Abstract
Telomeres, the protein–DNA complexes at the ends of linear chromosomes, are protected and regulated by the shelterin molecules, the telomerase complex, and other accessory factors, among which is Apollo, a DNA repair factor of the β-lactamase/β-CASP family. Impaired telomere protection in humans causes dyskeratosis congenita and Hoyeraal–Hreidarsson (HH) syndrome, characterized by premature aging, bone marrow failure, and immunodeficiency. We identified a unique Apollo splice variant (designated Apollo-Δ) in fibroblasts from a patient with HH syndrome. Apollo-Δ generates a dominant negative form of Apollo lacking the telomeric repeat-binding factor homology (TRFH)-binding motif (TBM) required for interaction with the shelterin TRF2 at telomeres. Apollo-Δ hampers the proper replication of telomeres, leading to major telomeric dysfunction and cellular senescence, but maintains its DNA interstrand cross-link repair function in the whole genome. These results identify Apollo as a crucial actor in telomere maintenance in vivo, independent of its function as a general DNA repair factor.
Keywords: telomeric repeat-binding factor homology-binding motif, TRF2
The telomeres correspond to the ends of linear chromosomes and are composed, in vertebrates, of TTAGGG repeats. During cell division, conventional DNA polymerases are unable to replicate the end of chromosomes fully, leading to the progressive loss of TTAGGG sequences. If telomeres reach too short a length, cells stop proliferating and enter replicative senescence and/or apoptosis. In stem cells, some activated cells, and the majority of human cancer cells, the telomere shortening is counteracted by the telomerase TERT, which adds TTAGGG repeats after each cell division (1). The telomerase acts in a complex formed with dyskerin, TERC, and other accessory factors (2). TRF1, TRF2, RAP1, POT1, TPP1, and TIN2 are the six known factors that compose the shelterin complex. They are dedicated to protecting chromosome ends from degradation and/or fusion as well as to regulating the telomerase activity (3). TRF1 and TRF2 act as a molecular platform to recruit shelterin factors and DNA repair proteins at telomeres. Among them, the ATM and ATR kinases, the MRE11/RAD50/NBS1 complex, and the nuclease Apollo interact with TRF2, whereas the helicase BLM and the ATM and DNA-PKcs kinases interact with TRF1 (4). Defects in several DNA damage response (DDR) factors lead to telomere dysfunction in humans and mice, underlining the intricate links between telomeric homeostasis and general DDR (5). In humans, a defect in telomere maintenance causes dyskeratosis congenita (DKC), a rare inherited disorder characterized by bone marrow failure associated with other dysfunctions such as mucocutaneous abnormalities and cancer predisposition (2). Mutations in components of the telomerase complex (dyskerin, TERC, TERT, NOP10, and NHP2) and in the shelterin TIN2 are associated with DKC. Patients who have DKC exhibit the common feature of excessive telomere shortening, which likely causes the stem cell failure, resulting in a wide spectrum of clinical manifestations (2). The Hoyeraal–Hreidarsson (HH) syndrome, which associates cerebellar hypoplasia, microcephaly, immunodeficiency, intrauterine growth retardation, and other developmental defects with bone marrow failure, represents the rarest and most severe form of DKC (6, 7). TIN2, TERT, and dyskerin mutations have been described in some patients with HH syndrome, but the molecular cause of most cases of DKC and HH syndrome remains undefined (8, 9). With the aim of identifying previously undescribed genetic causes of HH syndrome, we analyzed the cellular and molecular phenotypes of primary fibroblasts from patients with HH syndrome with no mutation in known DKC-responsible genes. Here, we describe a unique transcript variant of Apollo from a patient with HH syndrome inducing severe telomere dysfunction. This study highlights the critical role of Apollo in telomere physiology.
Results
Cellular Senescence and Telomere Dysfunction in Fibroblasts from a Patient with HH Syndrome.
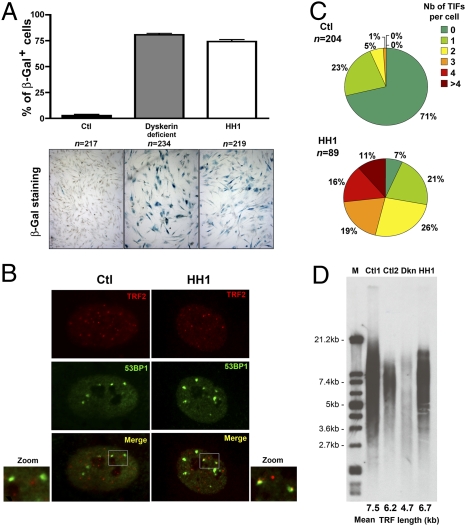
The patient investigated in this study (designated HH1) was a girl born to unrelated parents (10). She presented with clinical features characteristic of HH syndrome (6, 7), including severe intrauterine growth retardation, microcephaly, cerebellar hypoplasia, lack of B lymphocytes, and progressive aplastic anemia. Other clinical features were severe enteropathy associated with major villous atrophy and thin and sparse hair. She died at 4 years of age from disseminated infections consecutive to her severe bone marrow failure. A primary fibroblast line obtained from HH1 displayed a profound growth defect, even at early passages, as compared with primary fibroblasts from healthy controls. This was caused by an accelerated senescence revealed by senescence-associated (SA)–β-galactosidase activity (11), which was comparable to that of dyskerin-defective fibroblasts (Fig. 1A) (12). Cellular senescence of cells of HH1 was associated with the detection of telomere dysfunction-induced FOCI (TIFs) characterized by the specific recruitment of the DDR factor 53BP1 at telomeres (detected here by TRF2 staining; Fig. 1B) (13). A significant proportion of cells of HH1 (27%, n = 89) presented with four or more TIFs, whereas only 7% of the cells were TIF-negative. This contrasted with control fibroblasts, 71% of which were TIF-negative and none of which exhibited four or more TIFs (Fig. 1C). Because early senescence and increased TIFs are common features of cells with extremely short telomeres (13), we analyzed telomere length in cells of HH1 by telomeric terminal restriction fragment (TRF) assay (Fig. 1D). Although the estimated mean telomere length from dyskerin-defective fibroblasts was, as expected, shorter than that of two control cells (4.7, 7.5, and 6.2 kb, respectively) (14), the telomere length from fibroblasts of HH1 with a similar passage number was in the normal range (6.7 kb). Collectively, these observations indicate that primary fibroblasts of HH1 harbor several features of dysfunctional telomeres that are not associated with their abnormal shortening.
Fig. 1.
Cellular and molecular characteristics of fibroblasts of HH1. (A) Cellular senescence. Results are expressed as the percentage of SA–β-galactosidase (Gal)-positive cells. Primary fibroblasts are from a healthy control (Ctl; n = 217; black, passage 11), a dyskerin-deficient patient (n = 234; gray, passage 4), and HH1 (n = 219; white, passage 6). (B) Colocalization of 53BP1 foci with TRF2 representing TIFs can be detected in cells of HH1. (C) Quantification of TIFs. The fraction of fibroblasts from a control (Ctl; n = 204) and HH1 (n = 89) presenting with TIFs and the number of TIFs per TIF-positive cell were quantified. (D) Mean telomere length of primary fibroblasts from a healthy control (Ctl2), a dyskerin-deficient patient (Dkn), HH1, and control DNA from the TRF-kit (Ctl1; Roche) was estimated by the TRF method. All the results in this figure were obtained with cells having the passage number indicated in A.
Intraexonic Splice of Apollo in Cells of HH1.
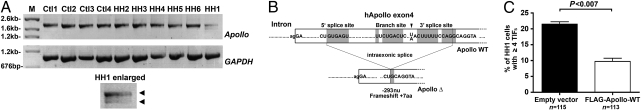
We next analyzed the known factors involved in telomere protection. No sequence alteration was found in the six shelterin-encoding genes. In contrast, the RT-PCR amplification of Apollo (15, 16) revealed an additional transcript of lower size in cells of HH1 that was not present in control cells or cells from other patients with HH syndrome with dysfunctional telomeres (Fig. 2A). This shorter Apollo transcript resulted from an intraexonic splice in exon 4 leading to an out-of-frame deletion of 293 (1247–1539) bp and a frameshift at E417, resulting in a premature stop codon (hereafter denoted Apollo-Δ) (Fig. 2B and Fig. S1). Importantly, the Apollo-Δ alternative transcript has not been described and is not registered in the EST database (17). Sequencing the two alleles (distinguishable through the presence of single-nucleotide polymorphisms) of the 9-kb long Apollo gene (comprising the introns and four exons) of HH1 and her parents did not reveal mutations. This suggests that the alteration of an uncharacterized element outside the Apollo gene is responsible for this splice variant. The Apollo-Δ is not the consequence of a general defect of the splicing machinery, because several other genes were correctly spliced (Fig. S1D). Although we could not identify the molecular origin of the Apollo splice variant in the cells of HH1, the known role of Apollo in telomere protection (15, 16) and the lack of the Apollo-Δ transcript in various databases and control cells prompted us to analyze further the impact of Apollo-Δ expression on telomere physiology.
Fig. 2.
Apollo intraexonic splice in cells of HH1. (A) RT-PCR amplification of Apollo transcripts with cDNA from primary fibroblasts from four controls (Ctl1, Ctl2, Ctl3, and Ctl4), five uncharacterized patients with HH syndrome (HH2, HH3, HH4, HH5, and HH6), and HH1. GAPDH primers were used as a control. (B) Sequence of the cryptic splice site (gray boxes) situated in the WT sequence of Apollo exon 4 and the product resulting from the intraexonic splice found in HH1. (C) Transduction of FLAG-tagged Apollo in primary fibroblasts of HH1 significantly reduces (P < 0.007) the number of cells with four or more TIFs. Each bar represents the mean value and SD of three separate determinations.
Complementation of Cell Defect in HH1.
First, we introduced a FLAG-tagged WT form of Apollo (FLAG-Apollo-WT) in primary fibroblasts of HH1. The ectopic expression of FLAG-Apollo-WT counteracts, at least in part, the telomere dysfunction in cells of HH1, as shown by the reduced proportion of cells with four or more TIFs (21.7–8.8%; P < 0.007), as opposed to cells transduced with an empty vector (Fig. 2C). These results suggest that the product of the Apollo-Δ transcript participates in the telomere dysfunction observed in cells of HH1.
Dominant Negative Effect of Apollo-Δ on Telomere Protection.
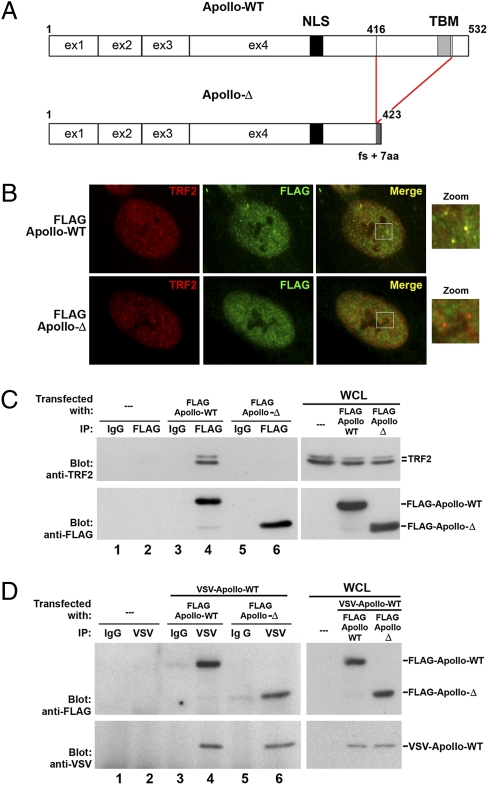
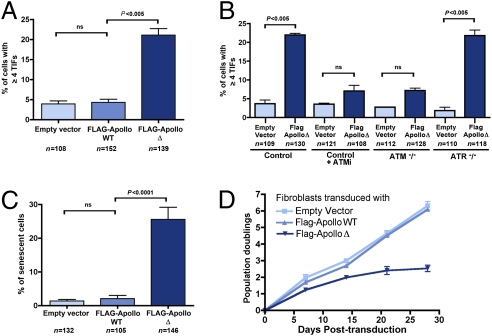
Next, we analyzed the consequences of ectopic Apollo-Δ expression in WT cells. The Apollo protein resulting from the intraexonic splice lacks the last 116 amino acids and has gained 7 amino acids generated by a frameshift. As a result, Apollo-Δ lacks the TRF homology-binding motif (TBM) critical for the interaction with the telomere shelterin factor TRF2 (18) (Fig. 3A and Fig. S1). The very low endogenous Apollo expression precludes any attempt at its detection by immunofluorescence or Western blot in cells of HH1 (15–17, 19). We thus analyzed the expression of FLAG-Apollo-Δ and FLAG-Apollo-WT on lentiviral transduction. Whereas FLAG-Apollo-WT colocalizes with TRF2 at telomeres as expected (15, 16, 19), FLAG-Apollo-Δ is diffuse in the nucleus (Fig. 3B). Consistent with its loss of TBM, the immunoprecipitation of FLAG-Apollo-Δ did not coprecipitate endogenous TRF2 (16) (Fig. 3C, lane 6). However, immunoprecipitation of a Vesicular Stomatitis Virus (VSV)-tagged WT-Apollo comparably coprecipitated FLAG-Apollo-WT or FLAG-Apollo-Δ (Fig. 3D, lanes 4 and 6), indicating that the previously described Apollo dimerization (16) is maintained between WT-Apollo and Apollo-Δ. We conclude that Apollo-Δ, which has lost its capacity to interact with TRF2, can still interact with WT-Apollo and may exert a dominant negative effect by titrating out the WT-Apollo molecules from telomeres. Indeed, colocalization of transduced FLAG-Apollo-WT with TRF2 is severely reduced in cells of HH1 (which express endogenous Apollo-Δ) (Fig. S2). The dominant negative effect of FLAG-Apollo-Δ is further sustained by the sharp increase of cells with four or more TIFs (21% vs. 4.3%; P < 0.005) on FLAG-Apollo-Δ transduction in WT primary fibroblasts (Fig. 4A). Interestingly, Apollo-Δ–mediated TIF induction was observed in ATR-deficient fibroblasts but not in ATM-deficient fibroblasts or in WT fibroblasts in the presence of ATM inhibitor (Fig. 4B), indicating that FLAG-Apollo-Δ–induced damaged telomeres rely on ATM-dependent DDR. FLAG-Apollo-Δ in control primary fibroblasts also significantly induced cellular senescence (Fig. 4C) and diminished cellular proliferation (Fig. 4D). Collectively, these results demonstrate that FLAG-Apollo-Δ recapitulates the cellular phenotype of cells of HH1 through its dominant negative effect on telomere protection when introduced in primary control fibroblasts.
Fig. 3.
Truncated Apollo (Apollo-Δ) resulting from the intraexonic splice does not interact with TRF2 but can interact with WT-Apollo. (A) Apollo intraexonic splice generates an Apollo protein (Apollo-Δ) truncated at the amino acid 416 position that lacks the TRF2 binding motif (TBM) but still harbors the nuclear localization signal (NLS). (B) Primary control fibroblasts were transduced with FLAG-Apollo-WT– or FLAG-Apollo-Δ–expressing vector. Immunofluorescence using anti-FLAG and anti-TRF2 antibodies shows that FLAG-Apollo-WT colocalized with TRF2 as opposed to FLAG-Apollo-Δ. (C) Proteins from whole-cell lysates (WCL) from cells transfected with the appropriate protein-expressing vectors were immunoprecipitated with either an irrelevant (IgG) or anti-FLAG antibody, blotted, and revealed by anti-TRF2 (Upper) or anti-FLAG (Lower) antibodies. (D) Proteins from WCL were immunoprecipitated with either an irrelevant (IgG) or an anti-VSV antibody, blotted, and revealed by anti-FLAG (Upper) or anti-VSV (Lower) antibodies.
Fig. 4.
Apollo-Δ exerts a dominant negative effect on telomere protection. (A) Primary fibroblasts from a healthy control were transduced either with an empty vector, a FLAG-Apollo-WT–expressing vector, or a FLAG-Apollo-Δ–expressing vector. The proportion of cells with four or more TIFs was counted. Mean values, SDs, and total events are indicated. ns, not significant. (B) Fibroblasts from a healthy control treated or not treated with the ATM inhibitor (ATMi) KU55933 (10 μM, 16 h), from ATM-deficient or ATR-deficient patients were transduced with an empty vector or a FLAG-Apollo-Δ–expressing vector. The proportion of cells with four or more TIFs was counted. Mean values and SDs are indicated. The results are representative of experiments performed with two different ATM-deficient cell lines. (C) Percentage of senescent cells measured after staining for SA–β-galactosidase 12 days posttransduction of primary control fibroblasts with an empty vector, a FLAG-Apollo-WT–expressing vector, or a FLAG-Apollo-Δ–expressing vector is indicated. (D) Diminished cell proliferation in Apollo-Δ–expressing cells. Cell numbers were measured at the indicated posttransduction time points.
Apollo-Δ Impairs the Proper Replication of Telomeres.
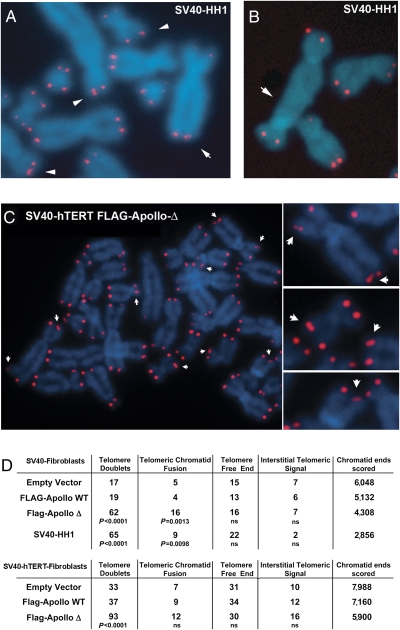
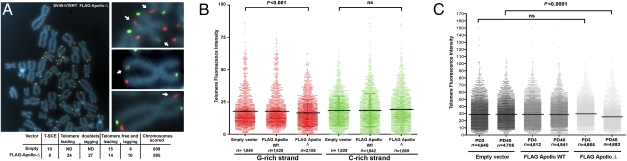
We transformed primary fibroblasts of HH1 with SV40 large T antigen (SV40) to overcome the p53-dependent senescence (20). Metaphase spreads from SV40-transformed fibroblasts of HH1 (SV40-HH1) exhibited a significant increase in chromatid ends harboring two distinct telomeric FISH signals (telomeric doublets; P < 0.0001; Fig. 5 A and D) and telomere–telomere fusions (P < 0.01; Fig. 5 B and D) as compared with SV40-transformed control fibroblasts. Ectopic expression of FLAG-Apollo-Δ in SV40-transformed WT cells also induced the formation of telomeric doublets whether the cells express human TERT (hTERT) or not (Fig. 5 C and D). Apart from this, FLAG-Apollo-Δ did not cause any other major telomeric aberration except for a statistically significant (P < 0.002) increase in telomere–telomere fusions in hTERT-negative cells (Fig. 5D). Chromosome orientation (CO) FISH used to distinguish G-rich (leading strand) from C-rich (lagging strand) telomeres (21) did not reveal a strand preference in the formation of telomeric doublets or increase in telomeric sister chromatid exchange (T-SCE) induced by FLAG-Apollo-Δ (Fig. 6A). Direct quantification of the CO-FISH signal (Q-CO-FISH) (21) in SV40-hTERT WT fibroblasts transduced with FLAG-Apollo-Δ revealed a significant decrease of the G-rich strand signal intensity as compared with cells transduced with FLAG-Apollo-WT or an empty vector (mean fluorescence intensity of 16.51, 17.77, and 17.80, respectively; P < 0.001) (Fig. 6B). This was not observed for the C-rich strand-specific signal, and therefore suggests that FLAG-Apollo-Δ compromises the completion of telomeric lagging strand DNA synthesis during replication. We reasoned that such an effect should result in progressive telomere attrition. Indeed, quantitative telomeric FISH revealed a telomere length shortening in FLAG-Apollo-Δ–expressing cells over time [mean fluorescence intensity of 29.78 at population doubling (PD) 3 to 25.35 at PD40; P < 0.0001] (Fig. 6C). This was further supported by TRF analysis, where mean telomere length decreased from 5.1 to 4.6 kb over 37 PDs in FLAG-Apollo-Δ–expressing cells, although remaining constant in control cells (Fig. S3). Telomere length shortening cannot be attributed to an inhibition of the telomerase activity per se, as demonstrated using a telomere repeat amplification protocol (TRAP) assay (Fig. S3). Although SV40 fibroblasts that do not express hTERT naturally display telomere length shortening following cell divisions, FLAG-Apollo-Δ further accelerates this attrition (Fig. S4). We conclude that FLAG-Apollo-Δ compromises telomere replication and leads, when cellular senescence is overcome, to accelerated telomere length shortening, telomeric doublet formation, and telomere–telomere fusion.
Fig. 5.
Telomeric doublets and telomere–telomere fusions in SV40-transformed cells of HH1 (SV40-HH1) and Apollo-Δ–expressing cells. SV40-HH1 cells exhibited telomere doublets (A) and telomeric fusion detected by telomeric FISH (B). (C) Telomere doublets (arrows) of SV40-hTERT fibroblasts transduced with FLAG-Apollo-Δ are detected by telomeric FISH. (D) Quantification of telomeric aberrations observed in SV40-HH1 fibroblasts and SV40 fibroblasts expressing or not expressing hTERT transduced by an empty vector, FLAG-Apollo-WT–expressing vector, or FLAG-Apollo-Δ–expressing vector is indicated. ns, not significant.
Fig. 6.
CO-FISH analysis of FLAG-Apollo-Δ–expressing cells. (A) Telomeric CO-FISH of SV40-hTERT fibroblasts transduced with FLAG-Apollo-Δ shows no strand preference for telomere doublets (arrows). (Lower) Quantification of telomere aberrations is indicated. ND, not done. (B) Telomere fluorescence intensity generated by CO-FISH probes was measured by quantitative CO-FISH in SV40-hTERT fibroblasts transduced with an empty vector, FLAG-Apollo-WT–expressing vector, or FLAG-Apollo-Δ–expressing vector. ns, not significant. (C) Quantitative FISH analysis of telomeric signals obtained from SV40-hTERT fibroblasts transduced with an empty vector, FLAG-Apollo-WT–expressing vector, or FLAG-Apollo-Δ–expressing vector after PDs 3 and 40. Individual and mean values are presented.
DNA Damage Repair Is Not Altered in Apollo-Δ–Expressing Cells.
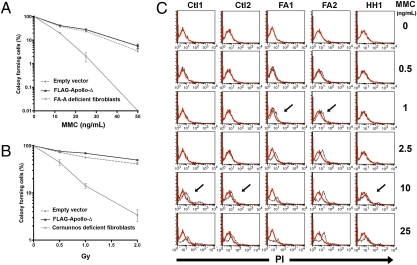
In addition to its function at telomeres, Apollo is involved in the repair of DNA lesions such as DNA interstrand cross-links (ICLs) generated by mitomycin C (MMC) and, to a lesser extent, in the repair of DNA damage produced by ionizing radiation (IR) (17, 22). SV40 fibroblasts expressing FLAG-Apollo-Δ did not show an increased sensitivity to either MMC or IR (Fig. 7 A and B). In addition, the MMC sensitivity of primary fibroblasts of HH1 was comparable to that found in primary fibroblasts from healthy controls (Fig. 7C). These results imply that FLAG-Apollo-Δ does not compromise DNA repair, contrasting with its dominant negative effect on telomere protection/replication.
Fig. 7.
No increased MMC and IR sensitivity of FLAG-Apollo-Δ–expressing fibroblasts. Survival of SV40-transformed fibroblasts transduced with empty vector or FLAG-Apollo-Δ–expressing vector after addition of MMC (A) and after IR up to 2 Gy (B) are shown. Results are expressed as the fraction of colony-forming cells in relation to untreated cells. Each point represents the mean value and SD of three separate determinations. Control MMC-sensitive SV40 fibroblasts are from a patient with Fanconi anemia (FA-A), and radiosensitive SV40 fibroblasts are from a Cernunnos patient. (C) Graphs representing the MMC sensitivity data in primary fibroblasts from two controls (Ctl1, Ctl2), two patients with FA (FA1, FA2), and HH1. On the x axis is plotted the MMC concentration at which dying cells were detected by propidium iodide (PI) intracellular uptake (arrows). Red and black lines represent PI uptake of untreated and MMC-treated cells, respectively. Results clearly demonstrate that the MMC sensitivity of cells of HH1 was comparable to that of controls, whereas FA cells exhibited a higher sensitivity to MMC treatment.
Coemergence of the TRF1/TRF2 Paralogues and TBM of Apollo.
Having found that Apollo requires its TBM to protect telomeres in human cells, we performed a phylogenetic analysis of this domain. Apollo (SNM1B), as well as its paralogues SNM1A and Artemis (SNM1C), is found in all genomes of metazoans, but only one equivalent is found in unicellular organisms (Fig. S5 and SI Text). Thus, the appearance of the three SNM1 paralogues, and therefore Apollo, is linked to the evolution toward multicellular organisms. We identified TBM in the C-terminal regions of the Apollo sequences in mammals as well as in chicken, fish, and amphibians (Fig. S6). In contrast, no TBMs or TBM-like motifs were identified in nonvertebrate species, strongly suggesting that the TBM is vertebrate-specific (Fig. S7 and SI Text). We thus wondered whether TRF2 and the TBM motif of Apollo could have coemerged in vertebrates specifically to direct interaction of Apollo with TRF2. Both TRF1 and TRF2 exist in vertebrates (Fig. S8), whereas only one TRF-like protein is found in several nonvertebrate eukaryotes (Fig. S8 and SI Text). TRF1 and TRF2 possess a TRFH domain for the dimerization and recruitment of various proteins to telomeres. Although TRF1 and TRF2 share a similar docking site in their TRFH domains, they bind different proteins, as shown by the differential recruitment of TIN2 and Apollo by TRF1 and TRF2, respectively (18). Thus, one may speculate that the coemergence of the TBM of Apollo and the two TRF paralogues TRF1 and TRF2 may be important to direct the specific binding of Apollo to TRF2 (Fig. S9).
Discussion
We describe telomere dysfunction in fibroblasts from a patient with HH syndrome (HH1), which express a unique Apollo transcript resulting from an intraexonic splice (Apollo-Δ). The expression of a WT form of Apollo in cells of HH1 partially reverts the telomere defect, and, in addition, the expression of Apollo-Δ in control cells recapitulates the telomeric dysfunction found in cells of HH1. These results indicate that Apollo-Δ, which lacks the TBM required to interact with the shelterin TRF2, exerts a dominant negative effect on telomere protection.
Fibroblasts of HH1 exhibited a phenotype comparable to that described in cells treated with Apollo siRNA (i.e., increased senescence, TIFs) (15, 16). SV40-transformed fibroblasts of HH1 and WT cells expressing Apollo-Δ both exhibited an increase in telomere doublets. Telomere doublet formation has been proposed to reflect impaired telomere replication, but the precise molecular process leading to this defect is unknown. Quantitative CO-FISH analysis of SV40-transformed fibroblasts transduced with Apollo-Δ shows a decrease of the FISH signal corresponding to the DNA synthesis of the lagging (C-rich) strand but not of the leading strand. This result supports the idea of impaired synthesis (or degradation) of the lagging strand DNA during telomeric replication in the absence of Apollo. As a consequence, SV40-transformed cells expressing Apollo-Δ exhibit a progressive telomere length shortening throughout cell divisions. These results demonstrate that Apollo is required both for proper DNA replication of telomeres and to avoid DDR at telomeres.
Human Apollo is a member of the PSO2/SNM1 family (comprising SNM1A, SNM1B/Apollo, and SNM1C/Artemis). Like SNM1A, Apollo was previously thought to be specifically devoted to the repair of ICL (17, 22). However, Apollo has recently been shown to be recruited to telomeres via a specific interaction with TRF2 (15, 16, 19). That Apollo-Δ–expressing cells, which are unable to protect telomeres, efficiently repair DNA injuries suggests a dissociation of Apollo functions between whole-genome maintenance and telomere protection/replication. Phylogenetic analysis revealed that the TBM domain of Apollo and the TRF1/TRF2 factors coemerged around the origin of vertebrates. One may speculate that the TBM of Apollo emerged to recognize the vertebrate-specific TRFH domain as well as to distinguish the paralogues TRF1 and TRF2, and thus mediate the specific interaction of Apollo with TRF2.
HH1 presented with all the features of HH syndrome, the most severe form of DKC. Most of these clinical manifestations are likely caused by the early onset of pronounced failure of progenitor cells from various lineages. In contrast to patients with DKC, acceleration of telomere length shortening was not observed in primary cells of HH1. This supports the important notion that DKC/HH syndrome is not invariably associated with excessive telomere loss.
In conclusion, based on the analysis of a unique Apollo splice variant expressed in a patient with HH syndrome, our work highlights the critical role of Apollo at telomeres via its interaction with TRF2. Although we did not identify the initial cause of this Apollo splice variant, our findings argue for the likely implication of Apollo-Δ in telomere dysfunction in HH1, and therefore make Apollo a pertinent candidate gene in other HH syndrome-related conditions.
Methods
Cells.
In accordance with the Helsinki Declaration, informed consent for our study was obtained from the families. This study was also approved by the Institutional Review Board of the Institut National de la Santé et de la Recherche Médicale (INSERM). Fibroblasts were obtained from skin biopsies. SV40-transformed and telomerase-immortalized cell lines were obtained as described (23).
Immunofluorescence Detection and Senescence-Associated β-Galactosidase Staining.
Immunofluorescence detection was performed as previously described (23). Cells were fixed for 5 min in 4% (vol/vol) paraformaldehyde in PBS, washed in PBS, and stained in β-galactosidase fixative solution in 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 2 mM MgCl2 in PBS.
FISH and CO-FISH.
Cells were arrested in metaphase with colcemid (KaryoMAX; Invitrogen) and incubated in hypotonic solution (75 mM KCl), fixed in methanol/acetic acid (3:1), and spread on slides. Air-dried slides were hydrated before fixation in 4% (vol/vol) formaldehyde, washed, and treated with pepsin. The formaldehyde fixation and washes were repeated, and the slides were dehydrated and air-dried. A hybridization mixture containing 70% (vol/vol) formamide, 0.3 μg/mL Cy3-(C3TA2)3 PNA probe (Panagene), and 1% (wt/vol) blocking reagent was added. DNA was denatured by heat. After hybridization, slides were washed with 70% formamide and 10 mM Tris and with 0.05 M Tris, 0.15 M NaCl, and 0.05% Tween-20. Slides were then dehydrated in ethanol, air-dried, and covered by antifade solution. CO-FISH was conducted as previously described (21).
Quantitative Analysis of Digital Images.
Blue (DAPI) and red (Cy3) fluorescence signals were captured by a CCD camera. Merged pseudocolor images were used to identify chromosomes based on DAPI. To estimate total telomere fluorescence intensity in FISH and CO-FISH experiments, 20–30 metaphases per preparation were captured and stored. National Institutes of Health software (ImageJ) was used for the quantitative analysis of images. Correction for uneven illumination of the slide was introduced using the pseudoflat field plug-in. The telomeric signal intensity was obtained by subtracting the mean pixel value associated with the interstitial regions of chromosomes from the mean pixel value for each telomeric spot (21).
Telomeric Restriction Fragment.
DNA (2 μg) was digested with HinfI and RsaI enzymes, resolved by a 0.7% agarose gel, and transferred to a nylon membrane. Hybridization was performed using EasyHyb solution (Roche) with digoxigenin (DIG)-labeled telomeric probe or γ-32P-labeled (TTAGGG)4 probe. After washes, membranes were exposed over a PhosphorImager (AGFA) or revealed by the DIG detection kit (Roche). PhosphorImager exposures of telomere-probed Southern blots were analyzed with the ImageJ program. The digitalized signal data were then transferred to Microsoft Excel and served as the basis for calculating mean TRF length using the formula L = (ODi)/(ODi/Li), where ODi = integrated signal intensity at position i and Li = length of DNA fragment in position i.
Expression Vectors and Western Blotting.
The cDNAs encoding full-length and truncated versions of human Apollo were amplified from cDNA of HH1 and cloned into a p3XFLAG-myc-CMV-26 expression vector (Sigma) and into a modified G-VSV-pcDNA3 vector. FLAG-Apollo and FLAG-Apollo-Δ coding sequences were cloned in a lentiviral vector, pTRIP. Coimmunoprecipitation experiments were performed as previously described (24).
Statistical Analyses.
For Q-CO-FISH and Q-FISH, the differences in mean fluorescence intensity (TFI) were analyzed using an unpaired Student's t test assuming a Gaussian distribution of the TFI values (verified by plotting a histogram of the different TFI values grouped into intervals) and considering the large number of values analyzed (>1,000 events for each condition). For telomeric aberrations observed by FISH and CO-FISH, the two-sided probablity values were obtained from a 2 × 2 contingency table analyzed using the χ2 test. Telomere doublets of each condition were compared considering the total number of chromatid ends (because one chromosome may exhibit more than one telomere doublet). The number of chromatid fusion with telomeric material was considered (because one chromosome can fuse with one or more telomeric ends).
Supplementary Material
Acknowledgments
We thank A. Londono and N. Arnoult for technical advice with Q-CO-FISH and discussion. We thank N. Vasquez for technical assistance. We thank P. Jeggo (University of Sussex, East Sussex, UK) for the kind gift of ATR-deficient fibroblasts. We thank S. Marcand and S. Latour for critical reading of the manuscript. This work was supported by institutional grants from the Institut National de la Santé et de la Recherche Médicale, Association pour la Recherche sur le Cancer, Institut National du Cancer (INCa)/Cancéropôle Ile de France, and CEA (LRC 40). F.T. received fellowships from the Fondation pour la Recherche Médicale. P.R. is a scientist from the Centre National de la Recherche Scientifique (CNRS).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914918107/-/DCSupplemental.
References
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Walne AJ, Dokal I. Dyskeratosis Congenita: a historical perspective. Mech Ageing Dev. 2008;129:48–59. doi: 10.1016/j.mad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 4.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 5.Blasco MA. Telomeres and human disease: Ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 6.Hoyeraal HM, Lamvik J, Moe PJ. Congenital hypoplastic thrombocytopenia and cerebral malformations in two brothers. Acta Paediatr Scand. 1970;59:185–191. doi: 10.1111/j.1651-2227.1970.tb08986.x. [DOI] [PubMed] [Google Scholar]
- 7.Hreidarsson S, Kristjansson K, Johannesson G, Johannsson JH. A syndrome of progressive pancytopenia with microcephaly, cerebellar hypoplasia and growth failure. Acta Paediatr Scand. 1988;77:773–775. doi: 10.1111/j.1651-2227.1988.tb10751.x. [DOI] [PubMed] [Google Scholar]
- 8.Walne AJ, Vulliamy TJ, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: Analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage SA, et al. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Revy P, et al. A syndrome involving intrauterine growth retardation, microcephaly, cerebellar hypoplasia, B lymphocyte deficiency, and progressive pancytopenia. Pediatrics. 2000;105:E39. doi: 10.1542/peds.105.3.e39. [DOI] [PubMed] [Google Scholar]
- 11.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 13.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 14.Heiss NS, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 15.Lenain C, et al. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 16.van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Demuth I, Digweed M, Concannon P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene. 2004;23:8611–8618. doi: 10.1038/sj.onc.1207895. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 19.Freibaum BD, Counter CM. hSnm1B is a novel telomere-associated protein. J Biol Chem. 2006;281:15033–15036. doi: 10.1074/jbc.C600038200. [DOI] [PubMed] [Google Scholar]
- 20.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 21.Arnoult N, Shin-Ya K, Londoño-Vallejo JA. Studying telomere replication by Q-CO-FISH: The effect of telomestatin, a potent G-quadruplex ligand. Cytogenet Genome Res. 2008;122:229–236. doi: 10.1159/000167808. [DOI] [PubMed] [Google Scholar]
- 22.Bae JB, et al. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene. 2008;27:5045–5056. doi: 10.1038/onc.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 24.Callebaut I, et al. Cernunnos interacts with the XRCC4 x DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.