Abstract
Among the large set of cell surface glycan structures, the carbohydrate polymer polysialic acid (polySia) plays an important role in vertebrate brain development and synaptic plasticity. The main carrier of polySia in the nervous system is the neural cell adhesion molecule NCAM. As polySia with chain lengths of more than 40 sialic acid residues was still observed in brain of newborn Ncam−/− mice, we performed a glycoproteomics approach to identify the underlying protein scaffolds. Affinity purification of polysialylated molecules from Ncam−/− brain followed by peptide mass fingerprinting led to the identification of the synaptic cell adhesion molecule SynCAM 1 as a so far unknown polySia carrier. SynCAM 1 belongs to the Ig superfamily and is a powerful inducer of synapse formation. Importantly, the appearance of polysialylated SynCAM 1 was not restricted to the Ncam−/− background but was found to the same extent in perinatal brain of WT mice. PolySia was located on N-glycans of the first Ig domain, which is known to be involved in homo- and heterophilic SynCAM 1 interactions. Both polysialyltransferases, ST8SiaII and ST8SiaIV, were able to polysialylate SynCAM 1 in vitro, and polysialylation of SynCAM 1 completely abolished homophilic binding. Analysis of serial sections of perinatal Ncam−/− brain revealed that polySia-SynCAM 1 is expressed exclusively by NG2 cells, a multifunctional glia population that can receive glutamatergic input via unique neuron-NG2 cell synapses. Our findings sug-gest that polySia may act as a dynamic modulator of SynCAM 1 functions during integration of NG2 cells into neural networks.
Keywords: polysialic acid, NG2 cells, glycosylation, polysialyltransferases, glycoproteomics
Glycosylation represents the most complex posttranslational modification, with an overwhelming diversity of oligosaccharide structures. Moreover, a single protein can be variably glycosylated giving rise to multiple glycoforms with distinct biological functions. Unraveling the impact of glycosylation on the structure and function of proteins is therefore often an arduous task. A striking example for the capability of an individual glycan structure to induce dramatic functional changes on the underlying protein is polysialic acid (polySia). In vertebrates, this linear α2,8-linked homopolymer of 5-N-acetylneuraminic acid (Neu5Ac) was first described as a developmentally regulated modification of the neural cell adhesion molecule (NCAM) (1–3). Polysialylation disrupts the adhesive properties of NCAM and appearance of the bulky polyanionic glycan on the cell surface generally increases the intercellular space (4, 5). Thus, polySia is a modulator of cell interactions involved in dynamic processes such as neural cell migration, neurite outgrowth, neural path finding, and synaptic plasticity (6–9). Although it is abundantly expressed during embryonic and early postnatal brain development, polySia is restricted to areas with ongoing neurogenesis and synaptic plasticity in adult brain (10–12).
In mammals, polysialylation is catalyzed by the Golgi-resident polysialyltransferases (polySTs) ST8SiaII and ST8SiaIV, and simultaneous ablation of both enzymes leads to a complete loss of polySia (13, 14). In contrast to Ncam−/− mice, which still contain residual amounts of polySia and manifest only a mild phenotype (15), St8sia2−/−St8sia4−/− double KO mice are characterized by postnatal lethality and severe malformations of major axon tracts (14, 16, 17). Lethality and brain wiring defects could be attributed to erroneous exposure of polySia-free NCAM (14, 18), highlighting the crucial role of polySia in masking the underlying protein and thereby preventing improper interactions.
Although NCAM is by far the most abundant polySia carrier in mammals, context-dependent polysialylation of a restricted set of other glycoproteins has been described. These are CD36 in human milk, the α-subunit of a voltage-gated sodium channel in adult rat brain, and neuropilin-2 in mature human dendritic cells (19–21). Based on the observation that Ncam−/− brains still contain low but clearly detectable amounts of polySia (ref. 15 and the present study), we performed a glycoproteomic approach to screen for respective polySia carriers. This led to the identification of the synaptic cell adhesion molecule SynCAM 1 as a target for polysialylation. SynCAM 1 is a member of the Ig superfamily composed of three Ig modules comprising six potential N-glycosylation sites, a variable stem region with several putative O-glycosylation sites, a single transmembrane domain, and a short carboxyl-terminal intracellular tail (22). Because of its identification in different tissues, SynCAM 1 (official gene name Cadm1) has various names: nectin-like protein 2 (Necl-2) (23), tumor suppressor in lung cancer 1 (TSLC-1) (24), spermatogenic Ig superfamily molecule (SgIGSF) (25), Ig superfamily 4 (IgSF4) (26), and RA175 (27). SynCAM 1 contributes to a variety of intercellular junctions by mediating Ca2+-independent cell adhesion through homo- and heterophilic interactions (22, 23, 28–30). In the brain, SynCAM 1 localizes to synapses, bridges the synaptic cleft by homo- and heterophilic transinteraction with SynCAM 2, and acts as a potent inducer of synapse formation (22, 31). Here we demonstrate that, in vivo, a subfraction of SynCAM 1 is selectively polysialylated at the third N-glycosylation site and expressed by a subset of NG2 cells. In vitro polysialylation by either ST8SiaII or ST8SiaIV completely abolished homophilic SynCAM 1 binding, implying that polysialylation affects SynCAM 1 functions and may serve as a crucial modulator of SynCAM 1 interactions during integration of NG2 glia into neural networks.
Results
Characterization of PolySia in Perinatal Brain of Ncam−/− Mice.
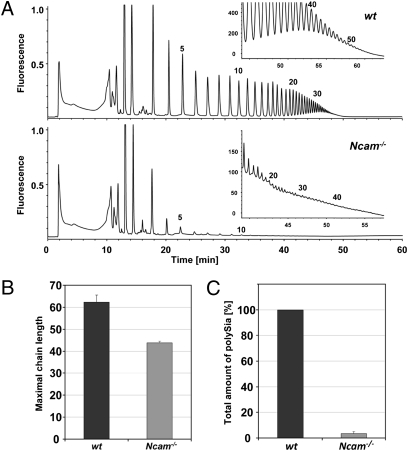
By immunostaining with an anti-polySia antibody, Cremer et al. identified residual amounts of polySia in brain of Ncam−/− mice (15). To analyze residual polySia, i.e., polymer length and total amounts in more detail, we applied the 1,2-diamino-4,5-methylenedioxybenzene (DMB)–HPLC method (13, 32, 33) to whole brain lysates of newborn Ncam−/− animals. Released polySia chains were fluorescently labeled and separated according to the degree of polymerization by anion exchange chromatography. Although the amount of all polymer species was drastically reduced compared with WT samples, polySia with more than 40 residues was still detectable in NCAM-deficient brain (Fig. 1 A and B). Quantification revealed that brain of Ncam−/− mice contained only 3.5% of the WT polySia level (Fig. 1C). Taking into account that visualization of long polySia chains strongly depends on the amount of material applied (34), one might assume that polySia synthesized in Ncam−/− and WT brain reaches similar chain lengths.
Fig. 1.
Chromatographic profiles of polySia from WT and NCAM KO mouse brains. (A) Delipidated brain homogenates obtained from WT and NCAM KO (Ncam−/−) mice (postnatal day 1) were directly derivatized with the fluorescence dye DMB and separated on an anion exchange column according to the number of sialic acid residues. In each case, 9% of the total brain homogenate (equivalent to 7 mg of original brain tissue) was injected. To determine the maximally detectable chain length, respective profiles were also generated with 86% aliquots (equivalent to 69 mg of brain tissue; Insets). The number of sialic acid residues is given for selected peaks on top of the profiles. (B) The average maximal chain length was determined from four independent experiments and amounted to approximately 62 and approximately 44 for WT and Ncam−/− mice, respectively. (C) Peak areas corresponding to polySia chains with more than eight sialic acid residues were calculated and summarized to obtain the total amount of polySia in brains of WT and Ncam−/− mice. Values are means of four independent experiments and were set to 100% for WT. The amount of polySia in Ncam−/− brains comprised only approximately 3.5% of the WT value.
Identification of SynCAM 1 as PolySia Carrier in Developing Mouse Brain.
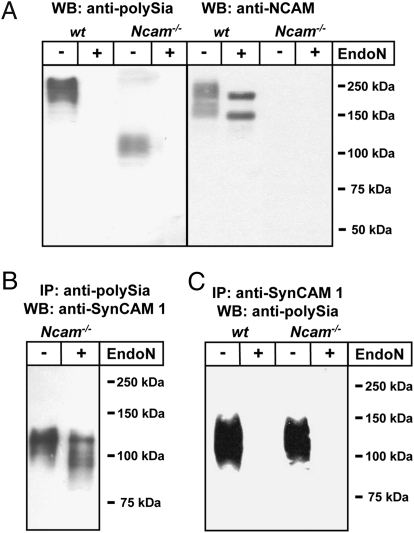
Polysialylated protein(s) in whole brain lysates of newborn Ncam−/− mice were characterized by Western blotting applying the tenfold amount of lysate compared with WT samples to compensate for the low polySia level. In WT samples, immunostaining with the polySia-specific mAb 735 revealed the typical broad polySia-NCAM signal at approximately 250 kDa, which was completely abolished after treatment with polySia-specific endosialidase N (endoN; Fig. 2A). Reprobing with anti-NCAM mAb H28 displayed a similar high molecular weight band which, after endoN treatment, gave rise to two focused bands representing the NCAM isoforms NCAM-140 and -180. In contrast, the main endoN sensitive signal observed with mAb 735 in lysate of Ncam−/− brain centered at approximately 110 kDa and no signal was obtained with mAb H28. To identify the underlying protein scaffold, polysialylated molecules were isolated from Ncam−/− brain extracts by affinity chromatography using mAb 735. After separation by SDS/PAGE, a gel slice spanning the molecular mass range of 100 to 150 kDa was used for tryptic in-gel digest. Analysis of the resulting peptides by peptide mass fingerprinting and mass spectrometric fragmentation analysis resulted in the identification of SynCAM 1 with significant probability scores of 78 and 186 (P < 0.05), respectively (Fig. S1). To verify this result, polysialylated proteins were affinity-isolated from Ncam−/− brain extracts and characterized by Western blotting with an anti-SynCAM 1 antibody. As shown in Fig. 2B, a broad band with an apparent molecular mass of 100 to 120 kDa was observed. After endoN digest, the signal broadened and bands with apparent molecular masses ranging from 85 kDa to 110 kDa could be distinguished. The fact that no discrete bands were formed is most likely because of the described heterogeneous glycosylation of SynCAM 1 by N- and O-glycans and/or the presence of different isoforms (35, 36). In a second experiment, SynCAM 1 immunoprecipitated with an anti-SynCAM 1 antibody was analyzed by immunoblotting with mAb 735 (Fig. 2C). Again, a polySia-signal in the molecular mass range of 100 to 150 kDa was observed, which was not detected after endoN pretreatment. The amount of total SynCAM 1 in perinatal Ncam−/− brain decreased only slightly after complete removal of the polySia-SynCAM 1 fraction by immunoprecipitation with mAb 735 (Fig. S2A). Thus, only a subfraction of SynCAM 1 is modified by polySia. Further analysis of the polySia-SynCAM 1 levels at postnatal d 2, d 21, and adult stage demonstrated a drastic decrease of polySia-SynCAM 1 during postnatal development, whereas no obvious change in the level of total SynCAM 1 was detected (Fig. S3).
Fig. 2.
Characterization of polysialylated proteins from WT and Ncam−/− mouse brains by SDS/PAGE and Western blotting (WB). Apparent molecular masses of standard proteins are indicated in kDa. (A) Brain homogenates of WT and Ncam−/− mice were separated by 10% SDS/PAGE using 4 μg (WT) or 40 μg (Ncam−/−) protein per lane with or without prior endoN pretreatment and immunostained using anti-polySia mAb 735 or anti-NCAM mAb H28. (B) PolySia proteins of Ncam−/− mice were immunoaffinity purified (IP) using mAb 735, separated by SDS/PAGE, blotted, and stained with rabbit polyclonal anti-SynCAM 1 antibody before and after endoN treatment. (C) Equal amounts of brain lysates (60 mg wet weight each) of newborn WT and Ncam−/− mice were used for immunoprecipitation with polyclonal anti-SynCAM 1 antibody. Immunoprecipitates were analyzed before and after endoN treatment by Western blot analysis with anti-polySia mAb 735.
Although our results clearly identified SynCAM 1 as a target for polysialylation in Ncam−/− mice, the question remained whether this is a compensatory response to the lack of NCAM. Consequently, polysialylation of SynCAM 1 was studied in perinatal brain of WT mice using 10 times the amount of brain extracts compared with Fig. 1A (Fig. S2B). Under these conditions, the polySia signal covered not only the dominating high molecular mass band of polySia-NCAM but also the mass range around 110 kDa, where polySia-SynCAM 1 migrates. To prove the presence of polysialylated SynCAM 1 in WT brain, immunoprecipitates obtained with an anti-SynCAM 1 antibody were stained with mAb 735 (Fig. 2C). In both WT and Ncam−/− brain, comparable amounts of polySia-SynCAM 1 were detected. Together these data demonstrate that SynCAM 1 is an NCAM-independent polySia carrier.
Polysialic Acid Chains Are Located on N-Glycans of the First Ig Domain.
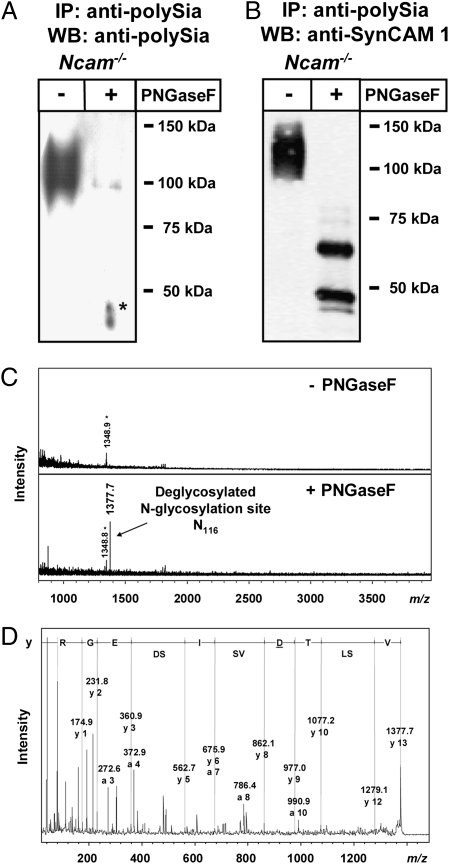
To determine whether polysialylation of SynCAM 1 occurs on N- or O-glycans, polysialylated SynCAM 1 immunoprecipitated from perinatal Ncam−/− brain was treated with N-glycosidase F (PNGaseF). As shown in Fig. 3A, PNGaseF digestion almost completely abolished mAb 735 staining, indicating that polySia chains were linked to N-glycans. The faint residual band in the range of approximately 90 kDa is presumably a result of incomplete PNGase F digestion. Parallel staining with anti-SynCAM 1 antibody (Fig. 3B) revealed that removal of N-glycans from polySia-SynCAM 1 resulted in two prominent bands. The molecular masses of approximately 48 and 65 kDa match with the masses described for unglycosylated and O-glycosylated SynCAM 1 variants, respectively (22, 35, 36).
Fig. 3.
Identification of the polysialylated glycosylation site in polySia-SynCAM 1 of Ncam−/− mice. (A and B) Western blot analyses of immunoaffinity purified (mAb 735) polySia–SynCAM 1 using anti-polySia mAb 735 (A) or polyclonal anti-SynCAM 1 antibody (B) with or without prior PNGaseF treatment. Apparent molecular masses of standard proteins are indicated in kDa. The asterisk indicates artifact. (C) MALDI-TOF MS spectra of immunopurified polysialylated glycopeptides before (−) or after (+) PNGaseF treatment. Monoisotopic masses of the pseudomolecular ions [M+H]+ are given. The asterisk indicates unknown contaminant. (D) Sequencing of the deglycosylated peptide by MALDI-TOF MS/MS. Sequence-specific ions are labeled according to previous studies (48) and the deduced amino acid sequence is shown. The Asp residue detected instead of Asn as a result of the known conversion of N-glycosylated Asn during PNGaseF release of N-glycans is underlined. After replacing Asp by Asn, the identified peptide sequence was used for database search (Mascot) verifying again SynCAM 1 with a significant score.
For allocation of the polySia chains to distinct N-glycosylation sites, the total fraction of polysialylated glycopeptides was immunoaffinity-isolated from whole brain homogenates and analyzed by MALDI-TOF MS. After de-N-glycosylation by PNGaseF, only one additional signal at m/z 1377.7 was detected (Fig. 3C), corresponding to the deglycosylated tryptic SynCAM 1 peptide comprising the third N-glycosylation site (Asn116) in which the glycosylated Asn has been converted to Asp as a result of PNGaseF action. Tandem MALDI-TOF MS analysis verified the sequence of this peptide as V112SLTDVSISDEGR124 (Fig. 3D), demonstrating that SynCAM 1 is polysialylated on N-glycans at Asn116. Thus, within the limit of detection, the presence of other proteins carrying polySia on N-glycans can be ruled out.
SynCAM 1 Is Polysialylated by ST8SiaII and ST8SiaIV in Vitro.
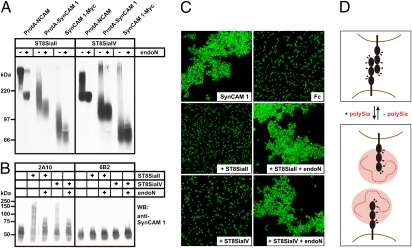
To investigate whether SynCAM 1 is a target for the two polysialyltransferases ST8SiaII and ST8SiaIV, an in vitro assay was performed using soluble SynCAM 1 as Protein A fusion protein or C-terminally tagged with a Myc-epitope. SynCAM 1 adsorbed to Sepharose beads was incubated with CMP-[14C]sialic acid in the presence of ST8SiaII or ST8SiaIV. A corresponding Protein A–NCAM chimera was used as positive control, and reaction products were analyzed before and after treatment with endoN. As shown in Fig. 4A, both ST8SiaII and ST8SiaIV were able to polysialylate SynCAM 1 as demonstrated by the appearance of radiolabeled protein that migrated significantly more slowly than the same protein after endoN treatment. To prove that polySia was added to SynCAM 1, reaction products obtained after polysialylation with nonradiolabeled substrate were analyzed by Western blotting with anti–SynCAM 1 antibody (Fig. 4B). Before polysialylation, soluble SynCAM 1 migrated with an apparent molecular mass of approximately 55 kDa (Fig. 4B Left), whereas after incubation with either ST8SiaII or ST8SiaIV, a broad smear ranging from 55 to 250 kDa appeared. This smear was sensitive to endoN, confirming that polySia was added to SynCAM 1. Notably, SynCAM 1 expressed in CHO-6B2 cells was not used as an acceptor molecule (Fig. 4B Right). Because of a lack of a functional CMP-sialic acid transporter, CHO-6B2 cells express exclusively asialo-glycoconjugates (37). Thus, the presence of terminally monosialylated glycans is a prerequisite for both ST8SiaII and ST8SiaIV to polysialylate SynCAM 1.
Fig. 4.
In vitro polysialylation of SynCAM 1 by ST8SiaII and ST8SiaIV. (A) Soluble Protein A (ProtA) fusion proteins of NCAM and SynCAM 1 as well as soluble SynCAM 1 with a C-terminal Myc-epitope were adsorbed to Sepharose beads and incubated with ST8SiaII (Left) or ST8SiaIV (Right) in the presence of CMP-[14C]sialic acid. Reaction products were separated by 7% SDS/PAGE before and after treatment with endoN and analyzed by autoradiography. (B) Soluble SynCAM 1-Myc was expressed in sialylation-competent CHO-2A10 (Left) and sialylation-deficient CHO-6B2 cells (Right). After adsorption to Sepharose beads, SynCAM 1 was incubated with CMP-sialic acid in the presence or absence of ST8SiaII or ST8SiaIV. Reaction products were separated by 8% SDS/PAGE before and after endoN treatment and analyzed by Western blotting (WB) with anti-SynCAM 1 mAb 3E1. (C) Homophilic SynCAM 1 binding is abrogated by polysialylation. Fluorescent Protein A beads were loaded with purified SynCAM 1-Fc chimera or with isolated Fc-fragments as control. Extensive clustering of SynCAM 1 coated beads was completely abolished by ST8SiaII and ST8SiaIV catalyzed polysialylation and was fully restored after removal of polySia by endoN treatment. (D) Proposed model for abrogation of homophilic SynCAM 1 binding by polysialylation. Ig-like domains are represented as black spheres, N-glycans are indicated by triangles, and polySia is shown as dashed red line with the hydrodynamic radius indicated by a red disk.
Homophilic SynCAM 1 Binding Is Abrogated by Polysialylation.
To study the impact of polysialylation on SynCAM 1–mediated interactions, we monitored the effect of this modification on homophilic SynCAM 1 binding. An Fc-chimera of the extracellular part of SynCAM 1 fused to the Fc region of human IgG was expressed in CHO-2A10 cells and isolated by affinity chromatography on protein G–Sepharose. Fluorophore-labeled beads were coated with purified SynCAM 1-Fc and bead aggregation was monitored before and after in vitro polysialylation (Fig. 4C). Extensive aggregation, leading to large bead clusters, was observed for the nonpolysialylated form of SynCAM 1. However, after in vitro polysialylation of SynCAM 1 by either ST8SiaII or ST8SiaIV, aggregation was abrogated and only monodisperse beads were visible. Subsequent removal of polySia by endoN restored SynCAM 1 binding, and reaggregation of the beads to large clusters was observed. Thus, polysialylation of SynCAM 1 inhibits homophilic binding in vitro, strongly indicating a functional role in modulating SynCAM 1 interactions in vivo (Fig. 4D).
PolySia-SynCAM 1 Is Expressed on NG2 Cells.
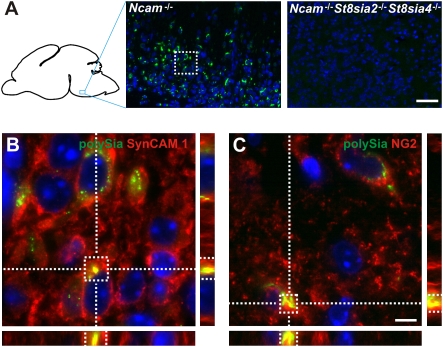
Analysis of serial brain sections obtained from newborn Ncam−/− mice revealed that polySia-positive cells were scattered throughout the gray matter but scarcely found in the white matter such as corpus callosum. PolySia staining was particularly abundant in the pontomedullary hindbrain and completely absent in brain sections of Ncam−/−St8sia2−/−St8sia4−/− triple KO mice (Fig. 5A). PolySia colocalized with SynCAM 1 and was restricted to a subpopulation of cells that are positive for the proteoglycan NG2 (Fig. 5 B and C and Fig. S4), a marker protein characteristic for a distinct type of glia cells. As NG2-negative cells that are wrapped by NG2-positive processes can be mistaken for NG2-positive cells, we confirmed our results by analyzing single cells in primary cultures from basal hindbrain of newborn Ncam−/− mice. Again, polySia was found colocalized with SynCAM 1 and associated with cells positive for NG2 and Olig2, a transcription factor frequently used as a second marker for NG2 cells (38) (Fig. S5). Consistent with described characteristics of NG2 cells (39), polySia-SynCAM 1–positive cells were negative for glial fibrillary acidic protein, β-III-tubulin, and microtubule-associated protein 2 (Figs. S4 and S5).
Fig. 5.
PolySia-SynCAM 1 is expressed on NG2 cells. (A) Representative immunofluorescence image showing abundant polySia expression (labeled by anti-polySia mAb 735; green) in the pontomedullary hindbrain of newborn Ncam−/− brain. Specificity of the staining was controlled on equivalent brain sections of Ncam−/−St8sia2−/−St8sia4−/− mice, where no polySia signal was detected. (B) Magnification of the boxed area in A. A 3D reconstruction of ApoTome z-stack images shows colocalization of polySia (labeled by mAb 735; green) and SynCAM 1 (labeled by mAb 3E1; red). Specificity of mAb 3E1 for SynCAM 1 was controlled by transfection experiments (Fig. S6). (C) Colocalization of polySia (green) and NG2 (labeled by NG2 antibodies). Nuclei are stained blue (DAPI stain). (Scale bars: 50 μm in A, 5 μm in B and C.)
Discussion
In the present study, we show that the synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in developing mouse brain. In vivo, polySia is selectively added to N-glycans at the third N-glycosylation site located within the first Ig domain, which is involved in homo- and heterophilic SynCAM 1 interactions (36). Addition of the bulky polyanionic carbohydrate polymer completely blocked homophilic binding in vitro. Although we cannot exclude differences in the extend of polysialylation under in vitro and in vivo conditions, this finding implicates that polySia serves as a potent regulator of SynCAM 1 interactions in vivo as it is known for NCAM (6). Compared with NCAM, which is composed of five Ig and two fibronectin type III modules, SynCAM 1 contains only three Ig-like domains. Both molecules comprise six N-glycosylation sites but only particular sites are used for polysialylation in vivo. Intriguingly, the polySia acceptor sites of both NCAM and SynCAM 1 are located in an Ig domain that is two domains apart from the membrane (Fig. S7). As polySTs are also transmembrane proteins, proper spacing might determine accessibility and in vivo selectivity for particular N-glycosylation sites as indicated by loss of site specificity in N-glycosylation mutants if both enzyme and acceptor molecule lack their transmembrane domain (Fig. S7).
In the perinatal brain, polySia-SynCAM 1 was found exclusively on a subset of NG2 cells. These glia cells (also known as polydendrocytes or synantocytes) are distinct from mature oligodendrocytes, astrocytes, and microglia, and make up 5% to 10% of all glia in the developing and mature CNS (39–41). They are scattered throughout the developing and adult brain and are considered as multipotential progenitor pool that can give rise to oligodendrocytes, astrocytes, and neurons. Remarkably, a subset of NG2 cells can promote presynaptic specialization in neurons, leading to unique synaptic association between NG2 cells and neurons (39, 41).
SynCAM 1 is known as a powerful inducer of synaptic differentiation. When coexpressed with glutamate receptors in nonneuronal cells, SynCAM 1 is sufficient to induce artificial synapses with cocultured neurons (22). To date, little is known what drives the assembly of neuron-NG2 cell synapses. To our knowledge, this is the first report of SynCAM 1 expression on NG2 cells, suggesting that this cell adhesion molecule could play a role in inducing this specialized neuron-glia synapse. By attenuating SynCAM 1–mediated functions, polysialylation of SynCAM 1 may have an important regulatory role in the formation of neuron-NG2 cell interactions. Moreover, polySia-SynCAM 1 has the potential to regulate the communication between NG2 cells and neurons, as it is known that polySia directly increases the probability of the open state of AMPA-type glutamate receptors (42), the receptor type through which NG2 cells receive synaptic inputs (43, 44).
Remarkably, only a subfraction of SynCAM 1 is polysialylated in perinatal brain. In contrast, at this developmental stage when both polySTs reach peak level and almost ubiquitous expression, NCAM is quantitatively converted to its polysialylated form (13, 45). As SynCAM 1 is broadly expressed in all brain regions perinatally, polysialylation might be restricted to particular glyco- and/or isoforms of SynCAM 1. Alternative splicing of three variable exons can theoretically give rise to eight transmembrane isoforms that differ only in the region between Ig3 and the transmembrane domain. The variably spliced peptides contain two to 17 putative O-glycosylation sites, leading to multiple glycoforms (35). In vitro, both polySTs were able to polysialylate soluble SynCAM 1 lacking the variable stem region, demonstrating that Ig1-3 modules are sufficient to mediate interaction with polySTs. However, it is still possible that, in vivo, only particular isoforms allow proper spacing and accessibility of the N-glycan acceptor site.
In summary, we characterized SynCAM 1 as a polySia acceptor in the developing brain and demonstrated that polySia-SynCAM 1 is restricted to a subpopulation of NG2 cells. NG2 cells form functional synapses in the postnatal brain and serve as the primary source of myelinating oligodendrocytes during development and myelin repair. Future experiments will be needed to determine the exact role of polySia-SynCAM 1 for NG2 cell functions.
Materials and Methods
Please refer to the SI Materials and Methods for details on mice, antibodies, and further methods.
Identification of SynCAM 1 as Polysialylated Glycoprotein and Intramolecular Localization of PolySia.
Isolation of polysialylated proteins, isolation and deglycosylation of polySia-glycopeptides, immunoblot, and DMB-HPLC anal-ysis were carried out as previously described (13, 33, 45). PolySia-SynCAM 1 was identified by in-gel tryptic digest, peptide mass fingerprint analysis using MALDI-TOF MS, MS fragmentation analysis, and database search. Isolated polySia-glycopeptides were chemically desialylated, treated with PNGase F and analyzed by tandem MALDI-TOF MS.
In Vitro Polysialylation and Bead Aggregation Assay.
SynCAM 1 lacking transmembrane domain and variably spliced stem region was produced in CHO cells either as a Protein A–SynCAM 1 chimera or C-terminally tagged with a Myc-epitope. After immunoadsorption to either IgG- or Protein G–Sepharose coupled with anti-Myc mAb 9E10, in vitro polysialylation was performed as described previously (46) with purified soluble ST8SiaII and ST8SiaIV. Homophilic SynCAM 1 binding was analyzed in a bead aggregation assay with purified SynCAM 1 fused to the Fc-part of human IgG1 (36).
Immunohistochemistry.
Dissection of brains from transcardially perfused mice, preparation of paraffin sections, immunofluorescence staining and microscopy were performed as described (13, 47).
Supplementary Material
Acknowledgments
We thank Rita Gerardy-Schahn for continuous support and helpful discussions, as well as Werner Mink and Siegfried Kühnhardt for expert technical assistance. This work was supported by Deutsche Forschungsgemeinschaft (Ge 527/3, MU 1774/3, and SFB 535, Project Z1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912103107/-/DCSupplemental.
References
- 1.Finne J, Finne U, Deagostini-Bazin H, Goridis C. Occurrence of alpha 2-8 linked polysialosyl units in a neural cell adhesion molecule. Biochem Biophys Res Commun. 1983;112:482–487. doi: 10.1016/0006-291x(83)91490-0. [DOI] [PubMed] [Google Scholar]
- 2.Rothbard JB, Brackenbury R, Cunningham BA, Edelman GM. Differences in the carbohydrate structures of neural cell-adhesion molecules from adult and embryonic chicken brains. J Biol Chem. 1982;257:11064–11069. [PubMed] [Google Scholar]
- 3.Edelman GM, Chuong CM. Embryonic to adult conversion of neural cell adhesion molecules in normal and staggerer mice. Proc Natl Acad Sci USA. 1982;79:7036–7040. doi: 10.1073/pnas.79.22.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson CP, Fujimoto I, Rutishauser U, Leckband DE. Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. J Biol Chem. 2005;280:137–145. doi: 10.1074/jbc.M410216200. [DOI] [PubMed] [Google Scholar]
- 5.Fujimoto I, Bruses JL, Rutishauser U. Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function and independence from neural cell adhesion molecule binding or signaling activity. J Biol Chem. 2001;276:31745–31751. doi: 10.1074/jbc.M104525200. [DOI] [PubMed] [Google Scholar]
- 6.Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- 7.Kleene R, Schachner M. Glycans and neural cell interactions. Nat Rev Neurosci. 2004;5:195–208. doi: 10.1038/nrn1349. [DOI] [PubMed] [Google Scholar]
- 8.Hildebrandt H, Mühlenhoff M, Weinhold B, Gerardy-Schahn R. Dissecting polysialic acid and NCAM functions in brain development. J Neurochem. 2007;103(suppl 1):56–64. doi: 10.1111/j.1471-4159.2007.04716.x. [DOI] [PubMed] [Google Scholar]
- 9.Gascon E, Vutskits L, Kiss JZ. Polysialic acid-neural cell adhesion molecule in brain plasticity: From synapses to integration of new neurons. Brain Res Rev. 2007;56:101–118. doi: 10.1016/j.brainresrev.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Seki T, Arai Y. Distribution and possible roles of the highly polysialylated neural cell adhesion molecule (NCAM-H) in the developing and adult central nervous system. Neurosci Res. 1993;17:265–290. doi: 10.1016/0168-0102(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 11.Angata K, et al. Human STX polysialyltransferase forms the embryonic form of the neural cell adhesion molecule. Tissue-specific expression, neurite outgrowth, and chromosomal localization in comparison with another polysialyltransferase, PST. J Biol Chem. 1997;272:7182–7190. doi: 10.1074/jbc.272.11.7182. [DOI] [PubMed] [Google Scholar]
- 12.Ong E, et al. Developmental regulation of polysialic acid synthesis in mouse directed by two polysialyltransferases, PST and STX. Glycobiology. 1998;8:415–424. doi: 10.1093/glycob/8.4.415. [DOI] [PubMed] [Google Scholar]
- 13.Galuska SP, et al. Polysialic acid profiles of mice expressing variant allelic combinations of the polysialyltransferases ST8SiaII and ST8SiaIV. J Biol Chem. 2006;281:31605–31615. doi: 10.1074/jbc.M606516200. [DOI] [PubMed] [Google Scholar]
- 14.Weinhold B, et al. Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J Biol Chem. 2005;280:42971–42977. doi: 10.1074/jbc.M511097200. [DOI] [PubMed] [Google Scholar]
- 15.Cremer H, et al. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 16.Hildebrandt H, Mühlenhoff M, Gerardy-Schahn R. Polysialylation of NCAM. Adv Exp Med Biol. 2010;663:95–109. doi: 10.1007/978-1-4419-1170-4_6. [DOI] [PubMed] [Google Scholar]
- 17.Angata K, et al. Polysialic acid-directed migration and differentiation of neural precursors are essential for mouse brain development. Mol Cell Biol. 2007;27:6659–6668. doi: 10.1128/MCB.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildebrandt H, et al. Imbalance of neural cell adhesion molecule and polysialyltransferase alleles causes defective brain connectivity. Brain. 2009;132:2831–2838. doi: 10.1093/brain/awp117. [DOI] [PubMed] [Google Scholar]
- 19.Yabe U, Sato C, Matsuda T, Kitajima K. Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J Biol Chem. 2003;278:13875–13880. doi: 10.1074/jbc.M300458200. [DOI] [PubMed] [Google Scholar]
- 20.Zuber C, Lackie PM, Catterall WA, Roth J. Polysialic acid is associated with sodium channels and the neural cell adhesion molecule N-CAM in adult rat brain. J Biol Chem. 1992;267:9965–9971. [PubMed] [Google Scholar]
- 21.Curreli S, Arany Z, Gerardy-Schahn R, Mann D, Stamatos NM. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem. 2007;282:30346–30356. doi: 10.1074/jbc.M702965200. [DOI] [PubMed] [Google Scholar]
- 22.Biederer T, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 23.Shingai T, et al. Implications of nectin-like molecule-2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J Biol Chem. 2003;278:35421–35427. doi: 10.1074/jbc.M305387200. [DOI] [PubMed] [Google Scholar]
- 24.Kuramochi M, et al. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 25.Wakayama T, Ohashi K, Mizuno K, Iseki S. Cloning and characterization of a novel mouse immunoglobulin superfamily gene expressed in early spermatogenic cells. Mol Reprod Dev. 2001;60:158–164. doi: 10.1002/mrd.1072. [DOI] [PubMed] [Google Scholar]
- 26.Gomyo H, et al. A 2-Mb sequence-ready contig map and a novel immunoglobulin superfamily gene IGSF4 in the LOH region of chromosome 11q23.2. Genomics. 1999;62:139–146. doi: 10.1006/geno.1999.6001. [DOI] [PubMed] [Google Scholar]
- 27.Urase K, Soyama A, Fujita E, Momoi T. Expression of RA175 mRNA, a new member of the immunoglobulin superfamily, in developing mouse brain. Neuroreport. 2001;12:3217–3221. doi: 10.1097/00001756-200110290-00015. [DOI] [PubMed] [Google Scholar]
- 28.Wakayama T, et al. Heterophilic binding of the adhesion molecules poliovirus receptor and immunoglobulin superfamily 4A in the interaction between mouse spermatogenic and Sertoli cells. Biol Reprod. 2007;76:1081–1090. doi: 10.1095/biolreprod.106.058974. [DOI] [PubMed] [Google Scholar]
- 29.Masuda M, et al. The tumor suppressor protein TSLC1 is involved in cell-cell adhesion. J Biol Chem. 2002;277:31014–31019. doi: 10.1074/jbc.M203620200. [DOI] [PubMed] [Google Scholar]
- 30.Furuno T, et al. The spermatogenic Ig superfamily/synaptic cell adhesion molecule mast-cell adhesion molecule promotes interaction with nerves. J Immunol. 2005;174:6934–6942. doi: 10.4049/jimmunol.174.11.6934. [DOI] [PubMed] [Google Scholar]
- 31.Sara Y, et al. Selective capability of SynCAM and neuroligin for functional synapse assembly. J Neurosci. 2005;25:260–270. doi: 10.1523/JNEUROSCI.3165-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue S, Inoue Y. A challenge to the ultrasensitive chemical method for the analysis of oligo- and polysialic acids at a nanogram level of colominic acid and a milligram level of brain tissues. Biochimie. 2001;83:605–613. doi: 10.1016/s0300-9084(01)01307-4. [DOI] [PubMed] [Google Scholar]
- 33.Galuska SP, Geyer R, Gerardy-Schahn R, Mühlenhoff M, Geyer H. Enzyme-dependent variations in the polysialylation of the neural cell adhesion molecule (NCAM) in vivo. J Biol Chem. 2008;283:17–28. doi: 10.1074/jbc.M707024200. [DOI] [PubMed] [Google Scholar]
- 34.Galuska SP, Geyer R, Mühlenhoff M, Geyer H. Characterization of oligo- and polysialic acids by MALDI-TOF-MS. Anal Chem. 2007;79:7161–7169. doi: 10.1021/ac0712446. [DOI] [PubMed] [Google Scholar]
- 35.Biederer T. Bioinformatic characterization of the SynCAM family of immunoglobulin-like domain-containing adhesion molecules. Genomics. 2006;87:139–150. doi: 10.1016/j.ygeno.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Fogel AI, et al. SynCAMs organize synapses through heterophilic adhesion. J Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckhardt M, Mühlenhoff M, Bethe A, Gerardy-Schahn R. Expression cloning of the Golgi CMP-sialic acid transporter. Proc Natl Acad Sci USA. 1996;93:7572–7576. doi: 10.1073/pnas.93.15.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligon KL, et al. Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci USA. 2006;103:7853–7858. doi: 10.1073/pnas.0511001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 40.Butt AM, Hamilton N, Hubbard P, Pugh M, Ibrahim M. Synantocytes: The fifth element. J Anat. 2005;207:695–706. doi: 10.1111/j.1469-7580.2005.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trotter J, Karram K, Nishiyama A. NG2 cells: Properties, progeny and origin. Brain Res Rev. 2010;63:72–82. doi: 10.1016/j.brainresrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaithianathan T, et al. Neural cell adhesion molecule-associated polysialic acid potentiates alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor currents. J Biol Chem. 2004;279:47975–47984. doi: 10.1074/jbc.M407138200. [DOI] [PubMed] [Google Scholar]
- 43.Bergles DE, Roberts JD, Somogyi P, Jahr CE. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature. 2000;405:187–191. doi: 10.1038/35012083. [DOI] [PubMed] [Google Scholar]
- 44.Ziskin JL, Nishiyama A, Rubio M, Fukaya M, Bergles DE. Vesicular release of glutamate from unmyelinated axons in white matter. Nat Neurosci. 2007;10:321–330. doi: 10.1038/nn1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oltmann-Norden I, et al. Impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid synthesis during postnatal mouse brain development. J Biol Chem. 2008;283:1463–1471. doi: 10.1074/jbc.M708463200. [DOI] [PubMed] [Google Scholar]
- 46.Mühlenhoff M, Eckhardt M, Bethe A, Frosch M, Gerardy-Schahn R. Autocatalytic polysialylation of polysialyltransferase-1. EMBO J. 1996;15:6943–6950. [PMC free article] [PubMed] [Google Scholar]
- 47.Schiff M, Weinhold B, Grothe C, Hildebrandt H. NCAM and polysialyltransferase profiles match dopaminergic marker gene expression but polysialic acid is dispensable for development of the midbrain dopamine system. J Neurochem. 2009;110:1661–1673. doi: 10.1111/j.1471-4159.2009.06267.x. [DOI] [PubMed] [Google Scholar]
- 48.Medzihradszky KF. Peptide sequence analysis. Methods Enzymol. 2005;402:209–244. doi: 10.1016/S0076-6879(05)02007-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.