Abstract
Gene regulation differs greatly between related species, constituting a major source of phenotypic diversity. Recent studies characterized extensive differences in the gene expression programs of closely related species. In contrast, virtually nothing is known about the evolution of chromatin structure and how it influences the divergence of gene expression. Here, we compare the genome-wide nucleosome positioning of two closely related yeast species and, by profiling their inter-specific hybrid, trace the genetic basis of the observed differences into mutations affecting the local DNA sequences (cis effects) or the upstream regulators (trans effects). The majority (∼70%) of inter-species differences is due to cis effects, leaving a significant contribution (30%) for trans factors. We show that cis effects are well explained by mutations in nucleosome-disfavoring AT-rich sequences, but are not associated with divergence of nucleosome-favoring sequences. Differences in nucleosome positioning propagate to multiple adjacent nucleosomes, supporting the statistical positioning hypothesis, and we provide evidence that nucleosome-free regions, but not the +1 nucleosome, serve as stable border elements. Surprisingly, although we find that differential nucleosome positioning among cell types is strongly correlated with differential expression, this does not seem to be the case for evolutionary changes: divergence of nucleosome positioning is excluded from regulatory elements and is not correlated with gene expression divergence, suggesting a primarily neutral mode of evolution. Our results provide evolutionary insights to the genetic determinants and regulatory function of nucleosome positioning.
Keywords: evolution, gene regulation, nucleosome positioning
Introduction
The large phenotypic differences between closely related species suggest that gene regulation is a major driving force of phenotypic diversity (King and Wilson, 1975). Widespread differences in gene expression were indeed identified when species with an almost identical set of genes were compared (Rifkin et al, 2003; Khaitovich et al, 2006; Tirosh et al, 2009). The genetic basis of most inter-species variations, however, remains largely unknown. In particular, only a small fraction of the observed differences could be traced to sequence divergence at transcription-factor binding sites (TFBSs) (Zhang et al, 2004; Tirosh et al, 2008). A compelling hypothesis is that expression divergence is due, at least partially, to changes in the chromatin environment and in particular the positioning of nucleosomes (Lee et al, 2006; Choi and Kim, 2008; Field et al, 2009).
The precise positioning of nucleosomes, in particular along gene promoters, can influence gene expression by restricting the accessibility of regulatory proteins for binding DNA (Yuan et al, 2005; Li et al, 2007). Consequently, promoter nucleosome occupancy was shown to correlate with the levels of gene expression (Lee et al, 2004, 2007; Liu et al, 2005; Pokholok et al, 2005; Hogan et al, 2006; Shivaswamy et al, 2008; Field et al, 2009; Zawadzki et al, 2009). The hypothesis that nucleosome positioning contributes also to the divergence of gene expression received some support from several analyses. In one study, inter-species divergence of gene expression was correlated with predicted changes in promoter nucleosome occupancy (Tirosh et al, 2008). An association study reported that a large portion of inter-strain expression variations are linked to a small number of chromatin regulators (Lee et al, 2006; Choi and Kim, 2008). Finally, differential expression of respiration genes in two distant yeast species was associated with differential nucleosome occupancy (Ihmels et al, 2005; Field et al, 2009). Yet, despite this potential role of nucleosomes, efforts to understand the genetic basis of regulatory evolution have focused on the divergence of TFBSs (Borneman et al, 2007; Doniger and Fay, 2007; Odom et al, 2007; Wray, 2007), whereas virtually nothing is known about the divergence of nucleosome positioning.
In this study, we used high-throughput sequencing to map nucleosome positioning in two yeast species, Saccharomyces cerevisiae and Sacchormyces paradoxus, as well as their inter-species hybrid. This data enabled us to characterize the divergence of nucleosome positioning between these closely related species, analyze the extent by which these changes can explain the evolutionary divergence of gene expression, and evaluate the relative contribution of different mechanisms that influence nucleosome positioning.
Results
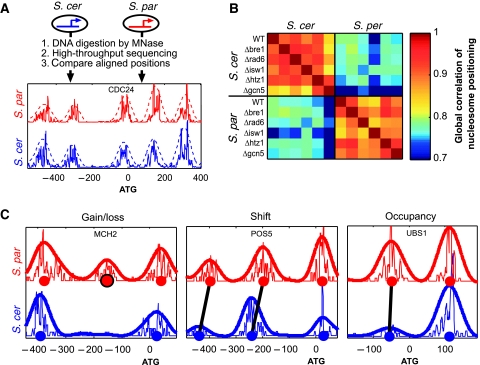
We mapped the genome-wide nucleosome positioning of the budding yeast S. cerevisiae and its close relative S. paradoxus (∼85% genome identity) by subjecting genomic DNA to MNase digestion, followed by high-throughput sequencing (Figure 1A). For each species, we profiled a wild-type strain and, in addition, five mutant strains each deleted of a key chromatin regulator (Figure 1B). The influence of these chromatin regulators on nucleosome positioning will be discussed elsewhere, whereas here we focus on the inter-species comparison of nucleosome positioning. Analyzing ∼4000 aligned promoters and coding regions, we identified reliable inter-species differences at ∼10% of the nucleosomes (Figure 1C; Supplementary Figures S1 and S2). Differences that appear to be due to MNase digestion bias were excluded from further analysis (Supplementary Figure S1) and the remaining differences were classified into three classes (see Materials and methods): nucleosomes whose occupancy was changed by at least twofold (∼2000 cases), nucleosomes that were shifted in their positions (∼300 cases), and nucleosomes that were completely lost or gained in one of the species (∼170 cases).
Figure 1.
Comparison of nucleosome positioning between two yeast species. (A) Mono-nucleosomal DNA was isolated from S. cerevisiae and S. paradoxus by digestion with MNase, pooled, and subjected to Illumina high-throughput sequencing. Reads were mapped to aligned positions in the two genomes and scores for nucleosome positioning were calculated with a Gaussian filter (dashed lines). The reads density and nucleosome scores are shown around the CDC24 start codon for the two species. (B) Correlations between nucleosome scores of the two species from the different experiments (WT and five mutant strains), calculated over all aligned positions. The correlations between different strains of the same species are typically much higher than that between strains of the two different species, indicating that deletions of chromatin regulators led to fewer changes in nucleosome positioning than those between the two species. (C) Examples of the three classes of differential positioning. Predicted center locations of nucleosomes are marked with circles. We identified ∼2000 occupancy changes, 300 shifts and 170 nucleosome gains/losses in comparison of the two species. Similar numbers of changes were found when comparing either wild-type or mutant strains of the two species, and approximately half were consistently observed at most (at least three) of these independent analyses.
Distinguishing cis versus trans effects on differential nucleosome positioning
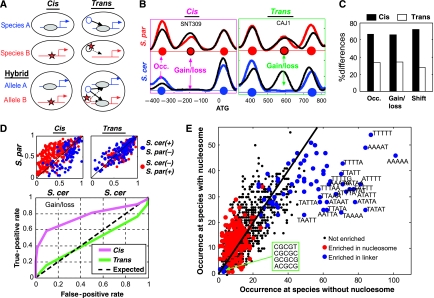
Nucleosome positioning is determined in cis by the local DNA sequences (Ioshikhes et al, 2006; Segal et al, 2006; Kaplan et al, 2009), and in trans through the activity of various factors such as chromatin-remodeling enzymes and the transcriptional machinery (Whitehouse et al, 2007; Hartley and Madhani, 2009; Kaplan et al, 2009; Zhang et al, 2009; Weiner et al, 2010). The relative contribution of cis and trans factors in determining nucleosome positioning is debated (Kaplan et al, 2009; Zhang et al, 2009). To classify the inter-specific differences in nucleosome positioning into those generated by mutations in cis or in trans, we mapped the allele-specific nucleosome positioning in the inter-species hybrid (Wittkopp et al, 2004; Tirosh et al, 2009). Within the hybrid, two orthologous alleles are regulated by the same trans environment; differences between the alleles must therefore be due to cis effects. In contrast, inter-species differences that disappear in the hybrid reflect trans effects (Figure 2A and B). This analysis classified ∼70% of the inter-species differences as cis (a similar difference observed also between the hybrid alleles) and the remaining ∼30% as trans (differences were not observed between the hybrid alleles) (Figure 2C).
Figure 2.
Cis- and trans-dependent changes in nucleosome positioning. (A) An inter-species difference in nucleosome positioning that is generated in cis will be preserved within the hybrid. In contrast, an inter-species difference in nucleosome positioning that is generated in trans will not be preserved in the hybrid as the two hybrid alleles are at the same nucleus and thus affected by the same proteins in trans. (B) Examples of changes classified as cis and trans, showing nucleosome scores (smoothed nucleosome occupancy) of S. cerevisiae (blue), S. paradoxus (red) and the corresponding hybrid alleles (black). Predicted center locations of nucleosomes are marked with circles. (C) Estimation of the percentage of cis and trans differences (see Materials and methods). (D) Differences in cis (but not trans) are predicted from sequence divergence. Nucleosome occupancy was predicted in the two species with a sequence-based model (Field et al, 2008). Upper panel: the predicted nucleosome occupancy of S. cerevisiae (x axis) and S. paradoxus (y axis) at positions of nucleosome gain/loss; blue and red dots refer to nucleosomes that are present only in S. cerevisiae and S. paradoxus, respectively. Lower panel: predictive power of the sequence model. For each possible threshold, we examined the frequency of nucleosome gains/losses with difference in predicted occupancy that pass the threshold (true positives) and compared it with the frequency of randomly selected positions (false positives) with difference in predicted occupancy that pass this threshold. (E) The number of times each 5-mer is found at positions of nucleosome gains/losses at the species without (x axis) or with nucleosome (y axis). Sequences are indicated for the most significantly biased 5-mers. Colors indicate whether a 5-mer is generally associated (in S. cerevisiae) with linker regions (blue) or nucleosomes (red).
Cis effects depend on the local DNA sequence and are thus expected to be relatively independent of genetic or environmental perturbations. In contrast, trans effects depend on protein activity, which is more likely to vary when conditions are changed. Predicted cis effects were indeed consistent between wild type and mutant backgrounds, whereas trans effects were variable (Supplementary Figure S3). Notably, only few of the trans effects could be attributed to one of the five regulators examined, suggesting a complex regulation by multiple factors (Supplementary Figure S3). A compelling hypothesis is that trans divergence is dominated by differences in environmental sensing and signal transduction, as we recently demonstrated for gene expression divergence (Tirosh et al, 2009).
To further examine the genetic basis for nucleosome divergence, we used a recent estimation for the contribution of each DNA 5-mer to nucleosome binding (Field et al, 2008). Most cis-dependent nucleosome gains/losses and changes in occupancy were correctly predicted by this model (Figure 2D; Supplementary Figure S4), whereas the model had no predictive power for trans-dependent nucleosome gains/losses. Notably, the predictive power of the model was due almost exclusively to the nucleosome-disfavoring property of AT-rich sequences, particularly poly(dA/dT) tracts (Iyer and Struhl, 1995; Segal and Widom, 2009) (Figure 2E; Supplementary Figure S4). In contrast, changes in other 5-mers that code for nucleosome-free DNA, including the binding sites for the RSC nucleosome-remodeling complex (Badis et al, 2008) did not contribute significantly to nucleosome divergence. Moreover, changes in sequences implicated as having a nucleosome-favoring activity were only weakly associated with diverged positions. Similarly, changes in dinucleotide patterns that are also implicated in the control of nucleosome positioning (Segal et al, 2006; Field et al, 2008) were not predictive of nucleosome gains/losses or reduced occupancy (Supplementary Figure S5).
Shift propagation through statistical nucleosome positioning
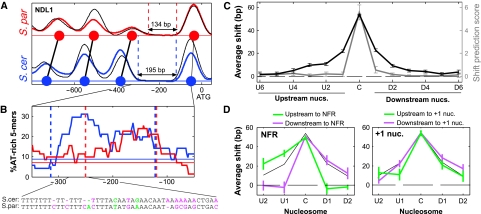
In contrast to the success of the sequence-based model in predicting nucleosome gains/losses or occupancy changes, it performed quite poorly in predicting shifts in nucleosome positioning (Supplementary Figure S4). Specific cases suggested that shifts propagate through several nucleosomes, thus explaining the poor predictive power of local DNA sequences (Figure 3A and B; Supplementary Figure S6). Shift propagation is indeed predicted by models of statistical positioning (Kornberg and Stryer, 1988; Mavrich et al, 2008; Zhang et al, 2009), which assumes that nucleosomes bind DNA mostly at random, but an apparent ordering is generated by some fixed border that creates a statistical preference for specific arrangements of adjacent nucleosomes. A shift in the position of the border elements will disrupt the preferred arrangement of multiple adjacent nucleosomes.
Figure 3.
Propagation of shifts supports the hypothesis of statistical positioning with NFR borders. (A) Shifts of three consecutive nucleosomes upstream of the NFR at the NDL1 promoter. Blue and red curves represent the nucleosome scores at S. cerevisiae and S. paradoxus, respectively, and thin black curves represent the hybrid nucleosome scores at the two corresponding alleles. (B) Enrichment of AT-rich 5-mers at the NDL1 NFR. Blue and red indicate the percentage of 5-mers composed of only A or T nucleotides at the S. cerevisiae and S. paradoxus NDL1 promoters, and the weak horizontal lines indicate the average percentage across the entire promoter. A segment of aligned sequences with higher AT-richness in S. cerevisiae than in S. paradoxus is shown on the bottom, with differences that increase or decrease AT-richness in S. cerevisiae in purple and green, respectively. (C) Propagation of shifts to adjacent nucleosomes. For each nucleosome with large shift, we examined the inter-species difference in nucleosome positions for all nucleosomes adjacent to it. Shifts at the same or opposite direction compared with the shift of the central nucleosome are defined as positive shift values or negative shift values, respectively. The average shift values are shown in black for the central nucleosome with maximal shift (C), and for upstream (Ui) and downstream (Di) nucleosomes. Gray line indicates the average shift prediction score of the sequence-based model. Shifts at positions of alignment gaps of >15 bp were excluded from this analysis. Shift of a single nucleosome could sterically interfere with the position of an adjacent nucleosome even without statistical positioning. However, steric effects should only influence nucleosomes at one direction (e.g. a downstream shift should propagate downstream but not upstream), whereas we find that shifts propagate in both directions to a similar extent, as expected for statistical positioning (Supplementary Figure S7). (D) Propagation of a subset of the shifts that are upstream (green) or downstream (purple) to NFRs (left panel) or to +1 nucleosomes (right panel). Thin black curve indicates the pattern at all shifts, taken from (C). Error bars were calculated by bootstrapping.
To more systematically examine the propagation of shifts, we focused on nucleosomes whose position shifted to a large extent between the species, and examined the flanking nucleosomes. Shifts propagated for approximately four flanking nucleosomes, decreasing in size from a maximum of ∼50 to ∼20 bps in the nearest nucleosomes. Propagation of shifts was similar for nucleosomes in both directions (upstream and downstream), indicating that it is not due to steric hindrance (Supplementary Figure S7). Notably, although sequence-based analysis had some power to predict shifts of the central nucleosomes (largest shift), it had no power to predict shifts in adjacent upstream (U) or downstream (D) nucleosomes (Figure 3C), supporting the notion that this propagation is indeed due to statistical positioning rather than changes in local sequences.
Shifts propagated away from nucleosome-free regions (NFRs) but did not propagate across NFRs (Figure 3D). For example, shifts of the −1 nucleosome typically propagate further upstream to the −2 and −3 nucleosomes but not through the NFR to the +1 and +2 nucleosomes (Figure 3A). In contrast, shifts adjacent to the +1 nucleosome, that is at the +2 nucleosome or at a −1 nucleosome in genes without NFR, did propagate across this nucleosome, even for +1 nucleosomes associated with strong nucleosome-favoring sequences (Figure 3D; Supplementary Figure S7). Thus, NFRs, but not +1 nucleosomes, seem to serve as stable border elements that establish the statistical positioning of adjacent nucleosomes.
Divergence of nucleosome positioning at regulatory elements
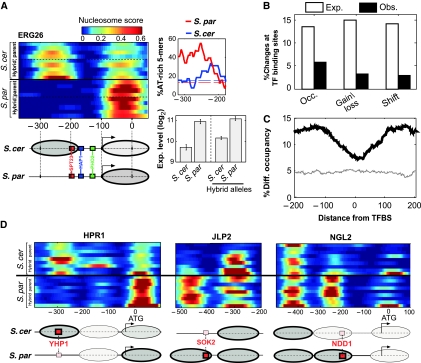
Our analysis so far focused on the genetic basis of nucleosome divergence. Next, we wished to examine the functional consequences of this divergence. Recent studies proposed that chromatin structure has an important role in generating gene expression divergence between closely related species (Lee et al, 2006; Choi and Kim, 2008), but this suggestion was not examined experimentally. Specific cases supported this proposal. For example, the −1 nucleosome of ERG26 covers two conserved TFBSs in S. cerevisiae but is absent in S. paradoxus (Figure 4A). This nucleosomal difference is also observed within the hybrid, and is associated with increased AT-richness in S. paradoxus (Figure 4A), suggesting a cis difference through exclusion of this nucleosome only in S. paradoxus. Consistent with the increased accessibility of the two S. paradoxus binding sites, expression of ERG26 is ∼2-fold higher in S. paradoxus than in S. cerevisiae, and this expression difference is also observed within the hybrid alleles, indicating a cis effect (Figure 4A).
Figure 4.
Divergence of nucleosome positioning is excluded from regulatory elements. (A) Expression divergence of ERG26 is correlated with divergence of nucleosome positioning. ERG26 promoter nucleosome scores are shown for all available strains of the two species and the respective hybrid alleles, indicating the absence of the −1 nucleosome at S. paradoxus. This nucleosome covers a conserved SPT23-binding site and borders a conserved HAP1-binding site. Consistent with this, expression of ERG26 is ∼2-fold higher in S. paradoxus and in the corresponding hybrid allele, than in S. cerevisiae. Also shown is the fraction of AT-rich 5-mers for the two species at the region of the −1 nucleosome. (B) The percentage of promoter changes for each class that overlaps conserved TFBSs (MacIsaac et al, 2006), compared with the expected percentage calculated by shuffling (P<0.05 for each of the three classes). (C) Percentage of positions with significant differences (P<0.05, t-test), as a function of the distance from conserved TFBS (MacIsaac et al, 2006), for comparison of all strains from the two different species (black) or for biological repeats of S. cerevisiae (gray). (D) Examples where nucleosome changes appear to be compensated by divergence of binding sites. Red boxes correspond to high-scoring binding sites, and smaller pink boxes correspond to weaker binding site (lower by at least two in LOD score based on MacIsaac et al). Ovals correspond to nucleosomes and their color represents occupancy (brighter ovals represent nucleosomes with low occupancy). In two of these cases (JLP2 and NGL2), expression is higher for S. paradoxus (not shown), suggesting that divergence of the motif had a more significant influence than divergence of the nucleosome. For the third case (HPR1), we do not have expression data.
We performed a systematic analysis of nucleosome positioning at TFBSs (MacIsaac et al, 2006), where differences are expected to be most influential. Although nucleosome changes are enriched at promoters, they are significantly underrepresented at conserved TFBSs (Figure 4B), and their occurrence gradually increases away from the positions of TFBSs (Figure 4C). Hence, divergence of nucleosome positioning is generally excluded from functional elements, which might indicate that it is mostly non-functional.
At positions of TFBSs that diverged between the species, differences in promoter nucleosome occupancy were more abundant than in positions of conserved TFBSs, but less abundant than in promoter positions without TFBSs. Divergence in nucleosome occupancy and in the sequences of TFBSs were generally not correlated (Supplementary Figure S8). However, at some promoters, we find that nucleosomal changes are coupled to the divergence of TFBSs, such that one species has a strong binding site that is covered by nucleosome whereas the other species has a much weaker binding site that is not covered by nucleosome (Figure 4D). These suggest compensatory evolution of nucleosome positioning and TFBSs, and thus also in these cases we expect that differences in nucleosome positioning did not drive expression divergence.
Divergence of nucleosome positioning does not explain differential expression
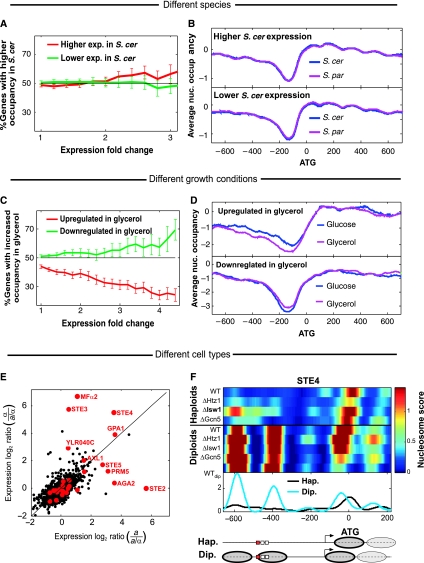
To directly examine whether inter-species divergence in nucleosome positioning can explain the divergence of gene expression, we used gene expression data generated for the same strains under the same conditions (Tirosh et al, 2009). This analysis showed that nucleosomal changes are generally not associated with inter-species expression changes, consistent with their exclusion from TFBSs. First, genes that are associated with diverged nucleosome positions are not more likely to diverge in expression (Supplementary Figure S9). We also considered the genes whose overall nucleosome occupancy along the promoter (or the ORF) diverged between the species, but these genes did not diverge in expression more than genes with conserved nucleosome occupancy (Supplementary Figure S9). Second, we found that genes that are differentially expressed are not more likely to diverge in nucleosome positioning (Figure 5A). Furthermore, the average nucleosome occupancy of genes with either higher or lower expression in S. cerevisiae, compared with S. paradoxus, was indistinguishable between the promoters, coding regions and 3′-UTRs of the two species (Figure 5B; Supplementary Figure S9). Finally, these results persisted when we tried to distinguish between genes that are activated or repressed by promoter regulatory elements (Supplementary Figure S9). Thus, our data indicate that most evolutionary changes in nucleosome positioning did not affect expression levels and are thus more likely to reflect neutral drift.
Figure 5.
Divergence of nucleosome positioning is not correlated with divergence of gene expression. (A) Percentage of genes with higher promoter nucleosome occupancy in S. cerevisiae (compared with S. paradoxus) among all the genes with significantly higher or lower expression in S. cerevisiae, as a function of the threshold for differential expression. Note that the slight (not statistically significant) trend toward higher nucleosome occupancy of genes with higher expression is opposite to the expected trend. (B) Average nucleosome occupancy for all genes with 1.5-fold higher expression in S. cerevisiae (top) or 1.5-fold lower expression in S. cerevisiae (bottom), compared with S. paradoxus. (C, D) Significant correlation between changes in nucleosome positioning and changes in gene expression for comparison of S. cerevisiae grown on glucose and glycerol. Figures are as described in (A, B) but for comparison of the same species in two conditions. (E) Differential expression between haploids and diploids for S. cerevisiae (a versus a/α) and S. paradoxus (α versus a/α). Genes with similar nucleosome positioning in haploids and diploids are shown in black and genes with the most significant differences in nucleosome positioning are shown in red (differences were averaged over the different species and strains, see Materials and methods for definition); 10 of the 28 genes (36%) with differential haploid–diploid positioning have at least fourfold higher expression in one of the haploids than the diploids and their gene names are indicated. In contrast, only 2% of the genes with similar positioning display such haploid-specific expression (P<10−16, binomial test). (F) Patterns of nucleosome positioning in haploids and diploids, for the haploid-specific gene STE4. Data are shown for four S. paradoxus haploid α strains, and for four hybrid and one S. paradoxus (WTdip) diploid a/α strains. Curves represent the average nucleosome scores of haploids (black) and diploids (cyan). Predicted nucleosome positions, TSS and binding sites (red for STE12 sites and white for others) are indicated below. Note that deletion of ISW1 decreases the difference between haploids and diploids, implicating ISW1 in this differential nucleosome positioning (see also Mfα2 and YLR040C at Supplementary Figure S10).
This lack of correlation between the divergence of nucleosome positioning and that of gene expression contrasts the situation within the same species: recent studies found significant, although modest, correlations between changes in nucleosome positioning and changes in gene expression when comparing S. cerevisiae at different growth conditions (Shivaswamy et al, 2008; Zawadzki et al, 2009) or cell-cycle stages (Hogan et al, 2006). We observed a similarly significant correlation when S. cerevisiae was transferred from growth on glucose to glycerol (Figure 5C and D).
Interestingly, in contrast to the lack of correlation between differential positioning and expression observed for the two species and the weak correlation observed for different growth conditions, we found a striking correlation for comparison of haploid and diploid cells. Nucleosome positioning in haploids differed from that of diploids in only a minority of the genes, but these were highly enriched (P<10−16) with haploid-specific genes (Figure 5E; Supplementary Figure S10). In fact, most (10 of 18) genes annotated as haploid specific (Galgoczy et al, 2004) displayed dramatic differences in nucleosome positioning between haploids and diploids, in a manner that was mostly correlated with the gene expression differences. For example, the mating gene STE4 has two promoter nucleosomes that cover multiple binding sites, which are specifically evicted in haploids, partially through the activity of ISW1 (Figure 5F). Furthermore, the +1 nucleosome is shifted downstream in the haploids thus exposing the transcription-start site (Figure 5F).
Discussion
Comparison of nucleosome positioning between two closely related yeast species allowed us to examine different models of nucleosome positioning from an evolutionary perspective. First, by analyzing the allele-specific nucleosome profiles in a yeast hybrid, we estimated that the majority (∼70%) of the inter-species differences in nucleosome patterns are caused by changes in the local DNA sequences (cis). Second, we find that cis differences that involve nucleosome gain/loss, as well as changes in occupancy are well explained by local divergence of AT-rich sequences, which are known to be disfavored for nucleosome binding (Iyer and Struhl, 1995; Segal and Widom, 2009). In contrast, sequences that are enriched at positions of bound nucleosomes have a marginal effect on the divergence pattern, suggesting that their enrichment might not be causal for nucleosome binding. Third, the pattern of shift propagation that we observe supports a significant role of statistical nucleosome positioning. Our results are consistent with a model in which nucleosome positioning is primarily determined by exclusion at specific regions such as NFRs and statistical positioning at adjacent sequences (Kornberg and Stryer, 1988; Mavrich et al, 2008).
Earlier studies have reported significant, albeit weak, correlations between changes in nucleosome positioning and gene expression in S. cerevisiae (Lee et al, 2004, 2007; Liu et al, 2005; Pokholok et al, 2005; Hogan et al, 2006; Shivaswamy et al, 2008; Field et al, 2009; Zawadzki et al, 2009), and we found a similarly weak correlation following changes in carbon source. In contrast, we describe a strong association for ploidy-dependent changes, in which most haploid-specific genes have dramatic changes in nucleosome positioning. These results may indicate that chromatin remodeling is used preferentially to prevent transcription of genes that are switched off (e.g. haploid-specific genes in a diploid), rather than fine-tune the levels of expressed genes. Consistent with this possibility, nucleosome remodeling by ISW2 has recently been shown to suppress transcription from cryptic sites (Whitehouse et al, 2007).
In contrast to differential positioning within S. cerevisiae, differences between the two species are, overall, not correlated with divergence of gene expression and are excluded from regulatory elements. These results suggest that transcription regulation is robust to many variations in nucleosome positioning, as long as they do not influence critical regulatory elements, and therefore that these neutral variations could accumulate during evolution with little influence of natural selection.
The central role of AT-rich sequences in excluding nucleosomes, the statistical positioning of nucleosomes, and the low correlation between changes in nucleosome positioning and in gene expression are consistent with earlier analyses in S. cerevisiae as well as in other organisms (Ozsolak et al, 2007; Field et al, 2008; Mavrich et al, 2008; Zawadzki et al, 2009). However, these might not be universally conserved, as recent reports suggest that in the fission yeast, Schizosaccharomyces pombe, AT-rich sequences are not enriched at NFRs, inter-nucleosomal distances are smaller (∼154 bps) and the correlation with gene expression is higher than in S. cerevisiae (Kristell et al, 2010; Lantermann et al, 2010). It would be interesting to examine how our results would generalize to other organisms such as S. pombe and multicellular organisms.
Materials and methods
Yeast strains and growth conditions
S. cerevisiae (BY4743), S. paradoxus (CBS 432 ho∷nat MATα) and their hybrid (BY4741 X CBS432 ho∷nat MATα) were grown to a log-phase growth at 30°C at rich media (YPD medium) or glycerol (YPglycerol). For each of the species and the hybrids, we profiled nucleosome positioning in the wild-type backgrounds and in five mutant strains (only the first three for the hybrid) deleted of the following chromatin regulators: ISW1, HTZ1, GCN5, BRE1 and RAD6.
High-throughput sequencing of mono-nucleosomal DNA fragments
Mono-nucleosomes were isolated from the two species (or from the hybrids) by standard protocols, pooled, and subjected to Illumina high-throughput sequencing (see Supplementary Information for more details). Reads that were unambiguously mapped to only one of the species were retained for further analysis. Note that our analysis is restricted to ∼87% of the genomic positions that differ between the species, which could induce a slight over-estimation of cis effects (see Supplementary Information). As reads are mapped to the ends of ∼150 bp fragments, we converted the mapped positions into the center positions of the ∼150 bp fragments, by adding or subtracting (based on the direction of each read) half the length of a typical fragment. This fragment length was estimated for each sample as the median distance between consecutive peaks of read density and was between 135 and 160 for all samples.
Estimation of nucleosome occupancy and positions
Nucleosome occupancy was generated by averaging the number of reads over windows of 150 bps. In addition, we defined nucleosome scores by smoothing the same data with a Gaussian filter with windows of 50 bps and s.d. of 25 bps. Nucleosome center positions were identified as peaks of nucleosome scores, with the lowest 10% of the peaks excluded. The raw data and normalized nucleosome scores are available at the GEO database (Accession number GSE18939).
Comparison of nucleosome positioning
At each aligned region of the two species, we identified nucleosomes that are gained/lost, shifted in position or changed in occupancy using strict criteria. These include at least twofold difference for occupancy changes, minimal number of reads (8) for gains/losses and shifts of at least 30 bp and statistical significance; in addition, each difference was evaluated based only on reads that mapped to the Watson strand and, separately, based only on reads that mapped to the Crick strand and only differences that were consistent in both analyses were retained (see Supplementary Information for full details).
Control experiments with naked DNA
To control for MNase and other biases, we conducted similar experiments with naked DNA from the two species and the hybrid (Supplementary Figure S1). Inter-species differences that were also observed in the control experiment were excluded from further analysis, as they might originate from MNase bias and not from evolutionary differences. This analysis also eliminates other systematic errors that generate differential occupancy in the two species, such as sequencing errors or alignment errors. Note that this control excludes ∼30% of the nucleosomes from our analysis, which could bias our results. Our analysis was also restricted to genomic positions that differ between the species (to allow species-specific read mapping, as described above), to genes that have one-to-one orthology relationships and complete sequence alignments (Kellis et al, 2003), and was focused on promoters and 2 kb of each gene. Relaxing these criteria did not have a major effect on our main conclusions (Supplementary Figure S11).
Classification to cis versus trans
The extent of differences between the two species (Δparents) was compared with that between the two hybrid alleles (Δhybrid). We expect to have Δhybrid approximately equal to Δparents in cis differences (i.e. ∣Δhybrid–Δparents∣ is low) and approximately zero in trans differences. We therefore classified cis changes as those where ∣Δhybrid–Δparents∣<∣Δhybrid∣ and trans changes as those where ∣Δhybrid–Δparents∣>∣Δhybrid∣. Intermediate cases, in which the difference between these values is small, were excluded from this analysis (see Supplementary Method).
Prediction by sequence models
To examine how much of the differences could be predicted from sequence analysis, nucleosome occupancy at aligned positions of the two species were predicted using a method described earlier (Field et al, 2008), which is primarily based on the contribution of each 5-mer to nucleosome positioning. The predictive power of this model was evaluated by comparing the differences in predicted nucleosome occupancy at locations of observed nucleosomal changes to the differences at randomly selected positions (control regions). For each difference in scores, we examined the frequency of control regions that have higher difference (false positive rate) and the frequency of nucleosome changes with higher score difference (true positive rate). For positions of shifts, the prediction score was defined as:
(Ocer,cer−Opar,cer)+(Opar,par−Ocer,par), where Oi,j is the predicted occupancy of species i at the location of the nucleosome of species j. We performed similar analysis with other sequence-based models and got similar results (not shown). Individual 5-mers were defined as nucleosome-enriched or linker-enriched if they have at least a twofold enrichment in the wild-type S. cerevisiae sample.
Carbon source-dependent changes in nucleosome positioning
Nucleosome positioning was measured for the wild-type strains of the two species (pooled) both in glucose and glycerol and processed as described above. We compared nucleosome positioning within S. cerevisiae among the two conditions by focusing on ∼4000 S. cerevisiae genes where inter-species expression differences were measured earlier (Tirosh et al, 2009) and where genomic differences allowed the species-specific mapping of at least 100 reads to the S. cerevisiae alleles. This analysis was not affected by MNase bias, as we are comparing the same species in different conditions. We compared the average nucleosome occupancy for genes upregulated or downregulated at glycerol (compared with glucose) by at least 1.5-fold, based on earlier microarray analysis (Tirosh et al, 2009). Similar results were obtained when we examined S. paradoxus or when we used different cutoffs for differential expression (not shown).
Ploidy-dependent changes in nucleosome positioning
To identify the differences in nucleosome positioning between haploids and diploids, we compared the nucleosome scores of four haploid samples from each species (WT, Δhtz1, Δgcn5 and Δisw1) to the corresponding alleles of the four hybrid dipolids (WT, Δhtz1, Δgcn5 and Δisw1), or to WT diploids of each species (see below). The haploid strains of each species were compared only with the corresponding alleles in the hybrid (the alleles of the same species). A measure of haploid–diploid nucleosome difference was defined as the Euclidean distance between nucleosome scores at −300 to +200, averaged across the four different sample pairs and normalized by the average Euclidean distance between sample pairs from the same group (haploid–haploid or diploid–diploid). This analysis was performed separately for S. cerevisiae (a versus a/α) and S. paradoxus (α versus a/α), and the average value of the two species was used for further analysis to identify all haploid–diploid differences, including those specific to a or to α cells. Similar results were obtained when we avoided the use of hybrid diploids and compared only WT haploids with the WT diploids of each species (not shown); we present the data from comparison of haploids to hybrid diploids (Figure 5E), as it is based on more samples and higher coverage than the analysis of the WT diploids. Half of the genes with the lowest haploid–diploid difference were considered as having similar nucleosome positioning, and the 28 genes with the highest haploid–diploid difference were considered as having different nucleosome positioning, as this number gave the most significant enrichment of haploid-specific genes (Galgoczy et al, 2004). Differences in nucleosome positioning were compared with differences in gene expression, which we previously measured for haploids and diploids of the two species (S. cerevisiae a versus a/α and S. paradoxus α versus a/α) (Tirosh et al, 2009).
Supplementary Material
Supplementary Methods, Supplementary Figures S1–S11
Acknowledgments
This work was supported by the Helen and Martin Kimmel Award for Innovative Investigations, the Crown Human Genome Center, the Israeli Ministry of Science and the European Research Council (Ideas).
Footnotes
The authors declare that they have no conflict of interest.
References
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M (2007) Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Choi JK, Kim YJ (2008) Epigenetic regulation and the variability of gene expression. Nat Genet 40: 141–147 [DOI] [PubMed] [Google Scholar]
- Doniger SW, Fay JC (2007) Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol 3: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E (2009) Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet 41: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E (2008) Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy DJ, Cassidy-Stone A, Llinas M, O’Rourke SM, Herskowitz I, DeRisi JL, Johnson AD (2004) Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 18069–18074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD (2006) Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet 2: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N (2005) Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309: 938–940 [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF (2006) Nucleosome positions predicted through comparative genomics. Nat Genet 38: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K (1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S (2006) Evolution of primate gene expression. Nat Rev Genet 7: 693–702 [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC (1975) Evolution at two levels in humans and chimpanzees. Science 188: 107–116 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Stryer L (1988) Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res 16: 6677–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristell C, Orzechowski Westholm J, Olsson I, Ronne H, Komorowski J, Bjerling P (2010) Nitrogen depletion in the fission yeast Schizosaccharomyces pombe causes nucleosome loss in both promoters and coding regions of activated genes. Genome Res 20: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee SI, Pe’er D, Dudley AM, Church GM, Koller D (2006) Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc Natl Acad Sci USA 103: 14062–14067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ (2005) Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E (2006) An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF (2008) A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, Macisaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E (2007) Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE (2007) High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol 25: 244–248 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP (2003) Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet 33: 138–144 [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J (2006) A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J (2009) Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR (2008) Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N (2009) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N (2008) On the relation between promoter divergence and gene expression evolution. Mol Syst Biol 4: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N (2010) High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T (2007) Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wray GA (2007) The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zawadzki KA, Morozov AV, Broach JR (2009) Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell 20: 3503–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K (2009) Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gu J, Gu X (2004) How much expression divergence after yeast gene duplication could be explained by regulatory motif evolution? Trends Genet 20: 403–407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Figures S1–S11