Abstract
The emergence of the pandemic 2009 H1N1 influenza virus has become a world-wide health concern. As drug resistance appears, a new generation of therapeutic strategies will be required. Here, we introduce a nanotechnology approach for the therapy of pan-demic and seasonal influenza virus infections. This approach uses gold nanorods (GNRs) to deliver an innate immune activator, pro-ducing a localized therapeutic response. We demonstrated the utility of a biocompatible gold nanorod, GNR-5′PPP-ssRNA nanoplex, as an antiviral strategy against type A influenza virus. In human respiratory bronchial epithelial cells, this nanoplex activated the retinoic acid-inducible gene I (RIG-I) pathogen recognition pathway, resulting in increased expression of IFN-β and other IFN-stimulated genes (ISGs) (e.g., PKR, MDA5, IRF1, IRF7, and MX1). This increase in type I IFN and ISGs resulted in a decrease in the replication of H1N1 influenza viruses. These findings suggest that further evaluation of biocompatible nanoplexes as unique antivirals for treatment of seasonal and pandemic influenza viruses is warranted.
Keywords: antivirals, surface plasmon resonance, innate immunity, interferon, retinoic acid-inducible gene I
A novel influenza A/H1N1 virus containing genome segments derived from avian, human, and porcine influenza viruses was first isolated in April 2009 and quickly spread globally, prompting the World Health Organization to declare a pandemic (1, 2). As of October 24, 2009, WHO reported at least 414,000 confirmed cases and nearly 5,000 deaths globally, although the actual number of total cases is likely to be manyfold higher, because current surveillance is focused only on severe and fatal cases (3). The United States government has declared the H1N1 pandemic a national emergency (4) with significant impact on public healthcare. Although vaccination programs form the backbone of public health intervention strategies, lengthy egg-derived H1N1 vaccine production timelines, suboptimal growth of vaccine strain viruses, and limited current manufacturing capacities delayed the availability of pandemic influenza vaccine (5, 6). Antiviral drugs are another public health tool for prophylactic and therapeutic interventions against influenza (7, 8). There are currently two classes of antiinfluenza virus drugs: the M2 ion channel blockers (amantadine, rimantadine) and the neuraminidase inhibitors (oseltamivir, zanamavir) (9). However, the emergence of influenza viral strains resistant to both of these classes of antiviral drugs is becoming increasingly common, highlighting the importance of devising new preventive and therapeutic strategies (10–13), particularly those that can be delivered effectively to severely ill patients together with appropriate clinical management and the use of lung protective strategies. One recent pharmacological approach has been the development of small molecules to augment the host innate immune response (14).
The innate immune system has evolved to recognize viral pathogens via the pathogen recognition receptors (PRRs) (15). Recognition of pathogen-associated molecular patterns (PAMPs) by PRRs results in rapid induction of antiviral cytokines, such as IFN-β, as well as cytokines responsible for the activation of adaptive immunity. Influenza viral RNA is detected by the cytosolic RNA sensor retinoic acid-inducible gene I (RIG-I) (16, 17). Following binding to RNA [double stranded (ds) or 5′PPP-single stranded (ss)], RIG-I undergoes a conformational change, allowing it to interact with IFN-β promoter stimulator 1 (IPS-1) (18–24). The interaction of IPS-1 and RIG-I leads to the induction of type I IFN genes and innate immune response cytokines (25). Hence, activation of RIG-I by its 5′PPP-ssRNA ligand is an attractive alternative to existing prophylactic treatments (9). Also, because innate immunity is evolutionarily conserved and crucial for host survival independent of viral strain, viral resistance to this therapeutic ap-proach is less likely to develop.
The major problem with using 5′PPP-ssRNA to activate RIG-I is the difficulty in delivering this ligand (26). In recent years, gold nanoparticles (GNPs) and nanorods (GNRs) have gained increasing interest as potential biocompatible and site-specific carriers of various diagnostic and therapeutic agents (27–31). Recently, we used GNR to deliver siRNA to silence genes that are associated with opiate drug addiction (32, 33). GNR surfaces can be easily modified to incorporate cationic charges, which facilitate their stable electrostatic interaction with anionic genetic materials, making them suitable delivery vehicles (34).
In this paper, we show GNR-mediated delivery of ssRNA as a unique therapeutic paradigm for treatment of seasonal and pandemic flu. Our hypothesis is that GNR-enhanced delivery of bioactive 5′PPP-ssRNA RIG-I ligand will result in up-regulation of type I IFN through stimulation of RIG-I. Increased type I IFN production will reduce concomitant viral replication. Our results demonstrate the successful internalization of GNR-5′PPP-ssRNA nanoplexes, up-regulation of antiviral responses, and reduction of replication of both a seasonal influenza A virus (A/Solomon Islands/03/06) and a 2009 H1N1 pandemic virus (A/California/08/09). These findings suggest that a nanotechnology-based approach to stimulate antiviral responses of the host innate immune system warrants further investigation.
Results
Electrostatic Binding of 5′PPP-ssRNA with GNRs to Form Biocompatible Nanoplexes.
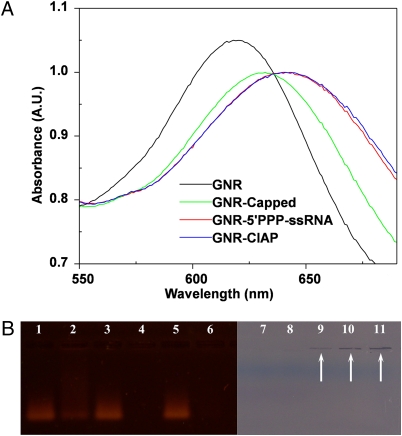
To determine successful complex formation of our gold nanorods to various nucleic acid constructs we used three different methods: surface plasmon resonance shifts, changes in ζ potential, and gel electrophoresis studies. Production of nanoplexes was accomplished by mixing the cationic GNR substrate with the anionic nucleic acid ligands. Determination of successful complex formation is dependent on two factors. First, efficient complex formation of the GNRs with RNA results in changes in the local refractive indexes around the GNRs, resulting in a red shift in the localized longitudinal surface plasmon resonance peak (Fig. 1A). 5′PPP containing ssRNA activates RIG-I-mediated antiviral response; however, synthetic RNAs that lack 5′PPP groups fail to activate the RIG-I pathway. Hence, in our studies we used in vitro transcribed ssRNA that contains 5′PPP moiety and, as negative controls, we removed the 5′PPP group by treating ssRNA with calf intestinal alkaline phosphatase (CIAP) or capped the 5′PPP group during synthesis so that the 5′PPP group was no longer available for RIG-I interaction. We observed a 14-nm shift between GNR alone and GNR upon complex formation with Capped-ssRNA (GNR-Capped). However, with bound 5′PPP-ssRNA (GNR-5′PPP) and CIAP-ssRNA (GNR-CIAP) we observed a 23-nm and a 25-nm shift, respectively. Thus, surface plasmon resonance becomes an important nanotechnological tool to determine if binding had indeed occurred between GNRs and RNAs.
Fig. 1.
Measurement of GNR-ssRNA complex formation and binding efficiency. (A) The localized longitudinal surface plasmon resonance peak of GNRs red shift upon complex formation with RNAs. (B) The nanoplexes that migrate on lanes 2, 4, 6, 9, 10, and 11 were prepared after premixing 900 ng of 5'PPP-ssRNA with 250 ng of GNRs (lanes 2 and 9), 500 ng of GNRs (lanes 4 and 10), and 750 ng of GNRs (lanes 6 and 11). The lanes 1, 3, 5, and 7 were loaded with 900 ng of 5'PPP-ssRNA, and lane 8 was loaded with 750 ng of GNRs. Lanes 1–6 were visualized by Et Br staining under UV light, and lanes 7–11 were visualized with white light.
Second, binding of RNA on the GNR surface reduces the overall net charge of the nanoplex. We observed that the ζ potential of free GNRs is +20.71 mV, and upon successful complex formation to 5′PPP-ssRNA, CIAP-ssRNA, and Capped-ssRNA, it decreased to −9.91, −9.61, and −8.23 mV, respectively (Table S1). These results suggest that binding of cationic GNRs to anionic nucleic acid material leads to a slightly negatively charged nanoplex and that complexing of genetic material to GNRs would increase uptake of the nanoplexes into the target cell due to evasion of the reticuloendothelial system and reduction in nonspecific interactions with proteins and other biomolecules as demonstrated by other studies (30, 35).
To identify the amount of GNR needed to completely bind a given amount of ssRNA, we conducted gel electrophoresis studies. Our results (Fig. 1B) show that addition of increasing amounts of GNR to a constant amount of 5′PPP-ssRNA leads to a decrease in the amount of free 5′PPP-ssRNA, visible by ethidium bromide staining, and increased nanoplex formation (Fig. 1B, lanes 1–6). Lanes 1, 3, and 5 were used as control lanes with free 5′PPP-ssRNA. To verify the presence of the immobile nanoplex in the gel, we visualized the gel under visible light (Fig. 1B, lanes 7–11). Increasing amounts of GNR-5′PPP nanoplex correlated to the increasing GNR added to the sample, as visualized by the purple lines in the wells marked by the arrows. Thus, on the basis of these electrophoresis studies we conclude that each microgram of our GNR preparation can bind ≈1.2 μg ssRNA.
Thus, the combination of plasmonic shift experiments, charge determination, and changes in electrophoresis migration confirmed the successful complex formation between GNRs and our RNA constructs.
GNR Nanoplexes Are Internalized by A549 Cells with Minimal Cytotoxicity.
The longitudinal surface plasmon oscillation of the GNRs gives a strong plasmonic scattering in the orange-red region of the optical spectrum (36). This phenomenon can be used to study the intracellular distribution of nanoplexes by dark-field microscopy. Here we examined the intracellular delivery of GNR conjugated to a fluorescently labeled siRNA (siRNAF) in A459 cells using dark-field imaging. Fig. S1 shows the dark-field and fluorescence images of A459 cells, with and without treatment with the GNR-siRNAF nanoplex. We used commercial siPORT (Ambion) as our positive control transfection agent. The rate of release of ssRNA species from the GNRs either in solution or after transfection into cells could not be determined due to the lack of a sensitive assay to determine the quantity of the ssRNA as it is not fluorescently labeled. Furthermore, free RNA species are degraded by RNases that are abundant in culture media. The intracellular delivery of the nanoplexes can be easily observed from the strong orange-red light scattering, a property of GNRs. Because it is not possible to determine intracellular localization with dark-field microscopy, we used confocal microscopy using Z-scans as well as transmission electron microscopy (TEM), which clearly demonstrate the uptake of GNRs, perhaps through micropinocytosis (37). Thus, another advantage of using nanotechnology in the delivery of therapeutics is that the unique properties of the nanoparticles also can be exploited to monitor their cellular entry and distribution.
We also measured fluorescence from cellular lysates following their treatment with free siRNAF, siRNAF complexed with GNRs, or siRNAF complexed with the commercially available gene-silencing agent siPORT to confirm our dark-field images. Our results indicate that the fluorescence from lysates of cells treated with GNR-siRNAF is ≈10% higher than that from lysates of cells treated with siPORT-siRNAF, indicating that the intracellular delivery efficiency of siRNA using GNRs is as good as that of a commercially available gene silencing agent (Fig. S2).
To specifically determine the uptake and intracellular distribution of our nanoplexes (GNR-5′PPP-ssRNA) in A549 cells we employed TEM. Cells were treated with nanoplexes for 24 h and viewed by TEM. Fig. 2 A–D shows the presence of these nanoplexes in endocytic vesicles. We postulate that the particles may be taken up by classical pinocytotic mechanisms of uptake but further confirmatory studies are required (38, 39).
Fig. 2.
Uptake of GNR-5′PPP-ssRNA by A549 cells. Cellular uptake and internalization of nanoplexes (GNR-5′PPP-ssRNA) in A549 cells has been visualized using transmission electron microscopy. GNR-5′PPP-ssRNA nanoplexes are clearly visible in endocytic compartments within the cell (indicated by arrows in A–D).
To determine the toxicity associated with the uptake of our GNR nanoplexes, we used a quantitative 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay 24, 48, 72, and 96 h posttransfection. The cell death detected after transfection with GNR, GNR-5′PPP, or GNR-Capped nanoplexes at all time points examined ranged from 0 to 0.8%, 7.8 to 8.8%, and 0.8 to 7.7%, respectively (Fig. S3).
Induction of IFN-β and RIG-I Expression by GNR-5′PPP-ssRNA.
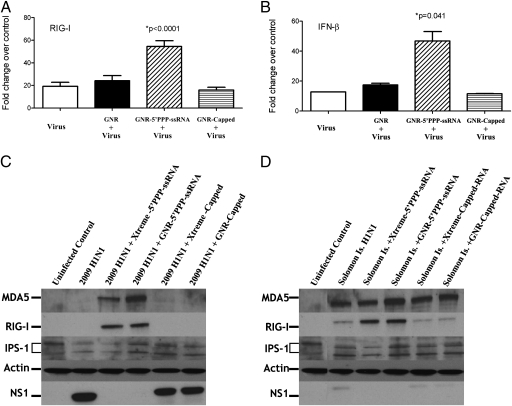
Although the nanoplexes clearly enter the cell (Fig. 2), we wanted to specifically address the ability of the RNA ligand to activate innate immune PRRs. Previous experiments in our laboratory have shown that using cationic lipids to transfect 5′PPP-ssRNA into A549 cells activated RIG-I and induced IFN-β expression (40). To determine whether GNR-based nanoplexes could similarly up-regulate the type I IFN response, we assessed changes in the message levels of RIG-I and IFN-β by quantitative RT-PCR. Transcription of IFN-β increased for at least 72 h following treatment of A549 cells with the GNR-5′PPP-ssRNA nanoplexes, reaching a maximum of ∼40-fold above untreated controls (Fig. 3A). Addition of GNR alone or GNR conjugated to capped-RNA or CIAP-RNA led to only marginal increases in IFN-β message levels. RIG-I expression was also increased by GNR-5′PPP nanoplexes but not by GNR alone, GNR-Capped, or GNR-CIAP nanoplexes (Fig. 3B). The increased RIG-I mRNA was correlated with a corresponding increase in RIG-I protein levels as assessed by Western blot analysis. At 48 or 72 h following transfection, strong bands corresponding to RIG-I can clearly be detected in the A459 cells, but not in the control cells that received GNR alone, GNR-Capped, or GNR-CIAP (Fig. 3C). Besides inducing the expression of RIG-I and IFN-β, GNR-5′PPP-ssRNA complexes also enhanced the levels of IFN-responsive genes, PKR, MDA5, IRF1, IRF7, MX1, CXCL10, ISG12, and others, whereas GNR alone or GNR-Capped had little or no impact on the expression of these genes (Fig. S4).
Fig. 3.
5′PPP-ssRNA-induced expression of RIG-I and IFN-β in A549 cells. A549 cells (3.5 × 105 cells/well) in a six-well tissue culture plate were mock transfected (control) or transfected with 3 μg/mL of RNA complexed with 2.5 μg of GNRs. GNR-5′PPP, GNR-Capped, and GNR-CIAP were used. After 24, 48, and 72 h of treatment, (A) IFN-β or (B) RIG-I expression was analyzed by quantitative RT-PCR. All data were normalized to β-actin, a housekeeping gene, and expressed as fold increases. Data shown represent the mean ± SD of three independent experiments and P values are given for the comparison of GNR-5′PPP with GNR alone. (C) A549 cells were transfected as above and 48 and 72 h posttransfection RIG-I protein expression was analyzed by immunoblot.
Antiviral Bioactivity of GNR-5′PPP-ssRNA.
We next determined whether the level of RIG-I activation achieved by treatment with GNR-5′PPP was sufficient to inhibit the replication of seasonal (i.e., A/Solomon Islands/03/06) or 2009 H1N1 pandemic (i.e., A/California/08/09) influenza virus strains. To do this, A549 cells were first treated with GNR nanoplexes and then infected with the appropriate influenza virus 48 h later. Samples were harvested and analyzed 24 h after viral infection.
Infection with A/California/08/09 virus failed to up-regulate RIG-I and IFN-β message (Fig. 4 A and B) or RIG-I protein (Fig. 4C); however, there was a significant increase in NS1 expression (Fig. 4C) and viral titers (Fig. 5). Nevertheless, pretreatment with GNR-5′PPP nanoplexes, but not with GNR-Capped or GNR-CIAP nanoplexes or GNR alone, increased IFN-β message (Fig. 4B) and protein levels (Fig. S5) and RIG-I message and protein levels (Fig. 4 A and C) over the levels seen with virus only. Furthermore, the treatment also reduced amounts of NS1 below the level of detection (Fig. 4C) and viral titers by ≈90% (Fig. 5). Similarly, pretreatment with GNR-5′PPP subsequently inhibited the induction of NS1 and up-regulated RIG-I expression postinfection with a seasonal influenza virus, A/Solomon Islands/03/06 (Fig. 4D). These findings suggest that nanoplex delivery of innate immune activators is sufficient to effectively impair the replication of both seasonal and pandemic H1N1 influenza viruses.
Fig. 4.
GNR-5′PPP-ssRNA enhances IFN-β, RIG-I, and MDA5 expression and inhibits NS1 expression following infection with 2009 pandemic H1N1 influenza viruses and Solomon Islands seasonal flu strain. A459 cells (3.5 × 105 cells/well) in a six-well tissue culture plate were mock transfected or transfected with 3 μg of RNAs complexed with 2.5 μg of GNRs per well for 48 h and then infected with A/California/08/09 or A/Solomon Islands/03/06 at an MOI of 1. Lysates to determine mRNA levels by qRT-PCR (A and B) and protein to assess the levels of NS1, RIG-I, MDA5, and IPS-1 (C and D) were collected 24 h later. Data shown in A–C are for cultures infected with A/California/08/09 and the data shown in D are for A/Solomon Islands/03/06. Results shown (A and B) are mean ± SD from two independent experiments.
Fig. 5.
GNR-5'PPP-ssRNA inhibits replication of 2009 pandemic H1N1 influenza viruses and Solomon Islands seasonal flu strain. A459 cells (3.5 × 105 cells/well) in a six-well tissue culture plate were mock transfected or transfected with 3 μg of RNAs complexed with 2.5 μg of GNRs per well for 48 h and then infected with A/California/08/09 or A/Solomon Islands/03/06 at an MOI of 1. The viral titers were determined from the supernatants collected 24 h later. Results shown are mean ± SD from two independent experiments and are expressed as viral titer (pfu/mL).
Discussion
The major objective in this research has been to evaluate the use of GNR nanotechnology to deliver 5′PPP-ssRNA, an innate immune activator with antiviral action against influenza virus infections. Gold-based nanoparticles and nanorods have gained increasing interest as a safe delivery system for therapeutic nucleic acids because of their biocompatibility and capacity to form stable nanoplexes. The lung is especially well suited for this unique therapeutic nanoplex delivery strategy as direct contact with the environment provides a portal for inhalation administration, avoiding parenteral injection. In particular, site-specific delivery of type I IFN or IFN inducers can potentially reduce systemic side effects (41, 42), in addition to having a beneficial therapeutic outcome of reducing influenza virus replication.
The recent spread of the 2009 H1N1 pandemic influenza viruses, as well as drug-resistant seasonal viruses, and the potential threat of highly pathogenic avian influenza viruses have intensified the search for new classes of antiviral drugs and therapeutic strategies. The major limitation of ssRNA therapy is the sensitivity of RNA to rapid degradation. Despite some of the initial successes in overcoming this lability, most current nucleic acid delivery systems have limitations based on cellular toxicity (e.g., cationic lipid complexes) or untoward immune responses and toxicity (e.g., virus-based systems) (26). Our findings clearly demonstrate that GNR complex formation enhances 5′PPP-ssRNA delivery to human bronchial epithelial cells and results in a biofunctional outcome with limited effects on cell viability. Complex formation of the nucleic acid to the GNR does not inhibit bioactivity of the 5′PPP-ssRNA as signaling through the RIG-I pathway that triggers the induction of type I IFNs is still active following successful delivery of the nanoplex. The RIG-I-induced type I IFN activation response is conserved among positive-sense single-strand RNA viruses, suggesting that 5′PPP-ssRNA induction of type I IFN can be extended as a treatment modality for these viruses. In addition to inducing the secretion of type I IFNs, 5′PPP-ssRNA also results in the induction of other innate immune cytokines, which may be critical for recruiting and activating leukocytes to the site of infection for viral clearance, initiating a successful adaptive immune response.
In summary, these findings demonstrate a unique therapeutic strategy based on nanotechnology-enhanced RNA delivery to potentially treat influenza, as well as other viral infections, where type I IFNs are part of a critical pathway to the resolution of the infection. Our findings clearly demonstrate the utility of a unique, noncytotoxic, antiviral strategy of employing GNR-5′PPP-ssRNA nanoplexes that can activate intracellular antiviral signaling path-ways in respiratory epithelial cells and can specifically inhibit both an H1N1 and a seasonal strain of influenza virus replication. Because innate immune response pathways are activated, this approach has potential application to prevent and treat diseases caused by other viruses. Furthermore, the ability of the viruses to develop resistance is remote as these pathways are evolutionarily conserved. This study clearly demonstrates the potential feasibility of employing biocompatible nanoparticle constructs of GNR complexed with specifically selected ligands (i.e., 5′PPP-ssRNA) to target cytosolic receptors that can trigger pathogen recognition pathways (i.e., RIG-I/MDA-5) to control and treat infectious disease.
Materials and Methods
Cell Lines.
A549 cells were grown in DMEM (Life Technologies) supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin.
Influenza Viruses.
Seasonal influenza virus, A/Solomon Islands/03/06 and the pandemic influenza virus, A/California/08/09 used in this study were obtained from the Influenza Division, Centers for Disease Control repository. All infections of A549 cells were carried out at a multiplicity of infection (MOI) of 1. Each treatment was carried out in duplicate cultures. After 24 h postinfection with viruses, cell culture supernatants were collected and stored at −80 °C for determination of viral titer by plaque assay as described previously, using Madin–Darby Canine Kidney (MDCK) epithelial cells (43).
Preparation of ssRNA.
The RNAs (5′PPP-ssRNA, Capped-ssRNA, and CIAP-ssRNA) used in this work were synthesized with the MEGAscript T7 High Yield Transcription Kit (Ambion), using a double-stranded DNA template made by annealing complementary oligonucleotides. The template was then digested with DNase I (New England Biolabs) and the RNA purified with TRIzol reagent (Invitrogen). Capped RNAs were made by substituting a 12:1 ratio of m7G(5′)PPP(5′)G cap analog:GTP for GTP in the transcription reaction. CIAP-ssRNA was made by removing the functional 5′PPP end with CIAP treatment. All kits and reagents were used according to the manufacturer's protocols. 5′PPP-ssRNA activates RIG-I and as controls we used the same 5′PPP-ssRNA from which the 5′PPP group is removed enzymatically with CIAP or blocked the 5′PPP group during synthesis by capping.
Supplementary Material
Acknowledgments
We thank our World Health Organization Global Influenza Surveillance Network partners for the viruses used in this study. We thank Mr. Alan Siegel, Department of Biological Sciences, University of Buffalo, for the help with transmission electron microscopy. This study was supported by National Institutes of Health Grant AI084410 (to P.R.K., P.N.P., and S.S.); National Cancer Institute Grant CA119397 (to P.N.P.); and National Institutes of Health Grants CA104492 (to E.J.B.), HL48889 (to P.R.K.), and AG031035 (to K.V.C.). This work was also supported by the Chemistry and Life Sciences Division of the Air Force Office of Scientific Research (P.N.P.) and a grant from the National Vaccine Program Office (to S.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914561107/-/DCSupplemental.
References
- 1.Dawood FS, et al. Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 2.WHO 2009. World now at the start of 2009 influenza pandemic. Available at http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html. Accessed September 8, 2009.
- 3.WHO 2009. Pandemic (H1N1) 2009 - update 71. Available at http://www.who.int/csr/don/2009_10_23/en/index.html. Accessed November 4, 2009.
- 4.Flu.Gov President Obama signs emergency declaration for H1N1 flu. 2009. Available at http://www.flu.gov/professional/federal/h1n1emergency10242009.htmlAccessed November 4, 2009.
- 5.WHO 2009. Pandemic influenza vaccine manufacturing process and timeline. Pandemic (H1N1) 2009 briefing note 7. Available at http://www.who.int/csr/disease/swineflu/notes/h1n1_vaccine_20090806/en/index.html. Accessed November 4, 2009.
- 6.Butler D. 2009. Pandemic flu vaccine yields worse than expected. July 13, 2009. Available at http://blogs.nature.com/news/thegreatbeyond/2009/07/pandemic_vaccine_yields_worse.html. Accessed November 4, 2009.
- 7.Hoelscher M, Gangappa S, Zhong W, Jayashankar L, Sambhara S. Vaccines against epidemic and pandemic influenza. Expert Opin Drug Deliv. 2008;5:1139–1157. doi: 10.1517/17425247.5.10.1139. [DOI] [PubMed] [Google Scholar]
- 8.WHO 2009. Recommended use of antivirals. Available at http://www.who.int/csr/disease/swineflu/notes/h1n1_use_antivirals_20090820/en/index.html. Accessed September 8, 2009.
- 9.Hayden FG, Pavia AT. Antivirals for influenza: Historical perspectives and lessons learned. Antiviral Res. 2006;71:372–378. doi: 10.1016/j.antiviral.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 10.ECDC Resistance to oseltamivir (Tamiflu) found in some European influenza virus samples. 2008 Available at http://www.ecdc.europa.eu/en/activities/sciadvice/Lists/ECDC%20Reviews/ECDC_DispForm.aspx?List=512ff74f%2D77d4%2D4ad8%2Db6d6%2Dbf0f23083f30&ID=206. Accessed July 14, 2009. [Google Scholar]
- 11.WHO 2009. Pandemic (H1N1) 2009 briefing note 1. Viruses resistant to oseltamivir (Tamiflu) identified. Available at http://www.who.int/csr/disease/swineflu/notes/h1n1_antiviral_resistance_20090708/en/index.html. Accessed September 8, 2009.
- 12.de Jong MD, et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med. 2005;353:2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- 13.Uyeki TM. Human infection with highly pathogenic avian influenza A (H5N1) virus: Review of clinical issues. Clin Infect Dis. 2009;49:279–290. doi: 10.1086/600035. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes J. Discovery of immunopotentiatory drugs: Current and future strategies. Clin Exp Immunol. 2002;130:363–369. doi: 10.1046/j.1365-2249.2002.02016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambhara S, Lehrer RI. The innate immune system: A repository for future drugs? Expert Rev Anti Infect Ther. 2007;5:1–5. doi: 10.1586/14787210.5.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Yoneyama M, Fujita T. RIG-I family RNA helicases: Cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Ranjan P, et al. Cytoplasmic nucleic acid sensors in antiviral immunity. Trends Mol Med. 2009;15:359–368. doi: 10.1016/j.molmed.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 20.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 21.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 22.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 25.Fujita T, Onoguchi K, Onomoto K, Hirai R, Yoneyama M. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie. 2007;89:754–760. doi: 10.1016/j.biochi.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today. 2006;11:67–73. doi: 10.1016/S1359-6446(05)03668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor EE, Mwamuka J, Gole A, Murphy CJ, Wyatt MD. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh P, Han G, De M, Kim CK, Rotello VM. Gold nanoparticles in delivery applications. Adv Drug Deliv Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Gold nanoparticles: Interesting optical properties and recent applications in cancer diagnostics and therapy. Nanomedicine. 2007;2:681–693. doi: 10.2217/17435889.2.5.681. [DOI] [PubMed] [Google Scholar]
- 30.Sonavane G, et al. In vitro permeation of gold nanoparticles through rat skin and rat intestine: Effect of particle size. Colloids Surf B Biointerfaces. 2008;65:1–10. doi: 10.1016/j.colsurfb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Stone JW, Sisco PN, Goldsmith EC, Baxter SC, Murphy CJ. Using gold nanorods to probe cell-induced collagen deformation. Nano Lett. 2007;7:116–119. doi: 10.1021/nl062248d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonoiu A, et al. MMP-9 gene silencing by a quantum dot-siRNA nanoplex delivery to maintain the integrity of the blood brain barrier. Brain Res. 2009;1282:142–155. doi: 10.1016/j.brainres.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonoiu AC, et al. Nanotechnology approach for drug addiction therapy: Gene silencing using delivery of gold nanorod-siRNA nanoplex in dopaminergic neurons. Proc Natl Acad Sci USA. 2009;106:5546–5550. doi: 10.1073/pnas.0901715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding H, et al. Gold nanorods coated with multilayer polyelectrolyte as contrast agents for multimodal imaging. J Phys Chem C. 2007;34:12552–12557. [Google Scholar]
- 35.Davis SS. Biomedical applications of nanotechnology—implications for drug targeting and gene therapy. Trends Biotechnol. 1997;15:217–224. doi: 10.1016/S0167-7799(97)01036-6. [DOI] [PubMed] [Google Scholar]
- 36.Sokolov K, et al. Real-time vital optical imaging of precancer using anti-epidermal growth factor receptor antibodies conjugated to gold nanoparticles. Cancer Res. 2003;63:1999–2004. [PubMed] [Google Scholar]
- 37.Sperling RA, Casals E, Comenge J, Bastús NG, Puntes VF. Inorganic engineered nanoparticles and their impact on the immune response. Curr Drug Metab. 2009;10:895–904. doi: 10.2174/138920009790274577. [DOI] [PubMed] [Google Scholar]
- 38.Huff TB, Hansen MN, Zhao Y, Cheng JX, Wei A. Controlling the cellular uptake of gold nanorods. Langmuir. 2007;23:1596–1599. doi: 10.1021/la062642r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Souza GG, Weissig V. Subcellular targeting: A new frontier for drug-loaded pharmaceutical nanocarriers and the concept of the magic bullet. Expert Opin Drug Deliv. 2009;6:1135–1148. doi: 10.1517/17425240903236101. [DOI] [PubMed] [Google Scholar]
- 40.Spiropoulou CF, et al. RIG-I activation inhibits ebolavirus replication. Virology. 2009;392:11–15. doi: 10.1016/j.virol.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 41.Gutterman JU. Cytokine therapeutics: Lessons from interferon alpha. Proc Natl Acad Sci USA. 1994;91:1198–1205. doi: 10.1073/pnas.91.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamidi M, Zarrin A, Foroozesh M. Novel delivery systems for interferons. Crit Rev Biotechnol. 2007;27:111–127. doi: 10.1080/07388550701503410. [DOI] [PubMed] [Google Scholar]
- 43.Guo Z, et al. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol. 2007;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.