Abstract
We used a model system of purified components to explore the effects of a downstream target on the signaling properties of a covalent modification cycle, an example of retroactivity. In the experimental system used, a bifunctional enzyme catalyzed the modification and demodification of its substrate protein, with both activities regulated by a small molecule stimulus. Here we examined how a downstream target for one or both forms of the substrate of the covalent modification cycle affected the steady-state output of the system, the sensitivity of the response to the stimulus, and the concentration of the stimulus required to provide the half-maximal response (S50). When both the modified and unmodified forms of the substrate protein were sequestered by the downstream target, the sensitivity of the response was dramatically decreased, but the S50 was only modestly affected. Conversely, when the downstream target only sequestered the unmodified form of the substrate protein, significant effects were observed on both system sensitivity and S50. Behaviors of the experimental systems were well approximated both by simple models allowing analytical solutions and by a detailed model based on the known interactions and enzymatic activities. Modeling and experimentation indicated that retroactivity may result in subsensitive responses, even if the covalent modification cycle displays significant ultrasensitivity in the absence of retroactivity. Thus, we provide examples of how a downstream target can alter the signaling properties of an upstream signal transduction covalent modification cycle.
Keywords: regulatory networks, retroactivity, sensitivity, signal transduction
Numerous cellular signal transduction systems consist of covalent modification cycles, in which signaling proteins are regulated by their reversible modification and demodification. In some cases, multiple covalent modification cycles are linked to form signaling cascades (Fig. 1A). Many cascade systems have a branched circuit, with different “downstream” targets under the control of a protein that is part of a covalent modification cycle (e.g., Fig. 1B). Signaling may also involve amplification of the concentration of a downstream target, so that the relative abundance of downstream targets changes upon signaling. To understand the function of these signal transduction systems based on cycles of reversible covalent modification, it will be important to learn how the downstream targets affect the functions of the upstream systems that pass signals to them. Signaling is typically considered to flow from the upstream stimuli that control the cycle to the downstream targets; that is, the layers of a signaling cascade are commonly considered to behave as independent modules. But recent modeling studies, and experiments with intact cells and embryos, suggest that sequestration of the substrate protein of a signaling cascade by downstream components may significantly alter the signaling properties of a covalent modification cycle (1–9), a form of “reverse signaling” known as retroactivity. In electrical engineering and hydrolic applications, retroactivity is a well known and quantifiable parameter, and insulation devices have been developed to mitigate retroactivity and by so doing allow modular behavior. Biological signaling systems should also be affected by retroactivity unless they contain insulation mechanisms to prevent it (2, Fig. 1A). Here, we use a model experimental system of highly purified components to investigate retroactivity and its underlying biochemical mechanisms in a reconstituted covalent modification cycle.
Fig. 1.
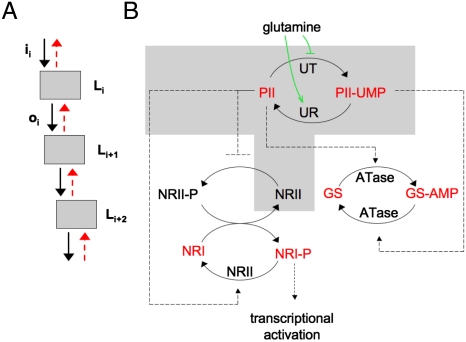
Retroactive effects in signaling systems. (A) Scheme representing a signaling cascade, each signaling level Li receives an input signal ii and provides an output signal oi that works as the input for the next level in the cascade. The interconnection between levels or modules gives rise to retroactive signals as indicated by the dashed red arrows. (B) Experimental system used in this paper, derived from the nitrogen assimilation control system in Escherichia coli. The UTase/UR-PII monocycle is depicted at the top; the UT and UR catalytic activities bring about the interconversion of PII and PII-UMP. Both PII and PII-UMP play a role in the regulation of ATase, which catalyzes the reversible adenylylation of GS and deadenylylation of GS-AMP. PII also controls the activities of NRII, which brings about the phosphorylation of NRI and dephosphorylation of NRI-P. We focused on the components inside the shaded box to examine the effect of the downstream target NRII on the signaling of the glutamine state by the UTase/UR-PII monocycle.
Our experimental system is derived from the nitrogen assimilation control system of Escherichia coli (reviewed in ref. 10). The initiation of transcription of nitrogen-regulated (Ntr) genes in E. coli is controlled by a cascade system with two linked cycles of covalent modification (Fig. 1B). In one cycle, the signal-transducing Uridylyltransferase/Uridylyl-removing enzyme (UTase/UR) brings about the uridylylation and deuridylylation of the PII protein. This interconversion is regulated by signals of nitrogen status (glutamine, α-ketoglutarate) as well as by signals of energy state (adenylylate energy charge) (11). In the second cycle, the two-component systems “transmitter” protein, NRII (NtrB) brings about the phosphorylation and dephosphorylation of the “receiver” protein NRI (NtrC) (Fig. 1B), which, when phosphorylated, is an enhancer-binding transcription factor that activates Ntr genes. The linkage between the two cycles is provided by the unmodified form of PII, which converts NRII from a form that brings about the phosphorylation of NRI to a form that brings about the dephosphorylation of NRI ∼ P (Fig. 1B).
In the experiments reported here, we used conditions where the system produces a continuously variable (graded) and reversible output in response to glutamine to focus our study on the effects of a downstream target (NRII) on glutamine signaling by the UTase/UR-PII cycle. In addition to controlling the activities of NRII, the PII protein controls additional receptors, such as the adenylyltransferase/adenylyl-removing enzyme (ATase) that regulates glutamine synthetase (GS) (Fig. 1B).
We examined two different versions of the experimental system, in which the downstream target, NRII, indirectly affected either one or both of the antagonistic activities of the UTase/UR enzyme. We found that at reasonable protein concentrations, NRII provided large effects on system sensitivity (apparent Hill coefficient) and/or S50 (concentration of glutamine required to achieve 50% of the maximal response). When both activities of the cycle were inhibited by NRII, the sensitivity of the system was dramatically reduced by NRII, but the S50 of the system was only modestly affected. Conversely, when only a single activity of the cycle was affected by NRII, significant effects on both sensitivity and S50 were obtained.
Results
Retroactivity Exerted by NRII Transformed an Ultrasensitive Response to Glutamine into a Subsensitive One.
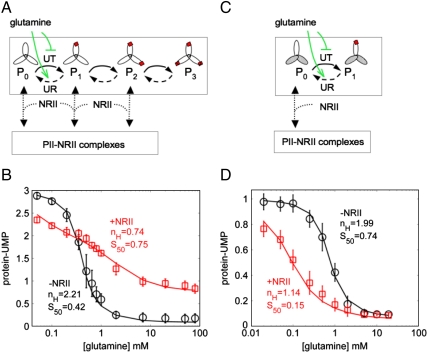
The PII covalent modification cycle and its regulation by glutamine are schematically depicted in Fig. 2A. The trimeric PII can be modified on each of its three subunits, thus its uridylylation state can vary between 0 and 3. We use a subindex in P to indicate the number of modified subunits, with P0 and P3 corresponding to the fully unmodified and fully modified forms, respectively. Glutamine controls the bifuntional UTase/UR enzyme, inhibiting PII modification and activating the hydrolysis of PII-UMP. NRII does not bind to modified PII subunits. Thus, it is unable to bind fully modified PII trimers (P3), whereas it can bind fully unmodified or partially modified PII, as indicated in the scheme in Fig. 2A. We provide biochemical evidence that NRII binds to partially modified PII trimers (P1,P2) in the SI Appendix, justifying the scheme in Fig. 2A.
Fig. 2.
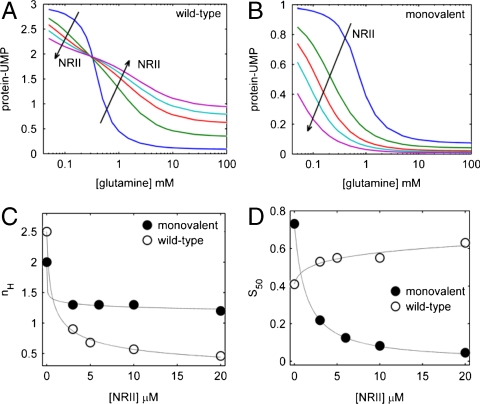
A downstream target affects the signaling properties of an upstream signal transduction covalent modification cycle. (A) Scheme representing the modification state of the trimeric PII protein, regulated by glutamine. The UT activity of the enzyme UTase/UR is indicated with solid arrows, whereas its UR activity is indicated with dashed arrows. UMP groups are represented by red squares over PII subunits. Note that glutamine inhibits all of the uridylylation steps and activates all of the deuridylylation steps; the action of glutamine was shown only on one of the uridylylation and deuridylylation steps for clarity. (B) Stimulus-response curves for the system in (A), where the stimulation is given by the level of glutamine and the response is the steady-state level of uridylylation of PII, which can vary between 0 and 3 uridylyl groups per PII trimer. This level is computed as the concentration of protein bound to UMP groups, normalized by the total concentration of PII protein in the experiment. Experimental conditions were described in Materials and Methods, with 3 μM PII, 0.8 μM UTase/UR, and NRII at 0 (black circles) or 10 μM (red squares), respectively. The values obtained when glutamine was absent were 2.97 ± 0.07 for -NRII and 2.71 ± 0.07 for +NRII. Error bars come from the determination of the steady state from several experiments. The lines superimposed over the data are the best-fit output of the detailed kinetic model described in the main text. The Hill coefficient (nH) and response point (S50) for each curve is shown. (C) Simplified system consisting of heterotrimeric, monovalent forms of PII. Non functional subunits are the shadowed ones. (D) Stimulus-response curves for the system in (C). Experimental conditions were described in Materials and Methods, with the PII heterotrimer mixtures containing 6 μM wild-type subunits (i.e., equivalent to 2 μM wild-type PII trimers) and 36 μM mutant Δ47-53 PII subunits (equivalent to 12 μM mutant PII homotrimers), 0.6 μM UTase/UR, and NRII at 0 (black circles) or 10 μM (red squares), respectively. The values obtained when glutamine was absent were 0.99 ± 0.10 for NRII and 0.84 ± 0.10 for +NRII. The lines superimposed over the data are the best-fit output of the detailed kinetic model for heterotrimeric PII as described in the main text.
The steady-state response to glutamine of the reconstituted UTase/UR-PII monocycle in the presence or absence of NRII is shown in Fig. 2B. NRII had a dramatic effect on signaling by the UTase/UR-PII cycle: it decreased the modification state of PII at low-glutamine concentrations, and increased the modification state of PII at high-glutamine concentrations (Fig. 2B). The sensitivity of the responses was determined by fitting the results to the Hill equation (SI Appendix), and also by determining the range of stimulatory effector concentrations required to move the system from 10 to 90% of the maximal response. Both methods gave essentially the same results; the system in the absence of a downstream target displayed a sensitivity corresponding to nH = 2.21 ± 0.07, whereas in the presence of 10 μm NRII the system was subsensitive (nH = 0.74 ± 0.01). (For calculation of errors, see SI Appendix). Conversely, the presence of NRII had only a minor effect on the glutamine S50, which was shifted from 0.42 ± 0.05 mM to 0.75 ± 0.01 mM glutamine.
A Simplified System to Study Retroactivity Exerted by NRII.
The wild-type UTase/UR-PII cycle is complicated by the trimeric nature of the PII protein and the consequent large number of species able to interact with NRII (Fig. 3A and SI Appendix). Thus, we also studied an alternative system in which PII was functionally monovalent and thus behaved as if it were a monomer. Heterotrimeric PII was assembled in vitro from wild-type subunits and mutant subunits unable to bind receptors, and the preparation consists of a mixture of trimeric species that includes small percentages of “divalent” and wild-type PII (ref. 12 and SI Appendix).
Fig. 3.
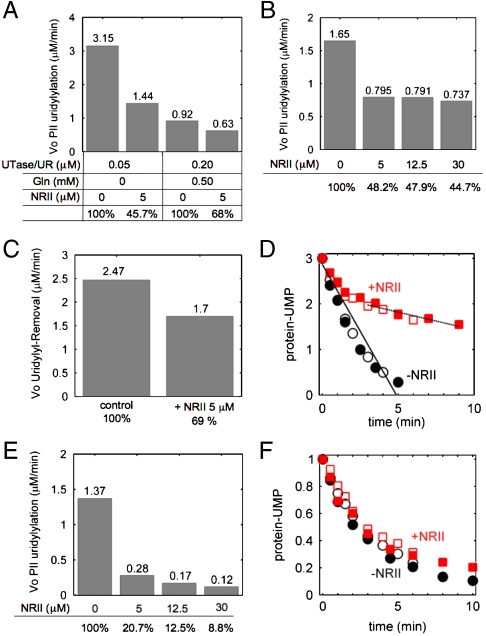
Enzymological basis for the regulatory properties. (A)–(D) show results with wild-type PII, and (E) and (F) show results with heterotrimeric PII. (A) Inhibition of the initial rate (Vo) of the UT reaction by NRII in the presence and absence of glutamine. Conditions were described in Materials and Methods, with 3 μM PII, 0.2 mM α-ketoglutarate, and enzyme, glutamine, and NRII as indicated. (B) NRII provided partial inhibition of the UT activity. Conditions were described in Materials and Methods, with 2 μM PII, 0.02 μM UTase/UR, 0.3 mM α-ketoglutarate, and NRII as indicated. (C) Inhibition of the UR activity of UTase/UR by NRII. Conditions were described in Materials and Methods, with 13.5 μM PII-UMP, 0.4 μM UTase/UR, 1 mM α-ketoglutarate, 10 mM glutamine, and NRII as indicated. (D) Burst kinetics analysis of the deuridylylation of wild-type, trimeric, PII-UMP. Conditions were described in Materials and Methods, with 8 μM PII-UMP, 3 μM UTase/UR, 1 mM α-ketoglutarate, 20 mM glutamine, and 0 or 10 μM NRII, respectively. Two independent experiments performed at 20 °C are indicated with open and filled symbols. The lines over the symbols are linear fits of each of the two kinetics phases. (E) Inhibition of the uridylylation of monovalent PII by NRII. Conditions were described in Materials and Methods, with monovalent PII formed from 2 μM wild-type and 12 μM PII-Δ47-53, 0.02 μM UTase/UR, 0.3 mM α-ketoglutarate, and NRII as indicated. (F) Burst kinetics analysis of the deuridylylation of monovalent PII-UMP. Conditions were described in Materials and Methods, with 1.18 μM monovalent PII-UMP, 0.47 μM UTase/UR, 1 mM α-ketoglutarate, 20 mM glutamine, and 0 or 10 μM NRII, respectively. Two independent experiments performed at 20 °C are indicated with open and filled symbols.
The scheme in Fig. 2C illustrates the simplified situation when monovalent PII was used. In this case, only the wild-type subunit is able to become uridylylated and deuridylylated and thus the modification state varies between 0 and 1. Because NRII does not bind to modified PII subunits, in this system it is only able to bind P0. The glutamine sensitivity of the reconstituted UTase/UR-PII cycle containing heterotrimeric PII, in the presence and absence of NRII, is shown in Fig. 2D. In this case, the effect of NRII was to decrease the modification state of PII at all glutamine concentrations. Unlike the results obtained with wild-type PII homotrimers, the response curves obtained in the presence and absence of NRII did not intersect. Sensitivities were determined as noted above; in the absence of NRII the system was ultrasensitive (nH = 1.99 ± 0.18), whereas a nearly hyperbolic response was obtained in the presence of NRII (nH = 1.14 ± 0.05) (Fig. 2D). In this system, the presence of NRII had a significant effect on the glutamine S50, which was shifted from 0.74 ± 0.15 mM to 0.15 ± 0.04 mM glutamine.
Note that the glutamine sensitivity of the systems with wild-type PII and monovalent PII was quite similar (nH = 2.21 and nH = 1.99, respectively). Thus, the trimeric structure of PII did not play a significant role in the glutamine sensitivity of the UTase/UR-PII cycle.
Enzymological Basis for the Regulatory Properties.
To understand the effects of NRII in the reconstituted systems, we investigated the effect of NRII on the individual uridylyltransferase (UT) and uridylyl-removing (UR) reactions, using well-established assay methods (13). For the system with wild-type PII, NRII inhibited the UT reaction when its concentration was in excess of the PII concentration, both in the presence and absence of glutamine (Fig. 3A). No amount of NRII, however, produced total inhibition of the UT reaction (Fig. 3B). The inhibition of the UT activity by NRII was not due to direct interactions of NRII with the UTase, as indicated by the failure of NRII to inhibit when the assays were conducted at the same conditions, but with PII present in large excess over NRII (SI Appendix). Together, these results suggest that NRII did not directly affect the enzyme but instead inhibited the UT activity by sequestering PII.
We found that NRII was an inhibitor of the UR activity (Fig. 3C), even though it is unable to bind to uridylylated PII subunits. To understand this inhibition more completely, burst kinetic analysis was performed. In this type of kinetic analysis, the catalytic rate is measured with enzyme and substrate at similar concentrations. In our experiments, fully modified PII-UMP (P3) was used as substrate, glutamine was in excess to activate the UR, and reactions were conducted at low temperature to slow the rate of catalysis. The deuridylylation of PII-UMP displayed unusual biphasic kinetics when NRII was present; initially NRII was a very poor inhibitor of the UR activity, but at later times, NRII provided strong inhibition (Fig. 3D). The first of these kinetic phases appeared to correspond to the removal of the initial UMP group from a fully modified PII trimer, whereas the second, strongly inhibited, kinetic phase appeared to correspond to the very slow removal of the remaining UMP groups.
Different results were obtained when the effects of NRII on the uridylylation and deuridylylation of monvalent PII were examined. In that case, NRII was an even more potent inhibitor of the UT activity than was observed when wild-type PII was used, and the degree of inhibition seemed to approach full inhibition at high concentrations of NRII (Fig. 3E). This ability of NRII to completely inhibit PII uridylylation may reflect the fact upon binding of NRII to the single functional subunit of monovalent PII, there are no other subunits in the complex that can be modified. Most importantly, when monovalent PII was used, NRII failed to provide significant inhibition of the UR activity (Fig. 3F). The small inhibition afforded by NRII may be attributed to the fact that in the monovalent PII preparation, a discernable concentration of divalent PII was present along with a trace of wild-type PII.
Together, the results of the analysis of the individual catalytic activities for the systems containing wild-type and monovalent PII provide an explanation for the effects of NRII on the steady-state output of the reconstituted cyclic systems. In the system containing wild-type PII, the inhibition of both the UT and UR activities by NRII provides an explanation for the effect that NRII had on the signaling of both the high- and low-glutamine states. By contrast, in the system containing monovalent PII, NRII only inhibited the UT activity but had little effect on the UR activity. Consequently, the modification state of PII was monotonically reduced, relative to the no-NRII control, explaining why the two curves in Fig. 2D did not cross.
Predictions of a Simple Model Allowing Analytical Solutions.
We used a simple model for a covalent modification cycle, identical to the model of Goldbeter and Koshland (14), where the substrate protein cycles between two states, W and W∗, under the control of a stimulus, S, that inhibits the formation of W∗ and activates the formation of W.
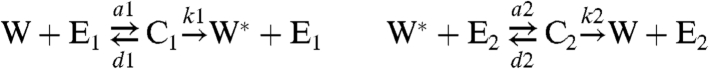 |
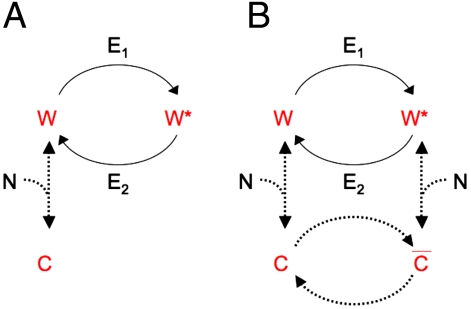
To this model, we added interactions of W and W∗ with a downstream target, N (Fig. 4).
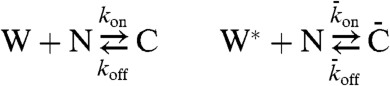 |
Fig. 4.
Schemes for toy models of a covalent modification cycle, where a downstream component interacts with one (A) or both (B) forms of the substrate of the cycle. E1 and E2 correspond to the two antagonistic converter enzymes of the covalent modification cycle, W and W∗ refer to the unmodified and modified forms of the protein that is the substrate of the cycle, N refers to the downstratem target of the cycle, C refers to the complex of N and W, and 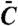 refers to the complex of W∗ with N.
refers to the complex of W∗ with N.
This simple model allowed analytical solutions for the steady-state output, the sensitivity, and the S50 of the system (SI Appendix). The relationship between the stimulus and the steady-state output (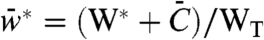 , where WT is the total amount of the substrate) is given by
, where WT is the total amount of the substrate) is given by
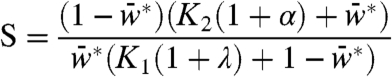 |
where K1 and K2 are the Km for the forward and reverse reactions divided by the substrate concentration, and λ and α are N divided by its Kd for W and W∗, respectively. When N bound to W only, α = 0. Systems where both W and W∗ interacted with N behaved differently than systems where only W interacted with N, as follows:
Effect on the steady-state output: When N interacted with W only, increasing N monotonically decreased the steady-state 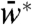 for all values of S (SI Appendix). But, when N interacted with both W and W∗, increasing N decreased the steady-state
for all values of S (SI Appendix). But, when N interacted with both W and W∗, increasing N decreased the steady-state 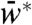 for low S and increased the steady-state
for low S and increased the steady-state 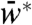 for high S (SI Appendix). These predictions correspond to the experimental results (Fig. 2 B and D).
for high S (SI Appendix). These predictions correspond to the experimental results (Fig. 2 B and D).
Effect on sensitivity: The sensitivity of the system is given by the response coefficient R defined as the amount of stimulus required to move the system from 90% of its maximal response to 10% of its maximal response (14). R is inversely proportional to the Hill coefficient (nH); specifically, nH = log(81)/ log R.
The sensitivity to the stimulus always monotonically decreased (R increased) as N was increased.
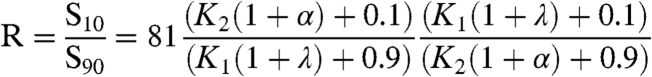 |
If N bound to W only, the sensitivity depended on the Km of W∗ for the demodification enzyme, and tended toward hyperbolic sensitivity (nH = 1) at very high N if this Km was large enough (SI Appendix). This situation corresponds to the results obtained in the experimental system with monovalent PII (Fig. 2D). When N bound to both W and W∗, sensitivity also approached a limit of nH = 1, independently of the values of the Km for the two opposing reactions of the cycle. However, subsensitive responses (nH < 1) were obtained if N bound to both W and W∗, forming complexes C and 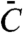 , and if covalent modification can occur between C and
, and if covalent modification can occur between C and 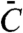 (Fig. 4B and SI Appendix). In the experimental system with wild-type PII, retroactivity by NRII brought about a subsensitive response (Fig. 2B), suggesting that such interconversions can occur. Further support for such interconversions of NRII-bound PII comes from the data in Fig. 3 A and D, showing that binding of NRII to wild-type PII could not completely inhibit the uridylylation of PII or deuridylylation of PII-UMP.
(Fig. 4B and SI Appendix). In the experimental system with wild-type PII, retroactivity by NRII brought about a subsensitive response (Fig. 2B), suggesting that such interconversions can occur. Further support for such interconversions of NRII-bound PII comes from the data in Fig. 3 A and D, showing that binding of NRII to wild-type PII could not completely inhibit the uridylylation of PII or deuridylylation of PII-UMP.
Effect on S50: The S50 is given by
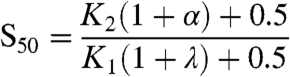 |
where K1, K2, λ, and α are as before. When N bound to W only (corresponding to α = 0), increasing N monotonically decreased S50. This corresponds to the results obtained with monovalent PII (Fig. 2D). By contrast, when N bound to both W and W∗, increasing N could monotonically raise or lower S50, or have no effect, depending on the dissociation constants of N for each of W and W∗ and the Km for the two enzymatic activities (SI Appendix). In the experiments with wild-type PII, NRII brought about a small increase in the S50 (Fig. 2B).
Detailed Kinetic Models for the UTase/UR-PII Cycle and the Effects of NRII.
To better understand the UTase/UR-PII cycle and its regulation by NRII, we developed detailed kinetic models for the systems containing wild-type and monovalent PII. For this, all known or predicted interactions and rates were explicitly described, as well as the inhibition of the UT activity and activation of the UR activity by glutamine. Previous modeling approaches have examined the E. coli nitrogen control system in detail, but used a modular approach that did not consider retroactive effects (15, 16). The schemes upon which our models are based are provided in SI Appendix.
We used a system of ordinary differential equations to describe the temporal evolution of the species (SI Appendix). The system was solved to get the steady-states corresponding to a given glutamine stimulation, optimizing the parameters involved so that the output of the model provided a good representation of the steady-state experimental data. Thus, the model does not deal with time-dependent behavior. Whenever possible, initial parameter values were from earlier studies. Comparison of the parameters from the fitting to experimentally measured parameters, then gave a sense of the validity of the modeling. As part of a reiterative process of modeling and experimentation, we made careful measurements of the UT basal activity, and included information from another study in our lab indicating that each NRII dimer bound to two PII trimers with high cooperativity (SI Appendix). We also examined the simulated kinetics of PII modification and demodification at several conditions, and compared these to experimental data. The correspondence of the modeled parameters to experimentally determined parameters is reasonable considering the diversity of approaches and materials used in the experiments.
The model describing the system with monovalent PII was simpler, of course (SI Appendix). For this system, only parameters related to PII-UTase/UR interaction were adjusted slightly (< 20%) relative to the parameters for wild-type PII, reflecting the possibility that the monovalent PII binds slightly differently to the enzyme and reacts at different rates. Because in the heterotrimeric PII preparation there is still a percentage of divalent and wild-type PII, we accounted for these minor species in the model. Acceptable fits were obtained with the experimental data, and the simulated distribution of subunits in the heterotrimeric species closely matched the expected distribution based on random assortment of subunits. The curves superimposed over the data in Fig. 2 B and D are the outputs of the model, using the parameters in SI Appendix.
Predictions of the Detailed Model.
The parameterized models were used to explore the relationship between NRII and glutamine in controlling system output. Simulation of the natural system containing wild-type PII, with different concentrations of NRII, is shown in Fig. 5A. A complex pattern of NRII effects was obtained, due to the influence of NRII on both the UT and UR activities, reminiscent of the results using the toy model. NRII had minor effects on the S50, which monotonically increased as NRII was increased. Notably, the simulations predicted that significant effects of NRII should be observed at concentrations of NRII well below the 10 μM concentration used in our experiments. We tested this by experimentally examining the effects of 2 and 5 μM NRII at high- and low-glutamine concentrations, and observed that the biphasic effects of NRII were clearly discernable. In simulations of the system utilizing monovalent PII, both NRII and glutamine reduced the extent of PII modification (Fig. 5B). NRII dramatically reduced the S50 and diminished the sensitivity of the response to glutamine, again reminiscent of the results with the toy model.
Fig. 5.
Predictions of the model. (A), (B) Stimulus-response curves obtained with the kinetic model for wild-type PII (A) or monovalent PII (B), when stimulated with glutamine, for increasing levels of NRII (0, 3, 6, 10, and 20 μM). (C) Effect of NRII on the sensitivity of the response to glutamine in the two systems. (D) Effect of NRII on the S50 of the glutamine response in the two systems.
The effects of NRII on the sensitivity of the systems to glutamine is replotted in Fig. 5C, and the effects of NRII on the glutamine S50 of the systems is replotted in Fig. 5D. When wild-type PII was used, the ultrasensitive system was converted by NRII to a subsensitive system with nH approaching 0.5 at saturating NRII (Fig. 5C). By contrast, NRII had a significantly less dramatic effect on the sensitivity of the system containing monovalent PII, where it was only able to reduce nH to about unity, consistent with the predictions of the toy model. With regard to the system S50, NRII had the greatest effect on the system with monovalent PII, reducing S50 about 17.5-fold at saturation, as observed (Fig. 2D). By contrast, in the system with wild-type PII, saturating NRII caused a small increase in the system S50.
Using the methods of our experimental study, the only variable that was experimentally measured was the total level of UMP groups that were covalently attached to protein. The detailed model let us explore the behavior of the different modification states of the PII trimers depending on both glutamine and NRII. Distinctions between these species could be physiologically significant, depending on the ability of partially modified forms of PII to activate or inhibit different cellular receptors. We observed that the distribution of the different modified forms of PII was affected by NRII (SI Appendix). Thus, retroactive effects of NRII has the potential to differentially affect the control of other receptors.
Discussion
We used model experimental systems and a quantitative approach to explore in detail the mechanisms by which downstream targets of a covalent modification cycle can influence the signaling properties of the cycle. We assembled a synthetic signaling system in which PII was functionally monomeric and all effects of NRII were limited to sequestering unmodified PII. In this model system, glutamine regulated PII modification state with sensitivity (nH ∼ 1.99) similar to that observed with the natural system (nH ∼ 2.21). Because in this synthetic system NRII only bound to the unmodified form of PII, NRII diminished the steady-state PII modification level throughout the range of glutamine concentrations, and reduced the sensitivity to glutamine to a hyperbolic response (nH ∼ 1.14). This effect of NRII corresponds to the retroactivity predicted from fundamental models (1, 2), such retroactivity should always be present if appropriate concentrations of downstream components are present. We observed significant effects of retroactivity on the steady-state output of a covalent modification cycle with a purified system, utilizing reasonable protein concentrations. Furthermore, both sensitivity and S50 of the system were affected by NRII. Thus, the downstream target played a significant role in shaping the output of the upstream covalent modification cycle.
The natural system that included wild-type PII proved to be more complex than the synthetic system containing monovalent PII, and the effect of NRII on the natural system was even more dramatic. We observed that for the natural system, NRII provided indirect inhibition of both the UT and UR activities, and that under a wide range of conditions, the inhibition of the UR activity was more complete. Burst kinetic experiments suggested that NRII inhibition of the UR activity resulted from NRII binding to the unmodified PII subunit of partially modified PII trimers, and by so doing, slow down the deuridylylation of the modified subunits of the complex. The observation that the UR activity is inhibited more severely than the UT activity under many conditions is consistent with the hypothesis that the UT and UR activities of the UTase/UR take place at separate active sites on the protein, as predicted by domain homologies. Perhaps the UR active site is more sensitive than is the UT active site to the presence of NRII bound to other subunits of the PII trimer. Whatever the details of the mechanism, NRII inhibition of the UR activity antagonized signaling of the high-glutamine state. Thus, in the natural system, NRII antagonized signaling of both high- and low-glutamine states, and by so doing, had very dramatic effects on system sensitivity, converting an ultrasensitive response to glutamine (nH ∼ 2.21) to a subsensitive response (nH ∼ 0.74).
How does NRII affect the sensitivity of the UTase/UR-PII cycle? The sensitivity of this cycle to glutamine did not depend on PII functioning as a trimer. This is consistent with earlier observations that the UT and UR reactions appeared to be nonprocessive. We will show elsewhere that under similar conditions, the modest ultrasensitivity of the system was due to a combination of zero-order and multistep effects. Under conditions where no zero-order effects were obtained (at low PII concentrations), the system was still slightly ultrasensitive (nH ∼ 1.4). Because we observed here that NRII lowered the sensitivity below this level, we must conclude that NRII did more than simply move the system away from the zero-order regime; NRII must have also diminished the multistep effects.
Our experimental studies using reconstituted systems were intended to focus upon general principles, as opposed to the role on retroactivity in nitrogen regulation in E. coli. Nevertheless, it should be pointed out that another PII receptor, the ammonium permease AmtB, can provide dramatic sequestration of PII in vivo in certain genetic backgrounds, resulting in bistability (17). More importantly, retroactivity, and other forms of sequestration (18–20) may play a role in a wide range of signaling systems controlling fundamental life processes in all cell types. Our study shows that retroactive effects were readily observable in reconstituted systems. Yet, the signaling systems in nature have been selected for effective signal transduction. Consequently, either there must be subtle mechanisms for preventing retroactivity by downstream components of signal transduction systems, or this retroactivity provides an advantage and is used by the systems.
Materials and Methods
Purified Proteins and General Assay Methods.
Preparations of PII, NRII, and UTase/UR described previously were used (12). Measurement of steady-state levels of protein uridylylation was as described previously (12), and employed α-[32P]-UTP. Levels of protein modification were determined by absorption and precipitation of aliquots of reaction mixtures onto nitrocellulose filters, which were washed extensively to remove unincorporated labeled and counted by liquid scintillation (e.g., ref. 12).
UTase/UR-PII Monocycle Experiments.
Reaction mixtures contained 100 mM Tris-Cl, pH 7.5, 25 mM MgCl2, and 100 mM KCl, 3.0 μM PII (homotrimers) and 0.8 μM UTase/UR (monomers), 0.5 mM UTP, 0.5 mM ATP, 0.2 mM α-ketoglutarate, 1 mM DTT, and glutamine as indicated. Because the system is very sensitive to glycerol, and all of the proteins were stored in storage buffer that includes 50% glycerol, all reaction mixtures contained the identical amount of storage buffer. This was accomplished by balancing the addition of NRII with the addition of storage buffer for samples lacking NRII. Reaction mixtures were preincubated in the absence of the ATP and UTP (which was labeled), and reactions were initiated by addition of the nucleotides. Samples were removed at various times and processed to determine the level of incorporated label. Steady-state levels were obtained by simple averaging of the values for later samples in the time course where the reactions were assessed as having reached the steady state.
Measurement of UT and UR Activities.
Initial rates of PII uridylylation and PII-UMP deuridylylation were measured as described previously (13), general conditions were as described above for the monocycle experiments, with specific conditions for each assay listed in the figure legends. PII-UMP containing 32P was prepared as described (13). For burst kinetic analysis of the UR activity using wild-type PII-UMP homotrimers, conditions were 100 mM Tris-Cl, pH 7.5, 50 mM MgCl2, 100 mM KCl, 2.67 μM PII-UMP (homotrimers), 3 μM UTase/UR, 1 mM α-ketoglutarate, 20 mM glutamine, 0.5 mM ATP, 1 mM DTT, 300 μg/mL BSA, ± 10 μM NRII, as indicated. Reaction mixtures were assembled lacking the PII-UMP substrate, and prewarmed at 20 °C. Reactions were initiated by the addition of prewarmed PII-UMP, and samples were removed at the indicated times, spotted onto filters, and processed as for the standard UR assay (13). For the burst kinetic analysis of heterotrimeric, monovalent, PII-UMP, procedures were the same and the conditions were as above, but with PII-UMP (mixture containing heterotrimers) at 1.18 μM uridylylated subunits, and 0.47 μM UTase/UR.
Simulations.
Simulations were performed in MATLAB, Version 7.7.0 (Mathworks). Computations utilized the Nyx-AMD Opteron 64-bit cluster at the Center for Advanced Computing, University of Michigan.
Supplementary Material
Acknowledgments.
A.J.N. and P.J. were supported by National Institutes of Health National Institute of General Medical Sciences Grant GM059637 (to A.J.N.). S.D.M. was supported by the Burrough Wellcome Fund, Breast Cancer Reasearch Foundation, and National Institutes of Health Grant NIH-CA77612. A.C.V. was supported by grants from the Department of Defense Breast Cancer Research Program and the Center for Computational Medicine and Bioinformatics. D.D.V. was supported in part by Air Force Office of Scientific Research Grant FA9550-09-1-0211. L.V.W. was supported by the Cellular Biotechnology Training Program, University of Michigan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.D.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913815107/-/DCSupplemental.
References
- 1.Ventura A, Sepulchre J-A, Merajver S. A hidden feedback in signaling cascades is revealed. PloS Comput Biol. 2008;4:e1000041. doi: 10.1371/journal.pcbi.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Del Vecchio D, Ninfa AJ, Sontag ED. Modular cell biology: Retroactivity and insulation. Mol Syst Biol. 2008;4:161. doi: 10.1038/msb4100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauro HM. Modularity defined. Mol Syst Biol. 2008;4:166. doi: 10.1038/msb.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saez-Rodriguez J, Kremling A, Gilles ED. Dissecting the puzzle of life: Modularization of signal transduction networks. Comput Chem Eng. 2005;29:619–629. [Google Scholar]
- 5.Saez-Rodriguez J, Gayer S, Ginkel M, Gilles ED. Automatic decomposition of kinetic models of signaling networks minimizing retroactivity among modules. Bioinformatics. 2008;24:i213–i219. doi: 10.1093/bioinformatics/btn289. [DOI] [PubMed] [Google Scholar]
- 6.Kholodenko BN, Demin OV, Moehren G, Hoek JB. Quantification of short term signaling by the epiderman growth factor receptor. J Biol Chem. 1999;274:30169–30181. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]
- 7.Legewie S, Schoeberi B, Bluthgen N, Herzel H. Competing docking interactions can bring about bistability in the MAPK cascade. Biophys J. 2007;93:2279–2288. doi: 10.1529/biophysj.107.109132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiao L, Nachbar RB, Kevrekidis IG, Shvartzman SY. Bistability and oscillations in the Huang–Ferrell model of MAPK signaling. PLoS Comput Biol. 2007;3:e184. doi: 10.1371/journal.pcbi.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y, et al. MAPK substrate competition integrates pattern signals in the Drosophila embryo. Curr Biol. 2010;20:1–6. doi: 10.1016/j.cub.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninfa AJ, Jiang P, Atkinson MR, Peliska JA. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr Top Cell Regul. 2000;36:31–75. doi: 10.1016/s0070-2137(01)80002-9. [DOI] [PubMed] [Google Scholar]
- 11.Jiang P, Ninfa AJ. Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylylate energy charge in vitro. Biochemistry. 2007;46:12979–12996. doi: 10.1021/bi701062t. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, Zucker P, Ninfa AJ. Probing interactions of the homotrimeric PII signal transduction protein with its receptors by use of PII heterotrimers formed in vitro from wild-type and mutant subunits. J Bacteriol. 1997b;179:4354–4360. doi: 10.1128/jb.179.13.4354-4360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang P, Peliska JA, Ninfa AJ. Enzymological characterization of the signal-transducing uridylyltransferase-uridylyl-removing enzyme (EC2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 14.Goldbeter A, Koshland DE., Jr An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutalik VK, Venkatesh KV. A theoretical steady state analysis indicates that induction of Escherichia coli glnALG operon can display all-or-none behavior. Biosystems. 2007;90:1–19. doi: 10.1016/j.biosystems.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Bruggeman FJ, Boogerd FC, Westerhoff HV. The multifarious short-term regulation of ammonia assimilation of Escherichia coli: Dissection using an in silico replica. FEBS J. 2005;272:1965–1985. doi: 10.1111/j.1742-4658.2005.04626.x. [DOI] [PubMed] [Google Scholar]
- 17.Blauwkamp TA, Ninfa AJ. Antagonism of PII signalling by the AmtB protein of Escherichia coli. Mol Microbiol. 2003;48:1017–1028. doi: 10.1046/j.1365-2958.2003.03479.x. [DOI] [PubMed] [Google Scholar]
- 18.Bluthgen N, et al. Effects of sequestration on signal transduction cascades. FEBS J. 2006;273:895–906. doi: 10.1111/j.1742-4658.2006.05105.x. [DOI] [PubMed] [Google Scholar]
- 19.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from mul;tisite phosphorylation in protein kinase cascades. J Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fell DA, Sauro HM. Metabolic control analysis. The effects of high enzyme concentrations. Eur J Biochem. 1990;192:183–187. doi: 10.1111/j.1432-1033.1990.tb19212.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.