Abstract
Gene copy number variation has been discovered in humans, between related species, and in different cancer tissues, but it is unclear how much of this genomic-level variation leads to changes in the level of protein abundance. To address this, we eliminated one of the two genomic copies of 730 different genes in Saccharomyces cerevisiae and asked how often a 50% reduction in gene dosage leads to a 50% reduction in protein level. For at least 80% of genes tested, and under several environmental conditions, it does: protein levels in the heterozygous strain are close to 50% of wild type. For <5% of the genes tested, the protein levels in the heterozygote are maintained at nearly wild-type levels. These experiments show that protein levels are not, in general, directly monitored and adjusted to a desired level. Combined with fitness data, this implies that proteins are expressed at levels higher than necessary for survival.
Keywords: feedback, gene dosage, protein levels
Introduction
Recent studies have highlighted a wide range of differences in genomic sequences between related species (Dumas et al, 2007), individuals within species (Kidd et al, 2008), and even diseased and normal tissues in an individual (Giaever et al, 1999). Central to interpreting the consequences of these genomic changes on an organism's phenotype is the question of how changes in DNA copy number affect protein abundance. Does a two-fold change in copy number lead to a two-fold change in protein abundance or does feedback buffer genotypic, stochastic, and environmental changes?
Using microarray technology, studies of tumor cell lines (Pollack et al, 2002) and of constructed aneuploidies in S. cerevisiae (Torres et al, 2007) have shown that copy number variation semiquantitatively correlates with mRNA expression changes. Despite this correlation at the mRNA level, 13 of the 16 proteins examined in S. cerevisiae did not change in abundance when their copy number was doubled (Torres et al, 2007). This suggests that compensation for protein abundance could be common, and that protein abundance need not track mRNA level. Furthermore, in several examples, feedback regulation has been shown to ensure homeostasis at the level of protein abundance, through several distinct mechanisms (Cleveland et al, 1981; Pearson et al, 1982; Preker et al, 2002; Ravid and Hochstrasser, 2007).
Such feedback at the protein level has been suggested (Brooker et al) to underlie the general lack of haploinsufficiency observed in genetic studies in Drosophila melanogaster (Lindsley et al, 1972) and S. cerevisiae (Deutschbauer et al, 2005). These studies show that in most cases, a single copy of a gene is sufficient for fitness and normal development. In both of these species, only 3–5% of the deletions are haploinsufficient. Potential explanations for this phenomenon include: (1) feedback buffers protein levels; (2) cells normally produce at least twice as much protein as they need; or (3) many fitness changes are not experimentally measurable. Understanding the contributions of these mechanisms would dramatically affect both how we think about the relationship between genotype and phenotype and how we interpret differences between and within organisms.
It is clear from numerous studies that the abundance of almost every protein can be altered by putting genes on multi-copy plasmids or expressing them under heterologous promoters. Furthermore, drug-induced haploinsufficiency profiling (Giaever et al, 1999), where drug action can be inferred by hypersensitivity of a heterozygous strain to a drug, shows that gene copy number can affect fitness when assayed under the appropriate conditions. However, none of these results give insight into the question of whether widespread feedback occurs at the protein level. First, different genes under the control of the same promoter show a large range in protein abundance (Sopko et al, 2006), underscoring the fact that significant control mechanisms operate at the level of translation and protein turnover. Second, and counterintuitively, overexpression of proteins by multi-copy plasmids and differential sensitivity to drugs between copy number variants is expected even in the presence of strong feedback (Supplementary information). In the presence of strong compensatory mechanism that maintain protein levels, drug-induced haploinsufficiency would still be observed due to saturation of feedback mechanisms. We therefore set out to determine the relationship between genomic copy number and protein abundance in the context of variation in genomic copy number, by comparing the level of expression of 730 different green fluorescent protein (GFP)-fusion proteins in the budding yeast S. cerevisiae in wild-type and heterozygous diploid strains. We show that in most cases, and in multiple environments, protein abundance quantitatively matches gene copy number.
Results
Construction and identification of strains
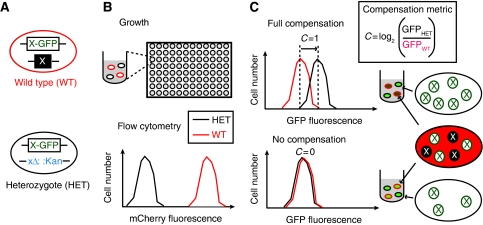
We constructed two libraries of diploid strains of S. cerevisiae from a haploid GFP-fusion library (Huh et al, 2003). One diploid library mimicked the homozygous wild type (X-GFP/X). The other library was heterozygous (X-GFP/Δx) and was constructed by mating the haploid GFP-fusion library to a matched library of haploid deletion strains (Winzeler et al, 1999) (Figure 1; Supplementary Figure S1). If protein expression levels are proportional to gene copy number, as both strains carry only one GFP-tagged gene, both should show the same level of fluorescence. In heterozygous strains that fully compensate for the gene deletion, however, the GFP fusion-tagged gene should be expressed at a two-fold higher level than that seen in the wild type. To assess the amount of GFP-fusion proteins, we used flow cytometry. Recent work that has established this technique accurately reports protein abundance and variance on a genome-wide scale (Newman et al, 2006).
Figure 1.
Systematic analysis of protein compensation in a heterozygous strains. (A) Pairs of diploid strains were constructed where one strain is ‘wild type,' containing a single copy of a GFP fusion and constitutively expresses mCherry (WT, red-bordered cell). The matching strain is a heterozygous deletion of the GFP-fusion gene, it contains one copy of the same GFP fusion but the second copy of the gene is deleted (HET, black-bordered cells). (B) The strains were co-cultured in 96-well plates. Fluorescence from many cells was simultaneously recorded for each strain by flow cytometry and segmented into HET and WT populations (shown as a histogram). (C) The mean GFP fluorescence was calculated for each population of cells (GFPHET and GFPWT). We defined a compensation metric, C=log2 (GFPHET/GFPWT). Two hypothetical examples are shown, the top panel depicts a situation where the heterozygous strain fully compensates for the deletion of one of the two copies of its gene–C=1. The bottom panel depicts a strain that does not compensate–C=0. Details of our calculations are elaborated in the Supplementary information. As depicted on the far right, in wild-type cells, half of protein X is GFP tagged (white circles with a green X), whereas the other half is untagged (black circles with a white X). All molecules of protein X in the heterozygote strain are GFP tagged. If the protein has full compensation, the total amount of protein X should be the same in the heterozygous and wild-type strains, and therefore the amount of X-GFP will be double than that in the wild-type strain. If there is no compensation, both strains will have the same amount of X-GFP as the wild-type strain.
Note that these GFP fusions all contain a heterologous 3′ UTR. Although regulation through the 3′ UTR has been identified, feedback regulation has not been widely reported, and most fusion proteins behave like their untagged cognates in terms of mRNA expression, localization, and degradation (Ghaemmaghami et al, 2003; Huh et al, 2003; Newman et al, 2006). Nevertheless, it is possible that experiments using these libraries may fail to capture some aspects of protein regulation that require the endogenous 3′ UTR.
X-GFP/X and X-GFP/Δx strains were cultured in the same well. The homozygous strains all constitutively express mCherry, allowing us to distinguish the cells of the two strains by flow cytometry at the same time as measuring GFP fluorescence (Figure 1; Supplementary information). By comparing the fluorescence of our ‘wild-type' diploid library (X-GFP/X) to a wild-type diploid library not expressing mCherry, or to the haploid GFP library (X-GFP), we were able to show that neither constitutive mCherry expression nor the presence of a second wild-type copy of the gene alters the expression of the GFP-tagged protein (Supplementary Figures S2 and S3). Of the ∼3350 paired strains in our library, we were able to accurately measure fluorescence levels for only ∼1600 pairs [similar to earlier studies (Newman et al, 2006)], due to low levels of signal. After eliminating strains likely to be aneuploid in the deletion collection (Hughes et al, 2000), we identified 730 pairs of strains in which the signal-to-noise ratio was high enough that we should be able to confidently detect variations in protein expression level of under two-fold in two different medias (rich and synthetic complete). We selected these strains for further study (Supplementary Table SI).
Changes in copy number correlate with protein levels
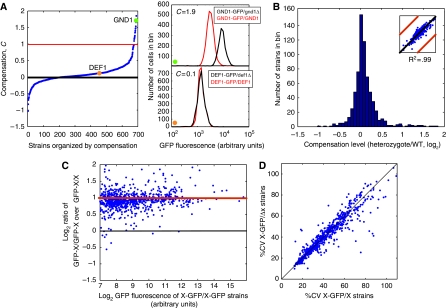
We quantitated the fluorescence in matched pairs of X-GFP/X and X-GFP/Δx strains grown in synthetic complete medium (SD) and found that the fluorescence level was predominantly the same in both libraries (Figure 2A and B; Supplementary information). As the X-GFP/X strain carries two X genes, one of which is not fused to GFP, whereas the heterozygous strain carries only the X-GFP gene, we infer that most proteins are expressed at half the level in the heterozygous strain. Active control of protein levels would result in the fluorescence level in the X-GFP/Δx strain being greater than the fluorescence level in the X-GFP/X strains. For most genes this is not the case.
Figure 2.
Quantitative flow cytometry reveals low levels of compensation for gene dosage. In these log2 plots, the black line indicates no change, whereas the red line indicates a two-fold difference between strains (in A and C) or replicate runs (in B inset). (A) The graph (left panel) shows the averaged compensation level for all replica runs for all strains in synthetic complete medium (SD) ordered by compensation level. The green circle and orange circle denote the compensation level of a compensator (GND1) and a non-compensator (DEF1), respectively. The histograms (right panel) show one experimental run for GND1 and DEF1 grown in SD medium. The heterozygous strain is show in black and the wild-type strain in red. If strains compensated fully, the ratios would be centered around the red line. Strains centered around the black line indicate no compensation. Subtle differences in the wild type versus deletion background most likely account for the 4% offset of whole population from no compensation (Supplementary information). (B) Histogram of the graph in (A) (left panel). The inset shows the reproducibility of replicate measurements. (C) Average mean expression of diploid strains where both alleles are fused to GFP (X-GFP/X-GFP) compared with diploid strains where only one of the alleles is fused to GFP (X-GFP/X). Strains centered around the red line have double the fluorescence when both alleles are tagged. (D) The percent coefficient of variance (CV) for the X-GFP/X strains is plotted against the percent CV of the X-GFP/Δx strains.
The number of genes that show close to complete compensation is very low: only 3% of the genes we studied show a mean fluorescence in the heterozygote that is at least 175% of the level of the wild type (Supplementary Table SII). In all, 14% of genes compensate to some degree, using a cutoff of 23% deviation from wild-type expression (twice the s.d. of the measurement error, corresponding to a 10–20% false discovery rate (Supplementary information)). A few genes (4%) actually decrease expression by >23% in the heterozygote (30–70% FDR). The degree of compensation in each strain was reproducible (average coefficient of variance, or CV, of 10%; inset Figure 2B; Supplementary information).
One trivial explanation for the lack of observed compensation is that our method cannot accurately detect a two-fold change in protein levels. To test this, we constructed a diploid library in which both alleles were GFP tagged (X-GFP/X-GFP). For the majority of strains, the diploid library with both alleles tagged with GFP has nearly double the fluorescence of the library with just one allele tagged with GFP (Figure 2C). Thus, the lack of compensation we observe cannot be explained by a failure to detect two-fold changes in protein expression levels. A second possibility is that if compensation were mediated through the 3′ UTR, we might not detect compensation because the genes in our collection have a shared non-endogenous 3′ UTR. We therefore tested seven N-terminal GFP fusions; for these genes, we found that the 3′ UTR is not a likely source of compensation (Supplementary Table SVII). Another possibility is that some, but not all, of the cells in the population show compensation. However, the population distribution of fluorescence is very similar for most of the matched X-GFP/X and X-GFP/Δx strains. Accounting for expression levels, there is no significant change in the coefficient of variation between paired X-GFP/X and X-GFP/Δx strains before or after filtering the data to eliminate the contribution of cell-size variance (Newman et al, 2006) (Figure 2D; Supplementary information). These data indicate that the level of fluorescence accurately reports on the level of protein expression. We therefore argue that our data conclusively show that active feedback control of protein levels is infrequent in the S. cerevisiae genome, at least under the growth conditions examined. For most genes, the average protein level is proportional to gene copy number and expression from each allele is independent, at least in a range close to wild-type expression.
The relationship between gene copy number and protein levels is maintained in different environments
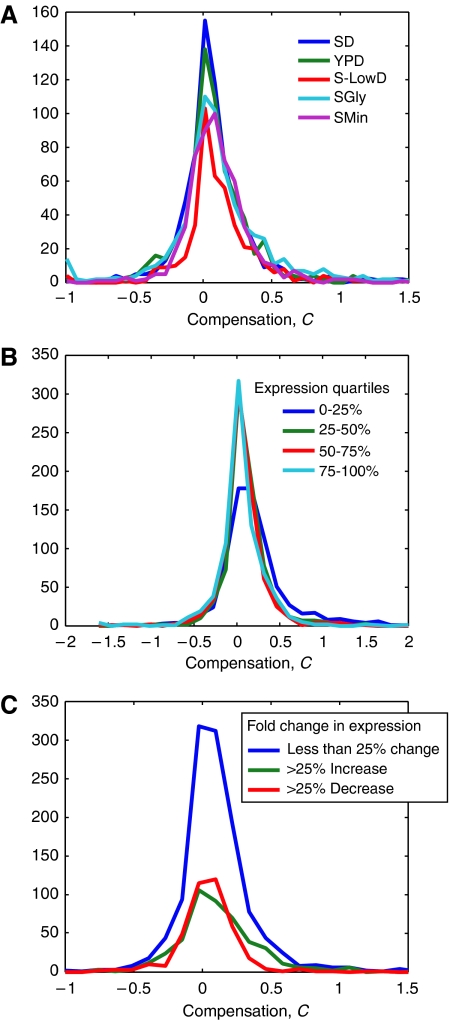
In our initial studies, we grew cells in synthetic complete media. However, a number of genes are only expressed under certain environmental conditions, and we reasoned that similarly compensation might also occur only under other, potentially more stressful, environmental conditions. Therefore, we looked for compensation in protein expression in a number of different media: synthetic minimal medium in which only essential amino acids are supplied, synthetic complete medium with low glucose, synthetic complete medium containing glycerol as the sole carbon source, and rich media (YPD). The protein abundance of a large number of genes changed under many of these conditions (Supplementary information). Nonetheless, both the overall frequency of compensation (Figure 3A) and the specific genes that compensated and exacerbated were similar in all the growth media (Supplementary Table SIII). Exacerbators are strains where the protein levels in the heterozygous strain decrease by more than two-fold. Furthermore, genes that changed protein abundance between two media were not more likely to compensate, nor were compensators more likely to change their protein abundance between these media (Figure 3B and C).
Figure 3.
Compensation is not strongly influenced by environment or expression. (A) Histogram showing the amount of compensation from five different growth media. YPD is rich media. SGlycerol is synthetic complete media with only glycerol as a carbon source. S-LowD is synthetic complete media with low glucose. Minimal is synthetic media containing only uracil, methionine, and glutamate (no other nitrogen source). (B) Histogram of the amount of compensation (averaged log2 of the ratio of X-GFP/X over X-GFP/Δx) broken up by expression range. Strains were divided into quartile expression bins (0–25% being the lowest expressers and 75–100% being the highest expressers) based on the absolute expression of the WT strain (X-GFP/X) in SD medium. (C) Histogram of the amount of compensation binned by fold expression change from growth in SD media. Strains were divided into three bins (decreased expression, increased expression, and no change) based on two times the s.d. of the measurement error for changes in expression (Supplementary information).
Compensators are represented in many gene classes
Given that compensators seem to be consistent across conditions, we asked whether there are recognizable similarities among the compensators. We examined the compensators for overrepresentation of any of the following characteristics: (1) common function, (2) duplicate genes, (3) high noise levels, (4) reduced growth when deleted, (5) common pathway, and (6) members of large complexes. We found compensators in all of these gene classes, but did not find a significant overrepresentation of compensators in any of them. (1) We were unable to find any apparent pattern in the functions of the compensating genes (Ashburner et al, 2000; Zeeberg et al, 2003) (using multiple different cutoffs for significance; Supplementary information). However, because our library did not include low abundance proteins, such as many signaling proteins and transcription factors, it is possible that these classes of proteins may behave differently. There was no correlation between expression level and compensators in our set (Supplementary Figure S4). (2) Compensators do not share any obvious correlations such as over/underrepresentation among duplicate genes, and (3) do not show enhanced deviation from expected noise levels (DM) (Newman et al, 2006). Interestingly, exacerbators do have higher than expected noise levels (P-value of 0.01). As exacerbators are proteins whose expression decreases by more than two-fold when gene dosage is decreased by two-fold, one might expect exacerbators to be involved in positive feedback regulation (Supplementary information). Some genes involved in positive feedback would be expected to be noisier than genes not involved in feedback regulation, perhaps explaining our observation (Supplementary information). (4) Compensators and exacerbators are overrepresented among genes that cause reduced growth when deleted, but only modestly so (Supplementary Table SIV). We also measured competitive fitness by flow cytometry (Breslow et al, 2008) for our strains in YPD and found there was no correlation between growth rate and compensation (Supplementary Figure S5). The majority of the strains queried had no growth defect as a heterozygous deletion, in agreement with earlier work (Deutschbauer et al, 2005). (5) We also examined many pathways for compensation at the pathway level. We found evidence for compensation in the lysine pathway, but no other pathways (Supplementary Table SV). (6) We also examined the role of complex assembly, as some proteins are unstable until they are assembled into larger complexes (Gorenstein and Warner, 1977) and several of these proteins appear to maintain protein levels in the face of aneuploidy (Torres et al, 2007). Although we were unable to find any correlation between compensation and members of protein complexes, our library was biased against a number of components of large protein complexes. To look at more indirect interactions, we extracted physical interaction data from BioGrid (Breitkreutz et al, 2008) (http://www.thebiogrid.org) and looked for proteins involved in feedback with a path length of five or smaller. Compensator and exacerbators were not enriched in this set. Of note, exacerbators are enriched among genes with longer mRNA half-life and significantly more likely to not express two-fold more GFP in a X-GFP/X-GFP strain as compared with a X-GFP/X strain (Supplementary Figures S7 and S8; Supplementary Table SIX). In total though, compensators and exacerbators are most notable for their lack of similarities.
Compensators are rare among essential genes and overexpressed genes
To test whether essential (Ess) genes behave differently from non-essential genes, we created 40 diploid strains where one copy of an essential gene was fused to GFP and the second copy was under the control of doxycycline (EssX-GFP/TetO7pr-EssX) (Mnaimneh et al, 2004). In the presence of 10 μg/ml doxycycline, production from the tetracycline promoter is blocked, creating a functionally heterozygous strain that can be compared directly with a wild-type strain (EssX-GFP/EssX). Two of the 40 genes compensated (Supplementary Table SVI), a similar proportion to that seen with non-essential genes. One gene showed altered expression in the absence of doxycycline, raising the possibility that in some cases there may be compensation for overexpression (Torres et al, 2007). The finding that most essential genes do not compensate indicates that essential genes behave similarly to non-essential genes.
To test whether protein levels compensate for overexpression, we created 65 strains where one copy was fused to GFP and a second plasmid-borne copy was under the control of a galactose-inducible promoter (X-GFP pGALpr-X) (Gelperin et al, 2005). When cells are grown in medium with raffinose as the sole carbon source, only the GFP fusion of protein is expressed; in medium with galactose as the sole carbon source, the GFP fusion of the protein is expressed and the plasmid-borne allele is overexpressed. If protein abundance is sensitive to overexpression, the GFP fluorescence should be suppressed in galactose medium but not in raffinose medium. In all, 8% of the GFP alleles showed suppression in overexpression conditions (Supplementary Table SVII). A caveat of galactose overexpression is that the degree of overexpression is often much larger than the absolute expression of the protein. As argued for drug-induced haploinsufficiency profiling (Supplementary information), this degree of overexpression may hide compensation. In general, protein abundance does not compensate for galactose overexpression.
Discussion
By making a series of 730 heterozygous deletion strains in S. cerevisiae, we showed that protein abundance quantitatively corresponds to gene copy number. This result was consistent in a variety of environmental conditions; cells grown in five different media produced similar results. We also confirmed this result for essential proteins and overexpressed proteins. Given that our data show a correspondence between gene copy number and protein abundance, our data also quantitatively support, on a single gene basis, the semiquantitative result that mRNA abundance tracks gene copy number (Torres et al, 2007) (Pollack et al, 2002). Although our results are not consistent with an earlier study that compared protein levels with gene dosage for 16 gene in aneuploid strains (Torres et al, 2007), this inconsistency most likely represents the accuracy of using GFP fluorescence over westerns to quantitate protein levels or our larger sample size.
Environmental perturbations often alter gene and protein expression. Homeostasis is an essential feature of life, and feedback regulation often attenuates (or exacerbates) the response to a new environment over time. A priori, one might therefore have expected that monitoring the relationship between protein levels and gene copy number would uncover many examples of feedback. In particular, one might expect genes involved in negative feedback to compensate for changes in gene dosage, and genes involved in positive feedback to exacerbate changes in gene dosage. The fact that compensation is observed so rarely suggests that, in S. cerevisiae, direct regulation is more common than feedback regulation (Levy et al, 2007) or that feedback regulation is relatively insensitive to protein levels. Supporting this, a number of human disorders are caused by haploinsufficiency in transcription factors (Seidman and Seidman, 2002). If feedback regulation were predominant, transcription factor haploinsufficiency in humans would be less common. Interestingly, this would suggest that transcription factors, a class of protein underrepresented in our study, are in many cases not present in humans at twice the necessary level.
Because of the constraints of flow cytometry, our study primarily included highly abundant proteins. It is therefore possible that lower abundance proteins such as transcription factors and signaling molecules, which were underrepresented in our study, may behave differently. Furthermore, proteins that are integrally packed in large macromolecular complexes are often difficult to successfully tag with GFP, which may bias us against discovering feedback in these cases. As the proteins in this study comprise the majority of total cellular proteins, in terms of bulk protein abundance, most of the cellular proteins are not controlled by feedback.
What are the consequences of the rarity of feedback at the protein level? The range of observed protein levels is determined by the rate at which selection eliminates strains with suboptimal protein expression and by the frequency of mutations that alter protein levels. The fitness of a given protein level is constrained by the benefit and cost of producing a given amount of a protein. Experimentally, our knowledge of these constraints is minimal. The cost of protein expression has been well studied for the lac system in Escherichia coli (Dekel and Alon, 2005), but even in this system, we do not know where the cost of protein expression arises (Stoebel et al, 2008), nor do we know whether these results are general for gene expression in bacteria, or whether they hold for eukaryotes (Gelperin et al, 2005; Sopko et al, 2006). It is entirely possible that in eukaryotes, excess protein production is a fitness advantage, as it can be used as a storage form for nitrogen. Balancing these selective forces is the large number of mutations that could affect protein stability or translational efficiency; such mutations can easily alter protein levels, especially in the absence of feedback. It is quite possible that the levels of many eukaryotic proteins can be neutral for fitness over a broad range, and that the levels of these proteins can change dramatically with little or no evolutionary consequences.
Combined with earlier studies and our own data that most heterozygous S. cerevisiae strains do not have altered fitness (Deutschbauer et al, 2005), our study suggests that a majority of yeast proteins are expressed at least twice the needed level. Other explanations for the prevalence of dominance, which are not necessarily mutually exclusive, include enzyme saturation (Fisher, 1928; Wright, 1934; Kacser and Burns, 1981), distributed sensitivities in pathways (Fisher, 1928; Kacser and Burns, 1981), and a requirement for high levels of protein expression under rare physiological conditions (Haldane, 1933).
Although variations in individual protein levels may not intrinsically lead to a fitness cost, they will often lead to quantitative differences in phenotype that may under some environments convey a selective advantage or disadvantage (Breslow et al, 2008; Hillenmeyer et al, 2008). Our results show that individual variations in gene copy number can be expected to cause concomitant variations in protein level. The effects of such variations on susceptibility to disease and the response to drugs will be a major area of future investigation.
Materials and methods
Strains and media
The strains used in this study were in the S288C background. All the libraries constructed were derived from three sources: the yeast GFP-fusion library (Huh et al, 2003), the MAT α yeast deletion collection (Winzeler et al, 1999), the yeast tetracycline inducible essential library (Mnaimneh et al, 2004), the yeast galactose overexpression library (Gelperin et al, 2005), and the SGA query strain (Tong and Boone, 2007). mCherry expressed under the TEF1 promoter and linked to a Kanamycin resistance cassette was integrated at the TRP1 locus in the SGA query strain. A second SGA query strain was made by integrating a Kanamycin resistance cassette at the TRP1 locus. Diploid libraries were made by crossing the MAT a GFP library to the yeast deletion collection, the SGA query strains, or other libraries made from haploid spores of these initial crosses. The libraries were grown in 96-deep well format (600 μl/well) at 23°C. Saturated overnight cultures were grown in YPD for each library. To compare two libraries, a second saturated overnight culture was made by pinning each of the libraries into the same 96-deep well plate in YPD. Each well thereby contained both a query strain and a reference strain. The night before an experiment, the cultures were diluted with a pinner into the media of interest. The cells were grown until OD600 0.3–0.7 at 23°C in a platform shaker, which took ∼12 h. More detail about strain construction and growth are in the Supplementary information.
Flow cytometry
Cells were pelleted by centrifugation at 3000 g for 3 min at RT. Cells were washed twice with TE (10 mM Tris, 1 mM EDTA pH 7.5). A BioMek FX liquid handling robot was used to transfer the cells from 96 well to 384 well plates. A flow cytometer with a high-throughput autosampler (LSRII with a HTS, Becton Dickinson) was used to record fluorescence from GFP and mCherry fluorophores as well as forward and side scatter (more detail are in the Supplementary information).
Data analysis
Analysis was performed largely as described by Newman et al (2006) except that a much less stringent size cutoff was used (only 10% of all cells were removed). Typically, 10 000–60 000 cells were counted per well. The presence or absence of mCherry fluorescence was used to differentiate the query and reference cells. The difference between the mean fluorescence between the control and experimental cells was on average two orders of magnitude; hence the separation was unambiguous. The GFP channel was recorded for each cell and the autofluorescence background was subtracted from each. This background was determined from running a series of cells, which did not express GFP. The logarithm of the ratio of the GFP expression of the experimental strain divided by the control strain was determined for each well. Replicate measurements were averaged and were used to determine the s.d. of the measurements. The s.d. between the replicate measurements was used to calculate the false discovery rate assuming no compensation. More extensive details of these calculations are given in the Supplementary information online.
Supplementary Material
Supplementary text, Supplementary tables SIII-IX, Supplementary figures S1–15
List of the names and well positions of all the strains in the compressed libraries
The averaged fluorescent values for the MSL1 and MSL3 from all the experimental media
Acknowledgments
We thank Ron Milo and Paul Jorgensen advice and discussions; Tamara Brenner, Itai Yanai, Uri Alon, Yitzhak Pilpel, and Becky Ward for commentary; Uri Alon and David Harmon for discussion of data analysis; Jan Ihmels and Nick Ingolia for reagents and help with cytometry; Fred Winston, Charlie Boone, Alex De Luna, and Pam Silver for reagents and strains; and the ICCB for help with robotics. This work was supported by the National Institutes of Health (NIH) 5R01 HD037277, HHMI, and the Helen Hay Whitney Foundation postdoctoral fellowship (M.S.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M (2008) The BioGRID Interaction Database: 2008 update. Nucleic Acids Res 36: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS (2008) A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods 5: 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker RJ, Widmaier EP, Graham LE, Stiling PD (2008) Biology, 1st edn. McGraw Hill, Boston, MA, USA [Google Scholar]
- Cleveland DW, Lopata MA, Sherline P, Kirschner MW (1981) Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell 25: 537–546 [DOI] [PubMed] [Google Scholar]
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592 [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G (2005) Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, Pollack JR, Sikela JM (2007) Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res 17: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1928) The possible modification of the response of the wild type to recurrent mutations. Am Nat 62: 115–126 [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW (1999) Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21: 278–283 [DOI] [PubMed] [Google Scholar]
- Gorenstein C, Warner JR (1977) Synthesis and turnover of ribosomal proteins in the absence of 60S subunit assembly in Saccharomyces cerevisiae. Mol Gen Genet 157: 327–332 [DOI] [PubMed] [Google Scholar]
- Haldane JBS (1933) The genetics society of America. Papers read at the Atlantic City meeting. Am Nat 67: 62–87 [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G (2008) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, Friend SH, Marton MJ (2000) Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25: 333–337 [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686. [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA (1981) The molecular basis of dominance. Genetics 97: 639–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R et al. (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Ihmels J, Carmi M, Weinberger A, Friedlander G, Barkai N (2007) Strategy of transcription regulation in the budding yeast. PLoS ONE 2: e250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Sandler L, Baker BS, Carpenter ATC, Denell RE, Hall JC, Jacobs PA, Miklos GLG, Davis BK, Gethmann RC, Hardy RW, Hessler A, Miller SM, Nozawa H, Parry DM, Gould-Somero M (1972) Segmental aneuploidy and the genetic gross structure of the drosophila genome. Genetics 71: 157–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng W-T, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF et al. (2004) Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Pearson NJ, Fried HM, Warner JR (1982) Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell 29: 347–355 [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO (2002) Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA 99: 12963–12968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Kim KS, Guthrie C (2002) Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M (2007) Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol 9: 422–427 [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C (2002) Transcription factor haploinsufficiency: when half a loaf is not enough. J Clin Invest 109: 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330 [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE (2008) The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178: 1653–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Boone C (2007) Methods in Microbiology, Vol. 36, 2nd edn. Elsevier, Maryland Heights, MO, USA [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wright S (1934) Physiological and evolutionary theories of dominance. Am Nat 68: 24–53 [Google Scholar]
- Zeeberg B, Feng W, Wang G, Wang M, Fojo A, Sunshine M, Narasimhan S, Kane D, Reinhold W, Lababidi S, Bussey K, Riss J, Barrett J, Weinstein J (2003) GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol 4: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text, Supplementary tables SIII-IX, Supplementary figures S1–15
List of the names and well positions of all the strains in the compressed libraries
The averaged fluorescent values for the MSL1 and MSL3 from all the experimental media