripts that overlap between conditions is given in Supplementary Figure 6.
The metabolites that significantly change their concentration upon entry into stationary phase (210–260 min) were subsequently compared with those whose levels changed within 10–40 min after the respective perturbation. Only 1 of the 12 pairwise comparisons of metabolic responses, (heat-stress response versus stationary phase of the oxidative stress) resulted in a significant similarity as based on the Fisher exact test (Figure 4B; see Supplementary information for a discussion regarding the overlap between stationary phase and heat stress). This indicates a high degree of dissimilarity (PPV=92%) between metabolic responses during growth cessation as induced through stationary phases or through various stress applications, which is in strong contrast to the high level of overlap reported for the response on the transcript level (Chang et al, 2002).
To assure ourselves that the difference described above between metabolite and transcript characteristics is not due to differences in experimental conditions, we performed the same comparison between the transcriptome changes observed during growth cessation due to stationary phase as compared with induced by stress application on our own data set. To this end, stationary phase samples from the oxidative stress experiment were analyzed for the transcriptome and compared against the transcriptome changes occurring as a result of stress application. With the exception of the cold stress response, a highly significant overlap between stationary phase-induced growth arrest and stress-induced growth arrest was observed (PPV=25%), thus further strengthening the significance of the observed disparate behavior for the metabolite response (Figure 4C).
The level of coordination between transcript and metabolite data is strongly influenced by the environmental conditions
As outlined in the Introduction, biological systems respond to changes in their environments by adjusting their entire physiology to the new condition involving different levels of the system. In this study, we have monitored responses in parallel on the transcriptome and the metabolite level thus allowing one to compare the level of coordination between both molecular readouts.
To perform this analysis, we followed two different approaches, an untargeted (holistic) co-clustering approach and a targeted approach using prior biological knowledge in conjunction with canonical correlation analysis (CCA).
In the co-clustering approach, metabolites and transcripts were jointly subjected to a k-means clustering. The resulting clusters were subsequently analyzed for overrepresentation of transcripts and metabolites from the same biochemical pathway (see Materials and methods section for details). When applying this approach to the entire data set, that is combining the measurements of all individual stress conditions, no co-clustering of metabolites and transcripts from the same pathway could be observed (data not shown).
Applying this co-clustering approach respectively to each growth phases of each stress condition separately (e.g. all time points from the oxidative stress condition), we were able to identify several metabolites and transcripts from the same pathway within the same cluster, although the overall enrichment is restricted to ≈10% of the derived clusters. Furthermore, several gene–metabolite pathway associations are not preserved and were found for only one of the conditions. Interestingly, the oxidative and cold stress conditions exhibit the largest number of associations (Supplementary Table 3 for a full representation of the results).
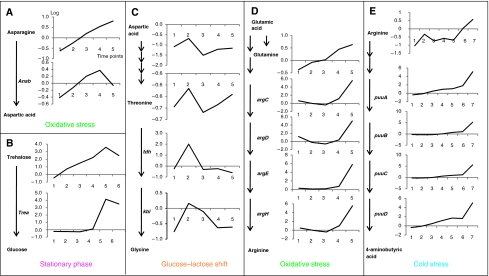
One striking observation immediately apparent was the overrepresentation of amino acids in the gene–metabolite associations and more specifically the association between amino acids and genes involved in amino-acid catabolism (cf. Figure 5 that shows in an exemplary manner a schematic view of the corresponding pathway and the representation of the corresponding transcript and metabolite levels). Thus, asparagine levels are highly associated with transcript levels of the asparaginase gene ansB threonine and its precursor—aspartic acid correlate with expression of the tdh and kbl genes, and arginine correlates with expression of genes involved in the arginine and ornithine degradation pathway. Glutamine levels correlate with a number of transcripts associated with arginine biosynthesis that might possibly indicate a common regulation by glutamate, which is a precursor for both arginine and glutamine synthesis.
Figure 5.
Co-clustering between metabolic changes and transcripts of corresponding pathway genes. Representative examples (A–E) of the condition specific co-clustering analysis are shown (for full list of associations see Supplementary Table 3). For the identified conditions (oxidative stress, glucose-lactose shift, cold stress), pathways with the respective genes and measured metabolites are shown in schematic way. Changes in transcript are shown next to the genes, metabolic changes next to metabolites across subsequent time points (x axis). Both changes are presented on log scale.
In contrast to the numerous associations between amino-acid catabolism genes and amino acids, only few associations are observable for amino acids and corresponding genes encoding biosynthetic enzymes. Examples for this type of association are observable between valine and one of the enzymes from the valine biosynthesis pathway—IlvC and between histidine and genes coding two enzymes involved in histidine biosynthesis HisB and HisC. The only association observed for a non-amino acid as a metabolite and a related gene is the co-clustering of trehalose and the gene treA encoding its degrading enzyme trehalase under stationary phase (Figure 5).
Most of the data described here and in other studies indicate that environmental changes are most profound in central metabolism especially with respect to the early response.
In a second approach, we therefore limited the analysis to particular pathways covering parts of central metabolism, which bears the further advantage of significantly reducing data complexity especially with respect to the transcripts, thus allowing other algorithms to be applied. More specifically, metabolites from glycolysis, the TCA cycle, the p.p.p., and anaerobic respiration were subjected to a CCA together with transcript data of all enzymes from those pathways as derived from EcoCyc. As we are also interested in general regulators, we further included several global transcriptional regulators, known to be involved in metabolism control (ArcA, ArcB, Cra, Crp, Cya, Fnr, Mlc). A complete list of all metabolites and transcripts covered is given in Supplementary Table 4.
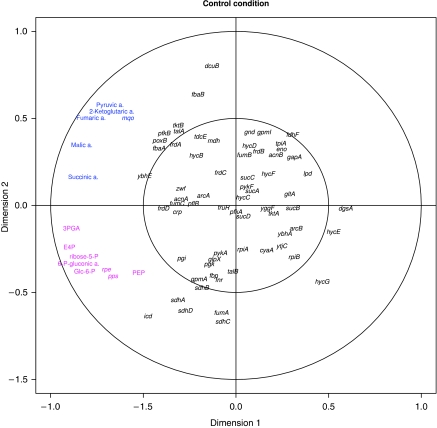
Figure 6 shows in an exemplary manner the canonical structure correlation plot as a result of the CCA, applied to the control condition data (see Supplementary Figure 8 for the remaining two conditions discussed in this section). The results for the three conditions are summarized in the form of projection onto pathways in Figure 7A–C.
Figure 6.
Visualization of the CCA results of metabolites and genes involved in primary metabolism under exponential growth. The canonical structure correlations of 69 genes and 11 metabolites covering p.p.p., glycolysis, TCA cycle, anaerobic respiration, and 8 transcriptional regulators involved in metabolic control with the first two canonical variates show two distinct groups of metabolite–transcript associations. The first group, colored in magenta, consists of metabolites from the oxidative pentose phosphate pathway (glc-6-P, 6-P-gluconic acid, ribose-5-P, and E-4-P) as well as all measured metabolites from the glycolytic pathway (3PGA and PEP in addition to glc-6-P) and the genes pps and rpe. The second group, colored in blue, consists of TCA cycle intermediates, that is 2-ketoglutaric, fumaric, malic, and succinic acids. In addition, the mqo gene encoding malate-quinone oxidoreductase (MQO) and pyruvic acid belong to this group.
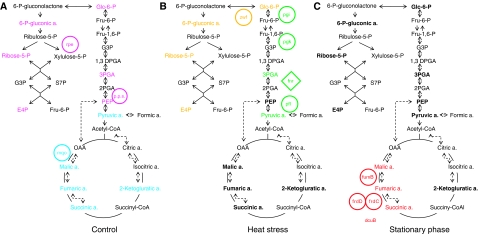
Figure 7.
Examples of the condition specific associations between metabolites and transcripts (A–C). Canonical correlation analysis (CCA) reveals condition-dependent association between response dynamics on the transcript and metabolite level: comparison of metabolite–transcript associations of central metabolism between control growth, heat stress, and stationary phase. Metabolites and genes displaying a close association in the CCA were extracted (Figure 6; Supplementary Figure 8) and projected on a schematic representation of the TCA cycle, glycolytic pathway, and p.p.p. Dotted lines indicate optional anaerobic pathways. Measured metabolites are indicated in bold. Biosynthetic genes are circled and regulatory genes are displayed in diamond shape. Transcripts and metabolites showing a close association in the CCA are indicated by the same color. With respect to heat stress, a selected part of the associations are shown.
When applying CCA to all conditions separately, multiple associations were observed only for three conditions: control growth, heat stress, and stationary phase. The visualization of the canonical structure correlations with the first two canonical variates (see Materials and methods section) shows a number of metabolites in close proximity to genes coding enzymes, which catalyze their biochemical conversions. For the remaining three conditions, cold stress, oxidative stress, and diauxic shift, very few or no intuitive associations were observed.
Under control conditions, two groups of highly associated metabolites and transcripts are observed (Figures 6 and 7A, colored in magenta and blue). The first comprises all measured metabolites from the oxidative p.p.p. (glc-6-P, 6-P-gluconic acid, ribose-5-P, and E-4-P) in addition to metabolites from the glycolytic pathway (3PGA and PEP in addition to glc-6-P) forming a strong association with two genes encoding pathway enzymes, that is rpe encoding ribulose phosphate 3-epimerase and pps encoding PEP synthase.
The high association of metabolites and transcripts from these two pathways is only observed under optimal growth conditions and is largely lost under all other conditions analyzed such as heat stress and during the stationary phase (see Supplementary Figure 8A and B). This tight coupling between glycolysis and the p.p.p. might reflect the strong demand of fast growing cells for synthesis of high levels of the nucleotide precursor ribose-5-P. It is known that exponentially growing cells metabolize glc-6-P into fructose-6-phosphate (fru-6-P) and 3PGA by glycolytic enzymes, and next use transketolase and transaldolase enzymes from p.p.p. to convert two molecules of fru-6-P and one molecule of 3PGA into three molecules of ribose-5-P (Berg et al, 2006). Finally, these data suggest that both rpe and pps could have a major regulatory function mostly exerted through transcriptional regulation of both genes.
The second group of coordinated metabolites and genes found under optimal growth conditions form part of the TCA cycle. Thus, the expression of the mqo gene encoding malate-quinone oxidoreductase (MQO) is associated with all TCA cycle intermediates measured: 2-ketoglutaric acid, fumaric acid, malic acid, and succinic acid. In addition, pyruvic acid, which is located at the key point between glycolysis and the TCA cycle, shows association with mqo. MQO catalyses the irreversible oxidation of malate to oxaloacetate (Kather et al, 2000) that in turn regulates the activity of citrate synthase, which is a major rate determining enzyme of the TCA cycle (Frederick and Roy Curtiss, 1996). Although the conversion of malate to oxaloacetate is also catalyzed by other enzymes including the NAD-dependent malate dehydrogenase (mdh), it was recently suggested that under optimal growth conditions, MQO is the major route of malate oxidation (van der Rest et al, 2000). The strong association between mqo gene expression and multiple members of the TCA cycle as well as pyruvate suggest mqo expression to have a major function for the regulation of the TCA cycle, which need to be experimentally validated.
The tight coupling between the oxidative p.p.p. and glycolysis is lost, however, under non-optimal growth conditions. Thus, during stationary growth, no association is observed between any metabolites and transcripts related to those pathways (Figure 7C). In contrast, under heat stress (Figure 7B; Supplementary Figure 8B), the expression of zwf gene encoding the glc-6-P dehydrogenase correlates with three intermediates of the p.p.p. including glc-6-P, 6-phosphogluconic acid, and E-4-P, suggesting a control of the flux through p.p.p. by changes in zwf expression. Expression of zwf gene that encodes the first key enzyme from p.p.p. is among others controlled by the SoxRS regulon in response to oxidative stress (Fawcett and Wolf, 1995). Correlation of expression of zwf and p.p.p. metabolites under heat stress indicates a similar redirection of p.p.p. under heat-stress conditions again emphasizing the similarity between heat and oxidative stress.
Analysis of the stationary phase data reveals among others the association of three metabolites of the TCA cycle including malic, fumaric, and succinic acid with the expression of several genes including fumarate reductase (frd C,D), fumarase B (emphfumB), and fumarate-succinate antiporter (dcuB). This is a most interesting observation as fumaric acid is known to serve as an alternative electron acceptor during anaerobic respiration further regulating the expression of genes associated with anaerobic respiration including the four genes mentioned above (Jones and Gunsalus, 1987; Zientz et al, 1998; Golby et al, 1999). The mechanism of this regulation includes activation of the DcuS-DcuR two component system by fumaric acid, which subsequently stimulates expression of target genes (Kleefeld et al, 2009). Our data confirm this model and in addition show that this regulation only holds true under stationary phase characterized among others by limiting oxygen availability. This model can be further extend based on the tight coordination between the expression of both fumarate reductase genes (frdC, frdD) also with malic and succinic acid that expression of these genes might be regulated by levels of all three metabolites, which is in agreement with previous studies (Kleefeld et al, 2009).
A complex picture different from both the stationary phase and the optimal growth conditions emerge from the analysis of the heat-stress experiment concerning the TCA cycle. Inspection of the canonical loadings shows among other associations a high similarity between the expression levels of pflB gene coding pyruvate formate-lyase (PFL) and concentration of pyruvic acid. Pyruvic acid further is strongly associated with the transcriptional regulator FNR (fnr). This association is in full agreement with a model developed for anaerobic conditions (which are approximated by heat stress), which suggests that expression of pflB is regulated in an FNR-dependent manner by pyruvic acid (Sawers and Bock, 1988). It is interesting to see that also two other genes from upper glycolysis (pgk and pgi) are in close proximity of fnr, pflB, pyruvic acid, and 3PGA (Supplementary Figure 8B). Both of these genes seem to have an important function in anaerobic metabolism. The expression of hpgk encoding phosphoglycerate kinase is induced under anaerobiosis (Nellemann et al, 1989), whereas a mutation in pgi was shown to reduce the expression of several anaerobically induced genes, including PFL, with glucose as the sole carbon source (Rasmussen et al, 1991). Interestingly, the effect of the pgi mutation could be overcome by addition of pyruvic acid (Rasmussen et al, 1991). This, together with our data, might suggest that the induction of PFL expression is dependent on the presence of glycolytic metabolic intermediates, whose synthesis is blocked in pgi mutant, most likely pyruvic acid (Leonardo et al, 1993).
This leads to the hypothesis that products of both pgk and pgi could have important functions under hypoxic conditions by controlling the levels of pyruvate, which is then converted by PFL in anaerobic respiration.
Conclusion
The time-resolved and combined analysis of the transcriptomic and metabolomic response of E. coli to four different stresses reveals conserved and specific responses on both levels of information processing. Different stress conditions have similar global impact on cell metabolism, which consists on energy conservation strategy as is evident on the transcript and metabolic level. Co-occurring responses on the transcript and metabolic level were observed as peaks of maximal changes directly post-perturbation irrespective of the stress applied. The co-occurrence of metabolic and transcript responses was observed for functionally related genes and metabolites and proposed to be an effect of strong co-regulation of both levels of response. Specificity of the response is higher on the metabolome as compared with the transcriptome level especially during early time points after perturbation. Stress-induced growth cessation is similar to stationary phase growth cessation when compared on the level of the transcriptome, but different when compared on the level of the metabolome.
Application of co-clustering and CCA on combined metabolite–transcript data identified a number of condition-dependent significant associations between metabolites and transcripts. The results obtained confirm and extend existing models about co-regulation between gene expression and metabolites demonstrating the power of integrated systems oriented analysis.
Materials and methods
E. coli culture conditions
For all experiments, E. coli strain MG1655 was used, which was obtained from the American Type Culture Collection (ATCC 700926). The minimal medium used for all experiments was a modification of MOPS (morpholinopropane sulfonate) minimal medium (Neidhardt et al, 1974) obtained from Teknova, CA (product number M2006), which contains 86 mM NaCl, 9.5 mM NH4Cl, 5 mM K2HPO4 and 0.2% glucose.
All cultures were grown aerobically in a thermostatically controlled 37°C culture room. Cultures (150 ml culture volume) were stirred by magnetic stirrers at 330 r.p.m. (Thermo Scientific Variomag Multipoint 6in) 1000 ml Erlenmeyer flask. Analysis of gene expression data for transcripts indicative for anaerobiosis showed the absence of any oxygen shortage under optimal growth conditions and rather in contrast showed a slight induction of genes associated with aerobic respiration for example ubiquinone oxidoreductase (nuoH, nuoN, nuoL). Induction of expression of genes associated with hypoxia was, however, observed after glucose-lactose shift, oxidative stress, and more pronounced during heat and stationary phase. Temperature and pH were carefully monitored during growth. Starting cultures were inoculated from a single colony and grown overnight. Each experimental culture was then inoculated from such an overnight culture at a dilution of 1:20 into 150 ml fresh MOPS minimal medium in a 1000 ml flask. Growth of cultures was monitored by measuring OD at 600 nm using an Eppendorf Biophotometer. All cultures were grown until early mid-log phase (OD 0.6), at which point each of the perturbations was applied.
Oxidative stress
A measure of 200 μg/ml of 30% pre-warmed hydrogen peroxide (Fluka) was added to 150 ml constantly stirred (330 r.p.m.) cultures kept in 1000 ml flasks. The amount of hydrogen peroxide used for the stress was calculated to cause a non-lethal 40 min lag phase. This was monitored by plating on solid LB medium and calculating viable cell number.
Cold stress
Cultures were transferred from 37°C into an ice cold water bath to lower the temperature, whereas stirring, to 16°C in <2 min. When 16°C had been attained, flasks were transferred to a 16°C water bath while constantly stirring (330 r.p.m.).
Heat stress
Cultures were transferred from 37°C to a 50°C water bath. While stirring, the temperature of each culture was raised to 45°C in <2 min. The constantly stirring (330 r.p.m.) cultures were then transferred to a 45°C water bath to maintain this temperature. In both temperature treatments, the temperature was constantly monitored ensuring both temperatures are constant.
Glucose-lactose shift
Carbon source concentrations of 0.15% lactose and 0.05% glucose were used (150 ml culture in 1000 ml flasks, 330 r.p.m. stirring). This meant that the growth lag phase was observed at OD 0.6.
Sampling
The first two time points were taken before stress at OD 0.5 and 0.6, in case of glucose-lactose shift additional time point before stress was taken at OD 0.3. After stress application, the subsequent sampling time points were at 10-min interval for up to 40 min (lactose shift and oxidative stress) or 50 min (cold, heat, and control). Rapid filtering using 2.5 cm diameter, 0.45 μm pore size Durapore filter disks (Millipore Corporation, MA) and a vacuum manifold and pump was used. Metabolite (1 ml) and transcript (3 ml) samples were taken simultaneously. Filters with adhering bacteria were rapidly transferred into 2 ml centrifuge tubes and flash frozen in liquid nitrogen. The whole process took <5 s (metabolites) or 10 s (transcripts) per sample from sampling to flash freezing in liquid nitrogen and has been shown to be superior to methods such as quenching or centrifugation (Bolten et al, 2007).
For GC-MS metabolite analysis, each of the filter discs with adhered bacteria was extracted in 500 μl Methanol (Merck) at 4°C, as this has previously been shown to be superior to hot methanol, hot ethanol, cold perchloric acid, hot alkaline, and cold methanol/chloroform extraction protocols (Maharjan and Ferenci, 2003). The extraction solution contained 0.1 μg/ml cholesterol as an analytical internal standard. Tubes were subsequently shaken at 4°C for 10 min at 1000 r.p.m. and again frozen in liquid nitrogen. This freeze-thaw cycle was repeated to ensure cell membrane rupture. Finally, filters were removed, samples centrifuged for 3 min at 14 000 r.p.m. at 4°C (Eppendorf model 5417R), and 450 μl of the supernatant transferred into new 2 ml centrifuge tubes. These samples were then dried to complete dryness in a rotary vacuum centrifuge device. Dried samples were subsequently stored at −20°C for a maximum of 2 weeks before analysis.
GC-MS analysis
Before GC-MS analysis, samples must be derivatized. A variation on the two-stage technique used by Roessner et al (2001) was used to firstly protect carbonyl moieties through methoximation, through a 90 min 30°C reaction with 5 μl of 40 mg/ml methoxyamine hydrochloride (Sigma-Aldrich) in pyridine (Merck), followed by derivatization of acidic protons through a 30 min 37°C reaction with the addition of 45 μl MSTFA (N-methyl-N-trimethylsilyltrifluoroacetamide) (Machery-Nagel). A measure of 1 μl of the derivatized sample was injected onto the column and analysis was commenced in non-split mode. GC-MS hardware comprised an Agilent 6890 series GC system fitted with a 7683 series autosampler injector (Agilent Technologies GmbH, Waldbronn, Germany) coupled to a Leco Pegasus 2 time-of-flight mass spectrometer (LECO, St Joseph, MI). Identical chromatogram acquisition parameters were used as those described earlier (Weckwerth et al, 2004). Chromatograms were processed using Leco ChromaTOF software (version 3.25) and analytical peaks determined using the method of Lisec et al (2006), with a modified peak picking algorithm that searches for local apex intensity from all mass traces in raw chromatograms. All data were normalized to cell number and the chromatographic internal standard.
General statistical analysis
All samples were normalized to the median of time points taken before stress to minimize technical influence. To ensure proper alignment of different biological replica, the expression of stress-specific marker genes was used as an anchor marking the actual moment of stress. In the case of the control culture, all time points were normalized to the average of time points taken at OD 0.5 and 0.6. HCA presented in Figure 1 is based on the log2 Euclidean distance measure and average linkage aggregation method, whereas this from Supplementary Figure 3 is based on Pearson correlation with complete linkage method. Both heat maps were created using MultiExperiment Viewer software available from http://www.tm4.org/. Dendrograms from Supplementary Figure 7 (log2 transformed, Euclidean distance measure and Ward's linkage method) were created using R software (Version 2.6.1, available from http://www.r-project.org) on the same data set.
To calculate the changes between neighboring time points (Figure 2A and B) multiple t-tests and ratios (fold change on a linear scale) between the time point of interest and the directly preceding one were calculated. The following significance thresholds were applied: P⩽0.05 and ratio ⩾2 for metabolic data and P⩽0.05, ratio ⩾3 for transcript data. To determine the overlap of responses between different conditions, the number of significant changes between time points from all stress conditions and parallel time points from control culture were calculated using the same strategy as described for neighboring time points, but additionally the direction of change (relative to control) was included. The number of significantly changed features in the same direction across different conditions was calculated, and the significance of overlaps between all pairwise comparisons was tested using the Fisher exact test (P⩽0.05) implemented in the R software package.
Responses to stationary phase and different stress conditions were compared in the following way. Metabolites and transcripts that change significantly (significance thresholds the same as above) within 10–40 min post-perturbation (relative to time points before perturbation), respectively, 210–260 min during stationary phase (relative to time points 90–150 min that reflect the resumption of growth) were compared and the significance of potential overlaps (same direction of the change) was tested using Fisher exact test with a significance level of P⩽0.05).
Transcript analysis
Array design
For transcript analyses, customized arrays were generated on the basis of the Agilent one-color microarray technology platform. The bases for the probe set design are the full genome and sequence annotation files of E. coli K12, which were downloaded from the NCBI genome FTP directory (ftp://ftp.ncbi.nih.gov/genomes/, 11 August 2006). The genome sequence file was parsed according to the annotation files to generate a full sequence list of coding and non-coding regions. The probes were designed using OligoArray 2.1.3 (Rouillard et al, 2002) covering all open reading frames. For each of the designed probes, a probe statistic was generated covering the position from 5′ end, the probe length, the melting temperature, the number of potential cross-hybridizations, the relative GC frequency within the probe, the longest homeomeric run, and the Agilent base composition (BC) score. On the basis of this list, probes with <10 overlapping nucleotides, a minimal sequence length of 50 nt and the best BC score by minimal differences to the arbitrary melting temperature of 88.5°C were selected and filtered. Only probe sets covering open reading frames were analyzed and used for quantification of signal intensity.
Sample preparation
RNA was extracted using the Qiagen RNeasy Mini Kit (74104) and mechanical cell disruption with glass beads but without enzymatic lysis. This was carried out in the Qiagen RNeasy kit lysis RLT buffer with β-mercaptethanol, according to the manufacturer's recommendations. Mechanical cell disruption was completed through shaking for 5 min using a Retsch mill (Retsch MM200) on maximum speed. RNA was subsequently cleaned on-column with an additional DNase treatment (Qiagen 79254). The quality of extracted RNA was determined with an Agilent 2100 bioanalyzer having used an Agilent RNA 6000 Nano Kit according to the manufacturer's recommendations. The labeling and hybridization of cDNA microarrays was performed by the out-sourced service provider imaGene GmbH (Berlin, Germany) and was based on Agilent technology.
Array data extraction and normalization
For further analyses, the processed signal intensities of all coding regions and RNA genes were extracted and used. Variance stabilization and normalization of the extracted intensities were performed with the vsn packages (Huber et al, 2002) of the R software environment (R Development Core Team, 2009) and back-transformed to normal intensity scale. For each probe set, for example all probes representing, for example, a single-coding gene, outliers were removed by boxplot statistics and the outlier-removed probe intensities were averaged in a robust way by computing the Tukey biweight. The complete transcript data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20305.
GO term enrichment analysis
The analysis of overrepresentation of GO terms describing biological processes was done using PageMan software freely available from the following web site: http://mapman.mpimp-golm.mpg.de/pageman/. The significance of overrepresentation of GO terms was assessed by Bonferroni corrected Fisher exact test (P⩽0.05).
Specificity of E. coli's response on metabolite and transcript level
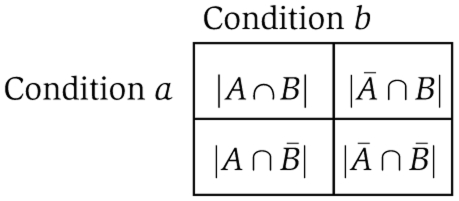
To compare the specificity of the response to perturbations between the metabolite and transcript levels, we rely on the variables (i.e. metabolites and transcripts), which show differential behavior over all examined conditions with respect to the control and over all time points. For a given time point, t, we then attempt to determine whether the overlap of the variables showing differential behavior arises by chance. To this end, for two conditions a and b, at time t, we build a dichotomous 2 × 2 contingency table, denoted by Tt. The contingency table Tt has the following entries:
Here, A denotes the set of variables with a differential behavior under condition a and analogously B corresponds to variables of condition b. Then, ∣A∩B∣ denotes the number of variables showing differential behavior under both conditions a and b. Furthermore, ∣Ā∩B∣ and ∣A∩B¯∣ denote the number of variables showing differential behavior only under condition b or a, respectively. Finally, ∣¯A∩B¯∣ represents the number of variables not changing under both conditions. Illustrations of Tt contingency tables can be found in the Supplementary information.
Let H0 denote the null hypothesis that the numbers of condition-specific variables with differential behavior for a and b are independent, that is, the overlap results by chance. By using Fisher exact test, we are able to either verify the null hypothesis or reject it in favor of the alternative hypothesis H1. Fisher's test gives the probability of the observed configuration for the contingency table under H0 regardless of the sample size (Agresti, 2002). This is important, because the sample size (equal to the sum of all entries in Tt) for the metabolites includes 191 variables, whereas the transcript sample consists of 288 variables. In our analysis, we consider a level of significance P⩽0.05. If the null hypothesis is valid, we will call the systems response to the conditions a and b specific.
Finally, we quantify the specificity of E. coli's response by the PPV of the 24 pairwise condition comparisons either on the metabolite or transcript level. Let TP denote the number of comparisons, where both conditions show an independent response and FP denote the number of dependent pairs of responses. We then define the PPV as PPVl=TPl/(TPl+FPl), where l denotes either the metabolite or transcript level. Note that our definition of the specificity differs from the classical statistical measure of the performance of a binary classification. This is attributed to the fact that there is no equivalent to negative realizations in this experimental setup: among all conditions, the number of changing genes or metabolites is always >0.
Co-clustering and pathway enrichment analysis of genes and metabolites
Here, we use a co-clustering approach to determine the extent to which genes and metabolites, showing differential expression under the investigated conditions, are involved in the same biochemical pathway. We simultaneously apply a k-means clustering algorithm to the combined metabolite and transcript level data for a specific condition, given in a form of an m × n matrix J (m is the total number of genes and metabolites and n is the number of time points).
To limit the effect of the absolute magnitude of concentration or expression levels on a used similarity measure, we normalized every row in J to have zero mean and unit variance (i.e. we perform a z-score transformation).
To supply a suitable estimate for the initial number of clusters (i.e. parameter k) for the k-means algorithm for every experimental condition, we used a graph-based approach to estimate a probabilistic data-dependent range for k (Klie et al, 2010). Briefly, this approach uses the topology of a graph-based representation of J to identify dense regions (i.e. clusters) of the data by means of a random walk. These dense regions are then enumerated by construction of a minimum spanning tree. Note that this range is dependent on the used similarity measure and was computed for Euclidean distance and Pearson's correlation coefficient, each resulting in an independent clustering of J. To further increase the robustness of the presented findings in section ‘The level of coordination between transcript and metabolite data is strongly influenced by the environmental conditions,’ we repeated the clustering procedure 100 times with randomized initial cluster centers for each k in the previously determined interval, for both similarity measures. Out of those 100 clustering runs, we selected the clustering that minimizes the root mean square error for a given k. This approach aims at compensating for the non-deterministic nature of the k-means algorithm.
Finally, over-representation of certain pathways on each cluster was determined analogous to finding enriched GO terms, using the hyper-geometric distribution as a null distribution (Rivals et al, 2007). The significance level was, again, set to 0.05 and the P-values are Benjamini–Hochberg corrected. We focus only on pathways that are enriched for both metabolites and genes, although the pathways enriched only for metabolites and only for genes can also be readily determined. To validate the significance of the observed co-clustering events of genes and metabolites resulting in an enrichment of a certain pathway, we applied a non-parametric bootstrap sampling procedure. Briefly, we sampled m genes and metabolites with replacement and uniform probability from the original condition-specific joint metabolite–transcript data set. The obtained bootstrap sample is then again subjected to k-means clustering to determine whether the previously observed co-clustering is significant or a random observation. A more detailed treatment of this analysis step can be found in the Supplementary information; the derived P-values and probabilities for co-clustering events can be found in Supplementary Table 3 together with the pathway enrichment results. We point out that all co-clustering events presented here are significant at the 1% level (after correcting for multiple testing).
In summary, we searched for pathway-over-enrichment in each combination of experimental condition, choice of k, and similarity measure.
CCA of genes and metabolites involved in primary metabolism
CCA is a statistical technique for studying associations between two sets of variables (Hotelling, 1936) measured under the same experimental units. CCA and its variants were previously applied to either compare data of the same source (e.g. microarray data) originating from different species (van den Berg et al, 2009) or to integrate different sources of data from the same system (e.g. complementing gene expression data with phenotypic data (Gonzlez et al, 2008) or integration of data originating from different ‘omics’ technologies (Le Cao et al, 2009)).
Given a set of genes and a set of metabolites, the principle idea of CCA is to find two linear combinations, one for the set of genes and one for the set of metabolites, which are maximally correlated. Here, the set of genes is described by the matrix X of dimension n × p, where rows correspond to the expression levels measured at n time points of p genes (columns) under one specific condition. Correspondingly, Y of dimension n × q represents the n measured concentrations of q metabolites under the same experimental condition. Furthermore, we denote the ith column of matrix X by Xi and correspondingly denote by Yj the jth column vector of Y. To avoid having many more variables than observations, we used the data from all three independent biological replicates individually, instead of taking the mean or median for the replicates. Moreover, it will be assumed that the columns of X and Y are standardized (i.e. a mean of 0 and a variance of 1), that p⩾q and X as well as Y are of full column rank p and q. Let a1=(a11;…;ap1)T and b1=(b11;…;bq1)T denote the two basis vectors (both of var (U1)=var (V1)=1), such that the correlation between the projections of the variables onto these basis vectors given by
and
are mutually maximized:
The derived linear projections U1 and V1 will be called the first canonical variates and ρ1 is referred to as the first canonical correlation. Higher order canonical variates can be found as a stepwise problem with the restriction to be orthogonal to the already determined set of linear combinations. Note that the successively computed canonical correlations satisfy ρ1⩾ρ2⩾…⩾ρq.
In this work, we use the results of the CCA on a subset of genes and metabolites involved in the primary metabolism (see section ‘The level of coordination between transcript and metabolite data is strongly influenced by the environmental conditions’ and Supplementary Table 4) as an explanatory tool to display associations between genes and metabolites that are less prominent by means of direct linear relationships (e.g. Pearson correlation) in the initial data.
Specifically, for the purpose of visualization, we use two-dimensional scatter plots for the genes and metabolites, which are also known as canonical loadings plots. Here, the axes define the canonical variates Uj and Uk with j≠k and both from the integer interval [1, q], for the gene set X. Coordinates of genes in X and metabolites in Y on each axis correspond to Pearson correlations of their initial representation (e.g. for gene i the corresponding column vector Xi) and the respective canonical variate Us, s∈j, k. This form of correlation is known as canonical structure correlations. An example of a canonical structure correlation vectors can be found in the Supplementary information (Supplementary Table 5).
As both genes and metabolites are assumed to be of unit variance, their projections on the plane (Uj; Uk) reside within a circle of radius 1 centered at the origin. Variables with a strong relation are projected in the same direction from the origin. Clearly, the greater the distance from the origin, the stronger is the relation. For clarity, a second circle with radius of 0.5 is shown to indicate associations of genes and metabolites, which are less strong and of limited importance for the conducted analysis. The CCA results presented in the paper rely on a regularized version of CCA, which is available in the CCA package (Gonzlez et al, 2008), which is available for the statistical software R.
Supplementary Material
Supplementary text, Supplementary tables S1–5, Supplementary figures S1–8
The raw metabolite peak height data (columns 9-205) are supplied together with all samples (rows). The basic description of all samples is given in Columns 1-8 and include: sample ID, experimental condition, time point, technical and biological repetition, internal standard peak height, and a list of samples prior to perturbation. For the further information about the GC-MS analysis and metabolic data processing see Materials and Methods section of the manuscript.Normalization.All metabolites were normalized to the optical density (Column 6) and the intensity of internal standard (Column 7). Additionally all time points were normalized to the average of the time points prior to perturbation which are depicted by "TRUE" in Column 8. The proper alignment of the time points from different biological repetitions of the same experiment was done based on the expression of stress specific marker genes (see manuscript main text for further details)
Acknowledgments
We to thank Aenne Eckardt, Andrea Leisse, Gudrun Wolter, and Antje Bolze for excellent technical assistance, Leonard Krall and Zoran Nikoloski for valuable discussions.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agresti A (2002) Categorical Data Analysis. New York: John Wiley & Sons [Google Scholar]
- Barker MM, Gaal T, Gourse RL (2001) Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for rnap. J Mol Biol 305: 689–702 [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L (2006) Biochemistry. New York: W. H. Freeman [Google Scholar]
- Bolten CJ, Kiefer P, Letisse F, Portais JC, Wittmann C (2007) Sampling for metabolome analysis of microorganisms. Anal Chem 79: 3843–3849 [DOI] [PubMed] [Google Scholar]
- Bradley PH, Brauer MJ, Rabinowitz JD, Troyanskaya OG (2009) Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae. PLoS Comput Biol 5: e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Yuan J, Bennett BD, Lu WY, Kimball E, Botstein D, Rabinowitz JD (2006) Conservation of the metabolomic response to starvation across two divergent microbes. Proc Natl Acad Sci USA 103: 19302–19307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Conway T (2002) Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol Microbiol 45: 289–306 [DOI] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ (2008) Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr SB, Kogoma T (1991) Oxidative stress responses in Escherichia-coli and Salmonella-typhimurium. Microbiol Rev 55: 561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett WP, Wolf RE (1995) Genetic definition of the Escherichia-coli zwf soxbox, the DNA-binding site for SOXS-mediated induction of glucose-6-phosphate-dehydrogenase in response to superoxide. J Bacteriol 177: 1742–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick CN, Curtiss R (1996) Escherichia Coli and Salmonella: Cellular and Molecular Biology, 2nd edn. Washington: ASM Press [Google Scholar]
- Gadgil M, Kapur V, Hu WS (2005) Transcriptional response of Escherichia coli to temperature shift. Biotechnol Prog 21: 689–699 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby P, Davies S, Kelly DJ, Guest JR, Andrews SC (1999) Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C-4-dicarboxylates in Escherichia coli. J Bacteriol 181: 1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L (1988) Posttranscriptional regulatory mechanisms in Escherichia-coli. Annu Rev Biochem 57: 199–233 [DOI] [PubMed] [Google Scholar]
- Gonzlez I, Djean S, Martin PG, Baccini A (2008) CCA: an r package to extend canonical correlation analysis. J Stat Softw 23: 1–14 [Google Scholar]
- Hagemann M (1996) Nacl acts as a direct modulator in the salt adaptive response: salt-dependent activation of glucosyglycerol synthesis in vivo and in vitro. J Plant Physiol 149: 746–752 [Google Scholar]
- Hengge-Aronis R (2000) The general stress response in Escherichia coli. In Bacterial Stress Responses, Storz G, Hengge-Aronis R (eds), pp 249–260. Washington, DC: ASM Press [Google Scholar]
- Hondorp ER, Matthews RG (2004) Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol 2: 1738–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H (1936) CCA: an r package to extend canonical correlation analysis. Biometrika 28: 321–377 [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 (Suppl 1): S96–104 [DOI] [PubMed] [Google Scholar]
- Irr JD (1972) Control of nucleotide metabolism and ribosomal ribonucleic-acid synthesis during nitrogen starvation of Escherichia-coli. J Bacteriol 110: 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HM, Gunsalus RP (1987) Regulation of Escherichia-coli fumarate reductase (frdabcd) operon expression by respiratory electron-acceptors and the fnr gene-product. J Bacteriol 169: 3340–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather B, Stingl K, Van der Rest ME, Altendorf K, Molenaar D (2000) Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate: quinone oxidoreductase. J Bacteriol 182: 3204–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefeld A, Ackermann B, Bauer J, Kramer J, Unden G (2009) The fumarate/succinate antiporter DcuB of Escherichia coli is a bifunctional protein with sites for regulation of DcuS-dependent gene expression. J Biol Chem 284: 265–275 [DOI] [PubMed] [Google Scholar]
- Klie S, Nikoloski Z, Selbig J (2010) Biological cluster evaluation for gene function prediction. J Comput Biol (e-pub ahead of print 8 January 2010; doi:10.1089/cmb.2009.0129) [DOI] [PubMed] [Google Scholar]
- Kultz D (2005) Molecular and evolutionary basis of the cellular stress response. Ann Rev Physiol 67: 225–257 [DOI] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A (2001) Role of inorganic polyphosphate in promoting ribosomal, protein degradation by the ion protease in E-coli. Science 293: 705–708 [DOI] [PubMed] [Google Scholar]
- Le Cao KA, Gonzlez I, Djean S (2009) Integromics: an r package to unravel relationships between two omics datasets. Bioinformatics 25: 2855–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemuth K, Hardiman T, Winter S, Pfeiffer D, Keller MA, Lange S, Reuss M, Schmid RD, Siemann-Herzberg M (2008) Global transcription and metabolic flux analysis of Escherichia coli in glucose-limited fed-batch cultivations. Appl Environ Microbiol 74: 7002–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo MR, Cunningham PR, Clark DP (1993) Anaerobic regulation of the adhe gene, encoding the fermentative alcohol-dehydrogenase of Escherichia-coli. J Bacteriol 175: 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lopez-Maury L, Marguerat S, Bahler J (2008) Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9: 583, Erratum in: Nat Rev Genet 2009; 10: 68 [DOI] [PubMed] [Google Scholar]
- Maharjan RP, Ferenci T (2003) Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem 313: 145–154 [DOI] [PubMed] [Google Scholar]
- Mandelstam J (1963) Protein turnover and its function in economy of cell. Ann N Y Acad Sci 102: 621 [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF (1974) Culture medium for enterobacteria. J Bacteriol 119: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellemann LJ, Holm F, Atlung T, Hansen FG (1989) Cloning and characterization of the Escherichia-coli phosphoglycerate kinase (pgk) gene. Gene 77: 185–191 [DOI] [PubMed] [Google Scholar]
- Nystrom T (2004) Stationary-phase physiology. Ann Rev Microbiol 58: 161–181 [DOI] [PubMed] [Google Scholar]
- Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli k-12. Mol Genet Genomics 272: 580–591 [DOI] [PubMed] [Google Scholar]
- Phadtare S, Inouye M (2004) Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol 186: 7007–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
- Rasmussen LJ, Moller PL, Atlung T (1991) Carbon metabolism regulates expression of the pfl (pyruvate formate-lyase) gene in Escherichia-coli. J Bacteriol 173: 6390–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals I, Personnaz L, Taing L, Potier MC (2007) Enrichment or depletion of a go category within a class of genes: which test? Bioinformatics 23: 401–407 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard JM, Herbert CJ, Zuker M (2002) Oligoarray: genome-scale oligonucleotide design for microarrays. Bioinformatics 18: 486–487 [DOI] [PubMed] [Google Scholar]
- Sawers G, Bock A (1988) Anaerobic regulation of pyruvate formate-lyase from Escherichia-coli k-12. J Bacteriol 170: 5330–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RA, Rubingh CM, Westerhuis JA, van der Werf MJ, Smilde AK (2009) Metabolomics data exploration guided by prior knowledge. Anal Chim Acta 651: 173–181 [DOI] [PubMed] [Google Scholar]
- van der Rest ME, Frank C, Molenaar D (2000) Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J Bacteriol 182: 6892–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R (2005) Genome-wide analysis of the general stress response network in Escherichia coli: sigma(S)-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Wenzel K, Fiehn O (2004) Process for the integrated extraction identification, and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4: 78–83 [DOI] [PubMed] [Google Scholar]
- Willetts NS (1967) Intracellular protein breakdown in non-growing cells of Escherichia coli. Biochem J 103: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientz E, Bongaerts J, Unden G (1998) Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J Bacteriol 180: 5421–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text, Supplementary tables S1–5, Supplementary figures S1–8
The raw metabolite peak height data (columns 9-205) are supplied together with all samples (rows). The basic description of all samples is given in Columns 1-8 and include: sample ID, experimental condition, time point, technical and biological repetition, internal standard peak height, and a list of samples prior to perturbation. For the further information about the GC-MS analysis and metabolic data processing see Materials and Methods section of the manuscript.Normalization.All metabolites were normalized to the optical density (Column 6) and the intensity of internal standard (Column 7). Additionally all time points were normalized to the average of the time points prior to perturbation which are depicted by "TRUE" in Column 8. The proper alignment of the time points from different biological repetitions of the same experiment was done based on the expression of stress specific marker genes (see manuscript main text for further details)