Abstract
In mammals, Up-frameshift proteins (UPFs) form a surveillance complex that interacts with the exon junction complex (EJC) to elicit nonsense-mediated mRNA decay (NMD). UPF3b is the component of the surveillance complex that bridges the interaction with the EJC. Here, we report the 3.4 Å resolution crystal structure of a minimal UPF3b-EJC assembly, consisting of the interacting domains of five proteins (UPF3b, MAGO, Y14, eIF4AIII, and Barentsz) together with RNA and adenylyl-imidodiphosphate. Human UPF3b binds with the C-terminal domain stretched over a composite surface formed by eIF4AIII, MAGO, and Y14. Residues that affect NMD when mutated are found at the core interacting surfaces, whereas differences between UPF3b and UPF3a map at peripheral interacting residues. Comparison with the binding mode of the protein PYM underscores how a common molecular surface of MAGO and Y14 recognizes different proteins acting at different times in the same pathway. The binding mode to eIF4AIII identifies a surface hot spot that is used by different DEAD-box proteins to recruit their regulators.
Nonsense-mediated mRNA decay (NMD) is an eukaryotic quality control process that subjects mRNAs with premature translation termination codons (PTCs) to rapid degradation. The pathway is active on PTC-containing transcripts that originate from genes that are mutated or are physiologically regulated by NMD, as well as from errors occurring during pre-mRNA synthesis or processing (reviewed in refs. 1 and 2). From a mechanistic standpoint, NMD factors are recruited to an mRNA when translating ribosomes recognize a stop codon as premature by sensing the presence of a second signal downstream (reviewed in refs. 1 and 3). Whereas the three central NMD effectors (the Up-frameshift proteins UPF1, UPF2, and UPF3) are conserved across species, the nature of the downstream signal varies (reviewed in refs. 4 and 5). The exon junction complex (EJC) is the most common and best-studied signal that is required for efficient NMD in mammals.
The EJC is a multiprotein complex that is deposited on spliced mRNAs in the nucleus, upstream of the position where an intron has been removed (6). Four proteins, including the heterodimer MAGO-Y14, the DEAD-box protein eIF4AIII, and Barentsz (BTZ, also known as MLN51), assemble in the presence of RNA and ATP to form a stable complex, which persists on the nucleic acid during and after export to the cytoplasm (7). The crystal structure of the EJC has been previously determined, showing how the complex maintains a stable grip on the RNA (8, 9). The current models of mammalian NMD envisage that the EJC associates in the nucleus with UPF3, which in turn recruits UPF2 at the cytoplasmic side of the nuclear envelope (10–14). In the cytoplasm, the NMD pathway is activated when translation termination at a premature stop codon occurs at least 30 nucleotides upstream of an EJC. The trigger is the formation of the UPF1-UPF2-UPF3 surveillance complex, which assembles when UPF1 (bound with release factors at the stalled ribosome) and the downstream EJC-UPF3-UPF2 assembly are in the correct configuration to interact (15, 16).
UPF3 is the physical link between the EJC core and the NMD machinery (13, 17). At the N terminus, UPF3 contains a conserved RNA recognition motif (RRM) domain that mediates the interaction with the C-terminal MIF4G (middle portion of eIF4G) domain of UPF2 (18). At the C terminus, human UPF3 contains a region that has been shown to interact with the EJC by coimmunoprecipitation assays and to induce NMD in tethering assays (19). This region has also been involved in modulating the NMD efficiency of UPF3b and UPF3a, the two variants of UPF3 that are present on chromosome X and chromosome 13 (also known as UPF3X and UPF3, respectively) (10, 19, 20). Three proteins in the EJC have been implicated in UPF3b binding by in vitro assays (17). However, how the interaction is achieved and why the whole EJC is required is unclear. In this manuscript, we set out to understand the mechanism of recognition between the EJC and UPF3b by determining the crystal structure of a MAGO-Y14-eIF4AIII-BTZ-UPF3b-RNA-AMPPNP (adenylyl-imidodiphosphate) core complex.
Results and Discussion
Structure Determination and Quality.
The human EJC core complex was reconstituted as described previously (7, 8) combining full-length MAGO bound to the RRM domain of Y14, full-length eIF4AIII bound to the SELOR domain of BTZ (Fig. 1), a U15 RNA, AMPPNP, and magnesium ions. A complex of the EJC and a peptide encompassing the previously identified NMD-functional domain of UPF3b (residues 421–434) (13) could be isolated by size-exclusion chromatography, but did not yield crystals. We proceeded by assembling the complex with full-length UPF3b instead of a peptide. Further addition of a UPF2 construct containing the UPF3b-binding domain (Fig. 1) led to a relatively homogeneous EJC-UPF3b-UPF2 sample (Fig. S1). The hexameric protein complex was subjected to limited proteolysis using elastase in a protein:protease ratio of 100∶1 (wt/wt), followed by gel filtration to separate the EJC-containing fractions for crystallization experiments (Fig. S1). Crystals from this sample preparation grew in a tetragonal space group with two molecules per asymmetric unit and diffracted to 3.4 Å resolution at the Swiss Light Source (SLS) synchrotron. Mass spectrometry analysis of dissolved crystals indicated that a peptide corresponding to the sequence DRPAMQLYQPGAR of UPF3b (residues 422–433) was present together with the EJC core proteins. The intensity for the UPF3b peptide encompassed 93% of the signal given by UPF3b fragments in the Orbitrap mass spectrometry analysis.
Fig. 1.
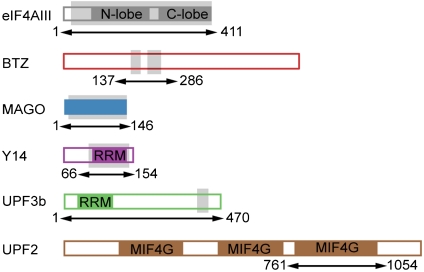
Minimal interaction domains in the EJC-UPF3b structure. (A) Schematic representation of the domain arrangements of the human proteins used in this study. Color-filled areas identify structural domains, which include the two RecA-like domains of the DEAD-box protein eIF4AIII (in dark gray), the MAGO protein fold (in blue), the RRM domains of Y14 and UPF3b (in pink and green, respectively), and the three MIF4G domains (middle domain of eukaryotic initiation factor 4G) of UPF2 (in brown). The arrows and corresponding residue numbers indicate the constructs used for the biochemical reconstitution. The portions of the polypeptides ordered in the three-dimensional structure are shaded in light gray.
The structure was solved by molecular replacement using the EJC coordinates as a search model and refined to an R free of 26.04%, R factor of 22.04%, and good stereochemistry (Fig. S2). The refined model consists of MAGO residues 4–146, Y14 residues 66–154, eIF4AIII residues 22–411, two segments of BTZ comprising residues 169–194 and 216–248, and the C-terminal region of UPF3b encompassing residues 418–432 (Fig. 1). Other portions of the proteins are not present in the structure, and are either poorly ordered or were separated upon proteolysis. The structure also includes MgAMPPNP and eight bases of polyU RNA. The two independent copies of the complex present in the asymmetric unit are very similar, and superimpose with a rmsd of 0.26 Å on 678 Cα atoms. Here we describe complex 1 unless stated otherwise. Complex 2 is engaged in lattice contacts near the UPF3b-binding site and thus localized differences in side-chain orientations at this region might be due to crystal packing. The electron density maps for regions of the molecules discussed in the text are shown in Fig. S3.
UPF3b Recognizes a Composite Surface on the EJC.
UPF3b stretches over the EJC in an extended conformation (Fig. 2A). Upon UPF3b binding, the overall structure of the EJC remains essentially unchanged from the core complex previously described (8, 9). In brief, the EJC assembles around the DEAD-box protein eIF4AIII (gray in Fig. 2A). The two RecA-like domains of eIF4AIII are arranged in a closed conformation, sandwiching ATP on one side and forming the binding site for single-stranded RNA on the other. BTZ interacts with eIF4AIII mostly without defined secondary structure elements (red in Fig. 2A). The N-terminal segment of BTZ visible in the electron density wraps around the second RecA-like domain of eIF4AIII, where it contacts MAGO and RNA. After a disordered linker of about 20 residues, BTZ extends onto the first RecA-like domain. MAGO and Y14 (Fig. 2A, blue and pink, respectively) form a structural unit whereby the Y14 RRM functions as a protein-binding domain rather than an RNA-binding domain (21). MAGO-Y14 interacts with the second RecA-like domain of eIF4AIII and with residues at the cleft formed with the first RecA-like domain, effectively locking the closed RNA-bound state of the DEAD-box protein (8, 9, 22).
Fig. 2.
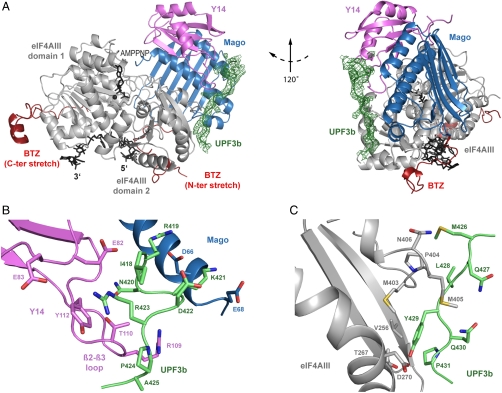
Structure of the EJC-UPF3b complex. (A) View of the structure of the EJC core (eIF4AIII in gray, BTZ in red, MAGO in blue, Y14 in pink, and RNA, AMPPNP, and magnesium in black) in complex with the EJC-binding domain of UPF3b (in green, shown together with the electron density map contoured at 0.9σ). UPF3b residues 418–432 bind in an extended conformation to a composite surface of the EJC comprising MAGO, the RRM of Y14, and the eIF4AIII C-terminal RecA-like domain (domain 2). This and all other ribbon drawings were generated using PYMOL (http://www.pymol.org). The structure is viewed in two orientations related by a counterclockwise rotation of 120° around a vertical axis. (B) Close-up view of the interactions between UPF3b (green) and MAGO-Y14 (blue and pink, respectively), showing the central position of Arg423UPF3b in the structure. The molecules are viewed in a similar orientation to that used in the left panel of A. (C) Close-up view of the interactions between UPF3b (green) and eIF4AIII (gray), showing the prominent interaction of Tyr429UPF3b. The molecules are viewed in a similar orientation to that used in the left panel of A.
As compared to the earlier EJC structures (8, 9), an additional ribonucleotide could be modeled at both the 5′ and 3′ ends, consistently with the presence of 7–9 nucleotides fragments in the RNase protection pattern of the EJC (6, 7). The RNA makes a sharp kink in the ribose phosphate backbone both at the 5′ end between nucleotides 1 and 2 and at the 3′ end between nucleotides 7 and 8. The first nucleotide contacts BTZ residues Lys185BTZ and Gly186BTZ, while the uracyl base of U8 stacks against eIF4AIII at Val146eIF4A3.
UPF3b binds the EJC with a stretch of 15 residues that bury approximately 1.2% (350 Å2) of the solvent-accessible surface area of the EJC. The EJC-binding segment of UPF3b ordered in the structure (residues 418–432) maps closely to the region which had been previously identified by tethering assays as the functionally important domain that is required to activate NMD [residues 421–434, (13)]. UPF3b interacts with MAGO-Y14 near the β2-β3 loop of the Y14 RRM and then stretches over the second RecA-like domain of eIF4AIII (Fig. 2A). UPF3b does not directly contact the RNA nor BTZ in the structure, consistent with previous results from RNase digestion and pull-down experiments (17). UPF3b also does not contact ATP nor the first RecA-like domain of eIF4AIII.
UPF3b-EJC Recognition: Conserved Interactions at MAGO-Y14.
UPF3b residues 418–425 bind MAGO-Y14. The N-terminal residues make a tight turn, with Arg419UPF3b and Lys421UPF3b contacting MAGO via long-range electrostatic interactions with Asp66MAGO and Glu68MAGO (Fig. 2B). The structural data are consistent with previous in vitro studies, showing that the interaction of UPF3b with an EJC Asp66MAGO/Glu68MAGO mutant is impaired in pull-down assays (17). Mutation of Asp66MAGO/Glu68MAGO and of the corresponding Arg419UPF3b and Lys421UPF3b also lead to a decrease in NMD activity in cell-based tethering assays (21, 23).
Another positively charged residue of UPF3b, Arg423UPF3b, contacts the β2-β3 loop of Y14 (Fig. 2B). The aliphatic side chain of Arg423UPF3b packs on one side against Tyr112Y14, engaging the guanidinium moiety in a typical amino-aromatic interaction with the π electrons of the phenyl ring. The hydrophobic environment of Arg423UPF3b is completed on the other side by Ile418UPF3b. The backbone of UPF3b in this region is stabilized by the side chains of Glu82Y14 and Arg109Y14 and the backbone of the Y14 β2-β3 loop is stabilized by the side chain of Asn420UPF3b. This extensive network of interactions is crucial for UPF3b recognition. Mutating Arg423UPF3b to Ala results in a loss-of-function effect in NMD tethering assays comparable to deleting the entire 421–434 segment of UPF3b (13). The structural data also rationalize why mutating the corresponding Glu82Y14/Glu83Y14 in the EJC impairs the interaction with UPF3b in in vitro assays (17).
UPF3b-EJC Recognition: Conserved Interactions at eIF4AIII.
Beyond Ala425UPF3b, UPF3b crosses over to reach eIF4AIII. In particular, Met426UPF3b and Leu428UPF3b contact the hydrophobic surface that connects MAGO-Y14 and eIF4AIII. The side chains of Tyr429UPF3b and Pro431UPF3b nestle into a hydrophobic pocket formed on the surface of eIF4AIII by Val256eIF4A3, Thr267eIF4A3, Met403eIF4A3, and Met405eIF4A3 (Fig. 2C). Met405eIF4A3 undergoes a prominent conformational change, moving from the position in the unbound EJC structure (where it would clash with UPF3b) to a conformation where it wraps around UPF3b. Here, the hydroxyl group of Tyr429UPF3b forms polar contacts with Thr267eIF4A3 and Asp270eIF4A3, explaining why a Tyr429UPF3b to Phe mutant has reduced NMD activity (13).
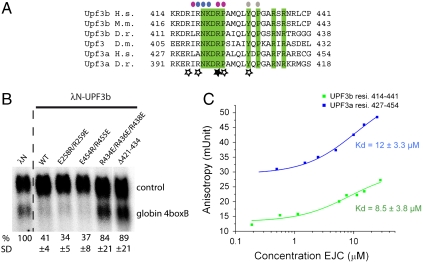
The last residue with interpretable electron density of UPF3b (Gly432UPF3b) is positioned at the beginning of an α-helix of eIF4AIII that features conserved and solvent-exposed negatively charged residues (Asp266eIF4A3, Asp270eIF4A3, and Asp273eIF4A3) (Fig. S3). The sequence downstream of Gly432UPF3b features a stretch with three conserved positively charged residues (Arg434UPF3b, Arg436UPF3b, and Arg438UPF3b) (Fig. 3A). Although no ordered electron density is present for this stretch of UPF3b, the structural and sequence features are compelling and point to a possible long-range electrostatic interaction. We therefore tested the importance of the conserved positively charged stretch of UPF3b. In tethering assays, a mutant containing three glutamic acid residues at positions 434, 436, and 438 has a drastic effect, comparable to the deletion of the entire 421–434 segment previously reported (13) (Fig. 3B). We conclude that the conserved positively charged stretch of human UPF3b between residues 434 and 438 is involved in eliciting NMD.
Fig. 3.
UPF3b conservation and affinity of the interactions. (A) Sequence alignment of the EJC-binding domain of UPF3b. The alignment includes the sequences of the two paralogues (UPF3a and UPF3b) present in Homo sapiens (H.s., Q9H1J1-1 and Q9BZI7-2, respectively) and Danio rerio (D.r., Q7SXF0-1 and B05733-1, respectively), of UPF3b from Mus musculus (M.m., Q3ULJ3) and Drosophila melanogaster (D.m., Q9W1H3-1). Conserved residues are highlighted in green. Above the sequences, colored circles identify residues of human UPF3b that interact with eIF4AIII (gray circles), with MAGO (blue circles) and Y14 (pink circles). Residues of human UPF3b shown by mutagenesis studies to affect NMD are indicated with a star (the filled star indicates the most important residue identified in ref. 13. (B) A conserved positively charged stretch of UPF3b is involved in NMD. A β-globin reporter with 4boxB in the 3′UTR was cotransfected in HeLa cells with a transfection control and plasmids expressing λN alone, wild-type λN-UPF3b, or different mutant versions of λN-UPF3b as indicated. Northern-blot of total RNAs probed with a specific body-radiolabeled DNA probes for β-globin is shown. β-globin 4boxB mRNA levels were normalized to that of the control (wt + 300 + e3, ref. 13). The normalized value of β-globin 4boxB mRNA level is set to 100% in cells transfected with λN vector. Mean values ± SD are indicated. (C) Binding of the EJC to UPF3b or UPF3a peptides (residues 414–441 and 427–454) in solution as measured by fluorescence anisotropy. The peptides were labeled with fluorescein at the carboxy terminus, and for labeling purposes a serine was introduced in place of the nonconserved cysteine residue at position 440 and 453 in UPF3b and UPF3a (Fig. 3A). The data were fitted to a binding equation describing a single-site binding model. The best fit was plotted as a solid line (green for UPF3b and blue for UPF3a).
Differences in UPF3b and UPF3a Binding to the EJC.
Human UPF3a has also been shown to induce NMD in tethering assays, albeit to a lesser extent than UPF3b (10, 19, 20). Using fluorescence anisotropy and surface plasmon resonance, we detect a reproducible but only subtle difference in affinity between the EJC-UPF3b and EJC-UPF3a interactions. Binding of the EJC in solution with a fluorescently labeled peptide encompassing residues 414–441 of human UPF3b results in a Kd of 8.5 ± 3.8 μM (Fig. 3 A and C), whereas the corresponding peptide encompassing residues 427–454 of human UPF3a results in a Kd of 12 ± 3.3 μM (Fig. 3 A and C). These results are consistent with the structural analysis, showing that the core EJC-interacting residues of UPF3b are conserved in UPF3a (Figs. 2 and 3A). Substitution of two of the core residues of UPF3b to alanines (Arg423 at the Mago-Y14 interface and Tyr429 at the eIF4AIII interface) results in a severe loss of affinity in fluorescence anisotropy measurements (Kd higher than 120 μM and 1 mM, respectively) (Fig. S4).
A similar trend in affinities between the two UPF3 variants is observed when measuring the binding of the EJC to full-length GST-tagged UPF3 proteins immobilized on BIAcore chips (Kd of 1.2 ± 0.4 μM for GST-UPF3b and of 2.7 ± 0.3 μM for GST-UPF3a) (Fig. S5). The quantitative analysis by surface plasmon resonance also shows that the interaction of the UPF3 proteins with the EJC is characterized by fast on and off rates. As a comparison, the UPF3-UPF2 interaction has a Kd in the low nanomolar range, with a slow on rate and very slow off rate (Fig. S6). Although the relative difference between the EJC-UPF3b and EJC-UPF3a interaction is comparable in the two experimental settings, the affinities measured by surface plasmon resonance are slightly higher than those obtained by fluorescence anisotropy. This might be attributed to the difference in methodology (for example, binding on a surface and in solution, respectively) and/or to the difference in the samples used (full-length proteins and peptides, respectively). In the latter case, peripheral residues in UPF3b outside of the 414–441 region would be expected to provide an additional, albeit minor, contribution to EJC binding. Interestingly, in this context, an additional segment of electron density separate from the 418–432 region of UPF3b became visible at the later stages of refinement in one of the two complexes in the asymmetric unit (complex 2, Fig. S3).
Although this additional segment of density is near lattice contacts, it is centered at a conserved residue (Arg329eIF4A3) at the end of the negatively charged α-helix of eIF4AIII discussed previously (with Asp266eIF4A3, Asp270eIF4A3, and Asp273eIF4A3). The features of the electron density and the chemical properties of the surface are consistent with a binding segment of three residues containing a negatively charged residue interacting with Arg329eIF4A3 and a contiguous positively charged residue interacting with the free carboxyl group at the C terminus of eIF4AIII (Fig. S3B). We tested reverse-charged mutations in NMD tethering assays of two compatible regions in human UPF3b (Glu454/Arg455 and Glu258/Arg259), but observed no difference as compared to wild-type UPF3b (Fig. 3B). It is possible that either three other residues of UPF3b or of BTZ from a symmetry-related molecule might dock onto this surface.
Surface Hot Spot on eIF4A Paralogues Binds Specific Regulators.
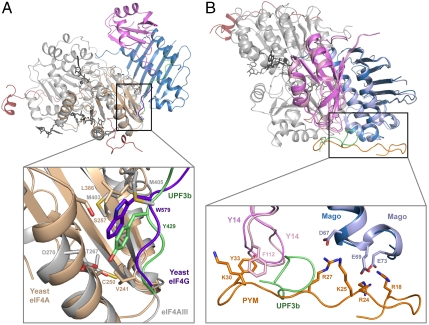
The second RecA-like domain of eIF4AIII interacts with UPF3b at a hydrophobic surface centered at Val256eIF4A3 that is well conserved in the paralogues eIF4AI and eIF4AII. However, eIF4AI and eIF4AII do not bind UPF3 and are not involved in NMD. Instead, they bind eIF4G and are involved in translation initiation. Comparison of the human EJC-UPF3b structure with that of the yeast eIF4A-eIF4G complex (24) shows that eIF4G and UPF3b bind at the equivalent surface of the paralogue DEAD-box proteins (Fig. 4A). In particular, Trp579eIF4G is in the same structural position as Tyr429UPF3b (Fig. 4A) (24) and is likewise crucial for binding (Fig. S4). This surface contributes only a portion of the intermolecular interactions observed in the eIF4A-eIF4G structure; eIF4G binds the second RecA domain at another region (centered at Arg312eIF4A, corresponding to Arg329eIF4A3) and binds also the first RecA domain (24). Thus, although the second RecA domain in eIF4A paralogues has a hot spot for interactions that has been evolutionary conserved, specificity in the binding of different proteins is brought about by other contributing surfaces.
Fig. 4.
Protein recognition at similar molecular surfaces. (A) The surfaces of eIF4AIII (gray) and yeast eIF4A (light brown) are shown (after optimal superimposition) bound to the corresponding interacting proteins, UPF3b (green) and eIF4G (dark blue). The close-up view shows the same structural position for Tyr429UPF3b and Trp579eIF4G. (B) The surface of MAGO-Y14 is used for binding both UPF3b (green) and PYM (orange). The dark and light shades of MAGO and Y14 correspond to the UPF3b-bound structure and to the PYM-bound structure, respectively. The molecules are shown after optimal superimposition in an orientation that is turned 70° around a vertical axis and 50° degree around a horizontal axis as compared to the orientation in A.
Interestingly, another DEAD-box helicase uses the same surface as a protein-binding site. The corresponding pocket in DDX6, a helicase that functions in translation repression and mRNA decapping, recognizes the FDF motif of the decapping activator EDC3. In particular, EDC3 positions Phe206EDC3 at the same structural position as Tyr429UPF3b (25) (Fig. S7). The DDX6-FDF structure also featured additional density at Arg375DDX6 (corresponding to Arg329eIF4A3) that is due to a bound CAPS (3-cyclohexylaminopropane-1-sulfonic acid) molecule from the crystallization buffer (25).
Common Surface of Y14 Binds Different NMD Proteins.
The highly conserved β2-β3 loop of Y14 is exposed to solvent and is a binding site for both UPF3b and PYM, a protein involved in EJC disassembly (26, 27) (Fig. 4B). The flanking negatively charged surface of MAGO, including the conserved and solvent-exposed Asp66MAGO/Glu68MAGO, is also used for binding both UPF3b and PYM (26) (Fig. 4B), consistent with results from in vitro pull-down (17) and coimmunoprecipitation studies (23). In the case of UPF3b, binding to MAGO-Y14 involves ∼200 Å2 and includes only half of the interactions necessary for stable association, the other half being contributed by eIF4AIII. In the case of PYM, binding to MAGO-Y14 involves ∼700 Å2 and includes all of the interactions necessary for a stable association. In the context of the EJC, PYM would clash with the surface residues of eIF4AIII that are used for UPF3b binding (Met403eIF4A3 and Met405eIF4A3) (8), and this structural prediction can be recapitulated biochemically (17). Thus, although UPF3b and PYM recognize the same surface of MAGO-Y14, UPF3b associates stably only in the presence of other EJC components, whereas PYM associates only in their absence.
Conclusions
The structure of the core EJC-UPF3b complex reveals how the C terminus of UPF3b interacts with a composite binding surface of the EJC consisting of MAGO-Y14 and of the second RecA domain of eIF4AIII. Although UPF3b does not contact the first RecA domain nor ATP or RNA, the appropriate geometry of the UPF3b-binding sites requires that the domains involved are in the conformation of the fully assembled EJC. The structure of the EJC remains essentially unaltered upon UPF3b binding. The EJC thus appears to function as a rather rigid platform for the recruitment of additional proteins. UPF3b binds to the EJC with relatively low affinity (in the low micromolar range) as measured by both fluorescence anisotropy and surface plasmon resonance. This is consistent with the structural analysis, showing that only a small fraction of the overall accessible surface area of the EJC is used to bind UPF3b. On one side, this implies that other EJC-binding proteins might be able to access the complex and exert their function in the presence of UPF3b. On the other, UPF3b binding occurs at a surface of MAGO-Y14 that is used to bind other proteins including PYM (26) and Importin 13 (28). This suggests a mechanism whereby mutually exclusive binding sites impart a sequential order in the interactions in the NMD pathway. In addition, UPF3b binds eIF4AIII at a hydrophobic pocket (centered at Val256eIF4A3) that is used by its paralogues eIF4AI/II and the more divergent helicase DDX6 to recruit their specific regulators. Comparison of the EJC-UPF3b structure with that of eIF4A-eIF4G (24) and DDX6-FDF (25) also points to the presence of a second hot spot for interactions on eIF4AIII (centered at Arg329eIF4A3). We speculate that this surface patch might be exploited to recruit other proteins to the EJC platform. Unraveling the complete interaction network centered on the EJC will be required to fully understand how this complex exerts its multiple roles on posttranscriptional regulation of mRNAs in metazoa.
Experimental Procedures
Protein Purification.
The EJC complex was reconstituted as described previously (8). Human full-length UPF3b (1–470) and human UPF2 (residues 761–1054) were expressed as recombinant His-tagged [Tobacco Etch Virus (TEV)-cleavable] proteins using Escherichia coli BL21-Gold (DE3) pLysS cells (Stratagene), Terrific Broth (TB) medium, and overnight induction at room temperature. The cells were lysed in 20 mM Tris·HCl pH 7.5, 250 mM KCl, 20 mM imidazole, 10% glycerol, 3 mM MgCl2, 1 mM ß-mercaptoethanol supplemented with protease inhibitors (Roche), and the His-tagged proteins were purified by affinity chromatography on nickel resin. After cleaving the His-tag with TEV protease during overnight dialysis in lysis buffer, UPF3b and UPF2 were further purified on nickel-beads and a Heparin Sepharose CL-4B column (GE Healthcare).
The EJC-UPF3b-UPF2 complex was formed by incubating the purified proteins at 4 °C in 20 mM Tris·HCl pH 7.5, 300 mM NaCl, 3 mM MgCl2, 10% glycerol, and 1 mM DTT (gel-filtration buffer), using a 1.5 M excess of UPF3b and UPF2. The complex was purified by size-exclusion chromatography (Superdex 200 GE Healthcare). For limited proteolysis, the complex was incubated 1.3 mg/mL for 1 h on ice in presence of elastase (Roche) at a 1∶100 (wt/wt) enzyme to protein ratio. The reaction was stopped with 0.01 mM 4-(2-aminoethyl)-benzylsulfonyl fluoride hydrochloride (Pefabloc, Roche). The mixture was loaded on a gel-filtration column and the fractions containing the EJC were pooled and concentrated to 30 mg/mL for crystallization.
For BIAcore experiments, full-length human UPF3b (residues 1–470) and UPF3a (1–476) were expressed as GST-fusion proteins using the same conditions described above and were purified on glutathione sepharose followed by a Heparin Sepharose CL-4B column and size-exclusion chromatography (Superdex 200 GE Healthcare) in gel-filtration buffer without glycerol.
Crystallization and Structure Determination.
Crystals were grown at 18 °C by vapor diffusion from drops formed by equal volumes of complex at 30 mg/mL and crystallization buffer (10% PEG 8000, 100 mM sodium cacodylate pH 6.5, 200 mM magnesium acetate). Small tetragonal crystals appeared after 2–3 weeks and were improved using microseeding, reaching a size of 80 × 30 × 30 μm. Crystals belong to the tetragonal space group P4(1)2(1)2 with cell dimensions a = b = 134.8 Å and c = 227.3 Å, contain two EJC-UPF3b418–432 complexes in the asymmetric unit, and have a solvent content of 53%. They were transferred to a stabilizing solution (15% PEG 8000, 100 mM sodium cacodylate pH 6.5, 200 mM magnesium acetate) containing 20% (vol/vol) glycerol as a cryoprotectant. Data were collected at the PXII beamline of the SLS to 3.4 Å resolution. Intensities were integrated with Mosflm and scaled with Scala [CCP4 (29)] (Fig. S2).
The structure was determined by molecular replacement using the program PHASER (30) and the EJC structure at 2.2 Å resolution (Protein Data Bank entry 2j0s) as a search model. Refinement was carried out with the program CNS (31), initially using strict noncrystallographic symmetry restraints between the two molecules of the asymmetric unit. Model building was carried out with the program COOT (32). The Ramachandran plot statistics for the final model in Fig. S2 were calculated with the program Molprobity (29).
Fluorescence Anisotropy (FA).
FA measurements were carried out at 22 °C on a Genios Pro (Tecan). Peptides corresponding to residues 414–441 of UPF3b and to residues 427–454 of UPF3a were labeled with 5-carboxy-fluorescein. The peptides were synthesized with a serine residue instead of Cys440 (UPF3b) and Cys453 (UPF3a). The peptides were diluted in the same buffer (40 mM Tris pH 7.5, 300 mM NaCl, and 3 mM MgCl2) to a final concentration of 13.5 nM in 50 μL-well plate. They were incubated with various EJC concentrations for 10 min prior to the measurements. Fluorescence anisotropy was measured by exciting at 485 nm and monitoring the emission at 535 nm. The integration time was 40 μs with 10 reads per well and repeated three times. FA data collection was controlled using the Magelan software. Measurements were made in triplicate and the G factor deviation was < 5%. The data were analyzed using nonlinear regression fitting of the data for a single site binding model using the Origin software.
Tethering Assays.
HeLa cells were grown in DMEM supplemented with 10% FCS and transfected with calcium phosphate by standard methods. For tethering assays, cells were cotransfected in 6 cm culture dishes with 10 μg β-globin 4boxB reporter (13), 1 μg of the β-globin control (wt + 300 + e3, ref. 13), and 10 μg of plasmids expressing λN-UPF3b fusion proteins. Total RNAs were extracted by the TriReagent method (Euromedex), separated on 1.2% denaturing formaldehyde agarose gels (10 μg/lane), and blotted onto positively charged membranes (Genescreen Plus, NEN Life Science). Specific body-radiolabeled DNA probes for β-globin were generated by unidirectional PCR using standard methods. Signals were quantified with a Fuji FLA-3000 apparatus.
Supplementary Material
Acknowledgments.
We would like to thank Clemens-Schulze Briese and the staff at SLS for excellent assistance with data collection. We also thank Jerome Basquin and the staff of the MPI-Martinsried Crystallization Facility, Stephan Uebel and Silva Andric for peptide synthesis, and Cyril Boulegue for mass spectroscopy analysis (MPI-Martinsried Microchemistry Core Facility). We also thank Debora Makino and members of our labs for help and discussions. This study was supported by the Max Planck Gesellschaft, the Sonderforschungsbereich SFB646, and the Gottfried Wilhelm Leibniz Program of the Deutsche Forschungsgemeinschaft (E.C.); by the Centre National de la Recherche Scientifique, l’Agence Nationale de Recherche, and the ATIP program from the Research Ministry (H.L.H). J.S. is the recipient of a fellowship from the Fondation pour la Recherche Médicale.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2xb2 and r2xb2sf).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000993107/-/DCSupplemental.
References
- 1.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behm-Ansmant I, Izaurralde E. Quality control of gene expression: A stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- 4.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr Opin Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 8.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 10.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 11.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 12.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 14.Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J. Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol Cell. 2007;27:780–792. doi: 10.1016/j.molcel.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Czaplinski K, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 18.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 19.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen KH, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring NH, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Schutz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tritschler F, et al. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Bono F, et al. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004;5:304–310. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 28.Bono F, Grunwald M, Ebert J, Conti E. Nuclear import mechanism of the EJC components Mago-Y14 revealed by structural studies of Importin 13. Mol Cell. 2010;37:211–222. doi: 10.1016/j.molcel.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 29.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.