Abstract
High-affinity, high-selectivity protein-protein interactions that are critical for cell survival present an evolutionary paradox: How does selectivity evolve when acquired mutations risk a lethal loss of high-affinity binding? A detailed understanding of selectivity in such complexes requires structural information on weak, noncognate complexes which can be difficult to obtain due to their transient and dynamic nature. Using NMR-based docking as a guide, we deployed a disulfide-trapping strategy on a noncognate complex between the colicin E9 endonuclease (E9 DNase) and immunity protein 2 (Im2), which is seven orders of magnitude weaker binding than the cognate femtomolar E9 DNase-Im9 interaction. The 1.77 Å crystal structure of the E9 DNase-Im2 complex reveals an entirely noncovalent interface where the intersubunit disulfide merely supports the crystal lattice. In combination with computational alanine scanning of interfacial residues, the structure reveals that the driving force for binding is so strong that a severely unfavorable specificity contact is tolerated at the interface and as a result the complex becomes weakened through “frustration.” As well as rationalizing past mutational and thermodynamic data, comparing our noncognate structure with previous cognate complexes highlights the importance of loop regions in developing selectivity and accentuates the multiple roles of buried water molecules that stabilize, ameliorate, or aggravate interfacial contacts. The study provides direct support for dual-recognition in colicin DNase-Im protein complexes and shows that weakened noncognate complexes are primed for high-affinity binding, which can be achieved by economical mutation of a limited number of residues at the interface.
Keywords: colicins, crystallography, disulfide-trapping, specificity, frustration
Specificity in protein-protein interactions (PPIs) is critical for the organization of macromolecular complexes involved in all aspects of cellular homeostasis and differentiation. Within the vast interaction networks in cells there is significant redundancy, with many proteins acting as “hubs” that recognize multiple binding partners (1), and yet also significant discrimination (2). The factors that tip the balance in favor of specificity or promiscuity in PPIs while not clear are coming under increasing scrutiny (3). Understanding this balance is critical both from fundamental and applied perspectives. Protein therapeutics are finding increasing use in medicine but off-target effects can have disastrous consequences (4). High-affinity, high-selectivity binding is therefore an essential goal in such engineered platforms. Advances in defining the molecular and thermodynamic basis for binding affinity have been made in a multitude of natural and engineered/designed PPIs (5–7), but our molecular knowledge of specificity remains rudimentary. In large part this is due to the lack of high-resolution structural information on weak and transient protein-protein complexes that are evolutionarily related to a cognate high-affinity complex but massively destabilized due to incorrect, noncognate contacts. We address this problem via entrapment of one such destabilized noncognate complex and comparison of its crystal structure to that of its ultra high-affinity counterpart. Viewed in the context of computational analysis of the interface, the structure provides significant insight into the mechanism by which 107-fold discrimination is engendered in a family of homologous protein-protein complexes.
The study of PPI specificity at the molecular level has roots in the crystal structure of the complex of bovine trypsin bound to bovine pancreatic trypsin inhibitor (8). Indeed, protease-inhibitor complexes are some of the most extensively studied PPIs, exhibiting both high-affinity (< pM) and a broad range of specificities, and dominate statistical surveys cataloguing the structural characteristics of PPIs (9–11). Extensive mutational analysis of cognate and noncognate protease-inhibitor complexes demonstrated that binding is additive to such an extent that binding affinities can be predicted through a sequence-to-reactivity algorithm (12). Such strong additivity is however not a property seen in many other PPIs where cooperative interactions prevail hence other systems are needed to probe the molecular basis for specificity.
Specificity in PPIs is typically studied through one of three approaches. The classical approach is to compare the structures and binding thermodynamics of families of closely related PPIs, e.g., cell-signalling SH2 domain-phosphopeptide complexes (2). More recently, computational (re)design (13 and 14) and directed evolution experiments (15 and 16) have been developed. The former provides a useful yardstick for gauging how well PPI specificity is understood; currently, the largest changes in specificity that have been designed into an interface are of the order of 102- to 103-fold (17–20). While this represents significant progress, when set against the > 1010-fold range of binding affinities that PPIs can exhibit in nature (21) it highlights how much remains to be uncovered.
We addressed this problem using as a model the complexes of colicin endonucleases (DNases) with immunity (Im) proteins. Colicins are plasmid-encoded, stress-induced protein antibiotics that specifically target Escherichia coli cells. Group E colicins, E2, E7, E8, and E9, kill through a C-terminal DNase domain that degrades the bacterial genome. The colicin nuclease is targeted to the cytoplasm through a series of PPIs. Since the nuclease is a potent cytotoxin only a specific (cognate) Im partner can protect the organism from endogenous and incoming colicin (22). The inhibitor protein binds at an Im protein exosite (IPE) on the DNase that is distal to the enzyme active site; the sequences involved in binding being the most variable in each of the two proteins (23). Colicin DNase-Im complexes are an ideal model for exploring specificity in PPIs since equilibrium dissociation constants (KDs) span the entire range of known heterocomplex stabilities (24 and 25); these can be as low as 10-4 M for noncognate complexes and 10-14–10-15 M for cognate complexes. We have proposed previously a dual-recognition mechanism for colicin DNase-Im specificity where binding affinity is dominated by a conserved hotspot, comprising, two tyrosine residues, Y54 and Y55, and an aspartic acid (D51) in helix III of the Im protein, with neighboring variable residues in helix II making positive, neutral, or negative contributions to specificity (26–28). Detailed presteady state kinetic studies show two-state association kinetics for cognate and noncognate complexes alike, which likely reflect rotamer dynamics in a transient intermediate complex (29). Only the cognate Im partner provides the pattern of contacts required to freeze out the high-affinity binding conformer. Consistent with this mechanism, a comparison of two cognate DNase-Im structures E9 DNase-Im9 and E7 DNase-Im7 reveals a 19° rigid-body rotation of the Im on the DNase surface centered on the hotspot tyrosine residues (30 and 31).
Here, through a disulfide-trapping approach, we present the structure of the noncognate E9 DNase-Im2 complex where we address the structural and energetic basis for high selectivity in this family of homologous high-affinity complexes.
Results
NMR-Based Docking and Disulfide-Trapping of the Noncognate Colicin E9 DNase-Im2 Complex.
Previous attempts to crystallize weak noncognate colicin complexes have been unsuccessful. Our strategy focused on disulfide linkage of a noncognate DNase-Im complex, an approach used in classical studies of protein-folding (32) and more recently applied to investigations of protein translocase topology (33) and G-protein-coupled receptor signalling (34). We selected the E9 DNase-Im2 complex for disulfide-trapping in order to understand why, despite sharing 68% sequence identity with Im9, binding of Im2 to E9 DNase (KD ∼ 0.1 μM) (29) is over seven orders of magnitude weaker than the cognate E9 DNase-Im9 complex. The position of trapping disulfides was based on NMR-based docking using chemical shift perturbation (CSP) data (SI Text). Given the lack of information on the structure of E9 DNase-Im2, and in light of kinetic analyses (29) indicating that noncognate complexes may be dynamic, NMR-based docking of E9 DNase-Im2 was undertaken to provide an interface-wide picture of which residues were involved in binding and help in the placement of disulfide-bond forming cysteine residues (Fig. S1 and Fig. S2). This analysis was aided by previous alanine-scanning mutagenesis data (26, 28, 35). As controls, we prepared combinations of Cys mutant proteins that were not expected to form a disulfide bond. Only one predicted pairing, E9 DNase G95C and Im2 E31C, readily formed a disulfide across the noncognate interface (see SI Text and Fig. S2). Isothermal titration calorimetry under reducing conditions confirmed the cysteine mutations had a negligible effect on binding (Fig. S3). The oxidized complex was then concentrated, purified by reverse-phase HPLC, crystallized and the structure determined to 1.77 Å resolution (Table S1).
Structure of the Colicin E9 DNase-Im2 Complex.
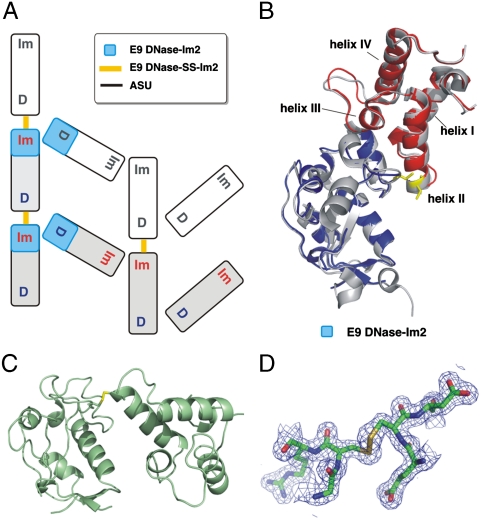
Surprisingly, crystallization of the disulfide-linked species yielded a noncovalent E9 DNase-Im2 complex. The E9 DNase G95C-Im2 E31C disulfide creates a scaffold where each Im is covalently attached to a DNase in an adjacent DNase-Im pairing due to 150° rotations around the disulfide bond, resulting in disulfide links between rather than within E9 DNase-Im2 complexes (Fig. 1). The disulfide thereby provides additional stabilizing interfaces between Im and DNase molecules which help trap the noncovalent complex. Hence the most significant interface in the crystal structure is not that between the two molecules of the target complex (E9 DNase-SS-Im2) but that between two molecules from adjacent complexes (Table 1).
Fig. 1.
E9 DNase-SS-Im2 acts as a scaffold trapping the noncognate E9 DNase-Im2 complex. A, Schematic of the three principal types of DNase-Im interface within the crystal (see Table 1). Disulfide links are formed between molecules in adjacent asymmetric units—Im2 (x, y, z), E9 DNase (x - 1/2, -y - 1/2, -z). B, Structure of the noncovalent E9 DNase-Im2 complex (blue and red, respectively; 2WPT) overlaid with E9 DNase-Im9 (1EMV). The helices of the Im protein are labeled. C, E9 DNase-SS-Im2 structure and D, disulfide electron density (contoured at 1.2σ).
Table 1.
Characteristics of the principal E9 DNase-Im2 interfaces observed in the crystal structure of E9 DNase G95C-SS-Im2 E31C and their comparison to two cognate complexes.
| DNase-Im interface * | ASA (Å2) | Direct H-bonds | Sc | Buried water molecules † |
| E9 DNase-Im2 | 1,566 | 15 | 0.75 | 8 |
| ASU | 996 | 5 | 0.60 | 4 |
| E9 DNase-SS-Im2 | 169 | 1 | 0.29 | 1 |
| E9 DNase-Im9 ‡ | 1,500 | 13 | 0.71 | 5 |
| E7 DNase-Im7‡ | 1,370 | 19 | 0.71 | 7 |
*Three principal interfaces are observed in the crystal structure of the E9 DNase G95C-SS-Im2 E31C complex; the noncognate E9 DNase-Im2 interface, the interface comprising the asymmetric unit (ASU) and the disulfided interface. A fourth minor crystal contact with ASA < 30 Å2 is not considered further.
†Buried waters, ASA and hydrogen bonds were analyzed using PISA (51) and protein surface complementarity using Sc (52).
‡Interface statistics for previously reported cognate complexes, E9 DNase-Im9 (pdb, 1EMV) and E7 DNase-Im7 (pdb, 7CEI).
Of the three principal DNase-Im interfaces in the crystal structure of E9 DNase-SS-Im2 (Fig. 1, Table 1) only the noncovalent E9 DNase-Im2 interface is characteristic of a stable PPI (9). The structure of E9 DNase-Im2 is almost identical to that of the cognate E9 DNase-Im9 (backbone rmsd = 0.39 Å; Fig. 1B) and, importantly, is very similar to the NMR-docked model on which it is based (Fig. S4). Binding is localized at the IPE on the DNase (22) and anchored by the interactions of conserved Im protein (Y54 and Y55) and DNase (F86) hotspot residues (Fig. 2A). Indeed, the pattern of hydrogen bonding at the E9 DNase-Im2 interface around the conserved hotspot residues is identical to that of the E9 DNase-Im9 complex, including those to five conserved interfacial water molecules (Fig. 2A; Table S2 and Table S3). Further authentication of the complex comes from its ability to rationalize previously unexplained mutational data. Mutation of Im E41 (conserved in Im2 and Im9) to alanine, destabilizes the E9 DNase-Im2 complex significantly more than the cognate complex (26). Our structure shows that Im2 E41 salt bridges with almost ideal geometry to two lysines (K89, K97) in E9 DNase while in E9 DNase-Im9 only K97 is within reach.
Fig. 2.
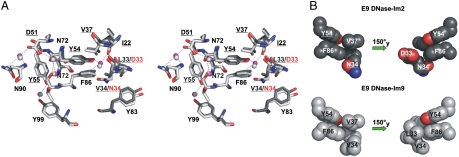
Conserved interfacial interactions anchor the frustrated E9 DNase-Im2 complex. E9 DNase-Im2, present work (2WPT) light gray; E9 DNase-Im9 (1EMV) dark gray. A, Stereoview of the interface core highlighting hotspot residues in both complexes (Im2 (underlined) D51, Y54, Y55, and E9 DNase F86) and the positions of the five conserved interfacial water molecules (gray) from the cognate structure, mirrored in the E9 DNase-Im2 complex (magenta). Table S2 and Table S3 list direct and water-mediated hydrogen bonds. B, Van der Waals representations of the noncognate (top) and cognate interface cores (bottom). Im2 Y54 forces unfavorable interactions of Im2 residues D33 (and to a lesser extent N34) with the E9 DNase hotspot residue F86.
The structural characteristics of the E9 DNase-Im2 complex listed in Table 1 are those of a bona fide DNase-Im interface. Indeed, defined purely in terms of interface statistics the complex appears as good if not better than the cognate complexes of E9 DNase or E7 DNase. Why then is this noncognate complex seven orders of magnitude weaker binding than E9 DNase-Im9? The following sections address this central question.
The E9 DNase-Im2 Complex is a Frustrated Protein-Protein Interaction.
The structure of the E9 DNase-Im2 complex provides direct support for the dual-recognition mechanism of colicin-Im binding. Fig. 2 confirms that the aromatic-stacking interaction of conserved Im hotspot residue Y54 with the E9 DNase hotspot F86 presents residues in the N terminus of Im2 helix II to the interface (29 and 31). F86 is the major hotspot and specificity site on E9 DNase; F86A reduces cognate complex stability by ∼4 kcal mol-1 (35). Im Y54 and Y55 are conserved residues whose mutation to alanine has even greater impact, each affecting binding by ∼5 kcal mol-1 (28). In the cognate complex, Im9 specificity residues L33 and V34 from helix II pack against E9 DNase F86. Substitution of these sites with charged/polar residues (Asp and Asn, respectively) does not steer Im2 towards an altered binding configuration as seen in the complex of the E7 DNase with Im7 (30). Instead, close and unfavorable packing between F86 and positions 33 and 34 in the Im is maintained (Fig. 2). F86 CZ is within 4.26 Å of N34 OD1 and 5.98 Å of D33 OD2. Statistical studies of sidechain contact preferences indicate Phe-Asp to be one of the most unfavorable of all sidechain interactions, with Phe-Asn also heavily disfavored (36). The E9 DNase F86-Im2 Y54 contact, in combination with the other Im2 hotspot residues D51 and Y55 and conserved interfacial water-mediated hydrogen bonds is so strong that these unfavorable sidechain interactions are locked-in and the complex experiences frustration. Frustration is a term used in protein folding to describe accumulation of nonnative or unfavorable contacts, with optimal folding resulting from minimal frustration (37).
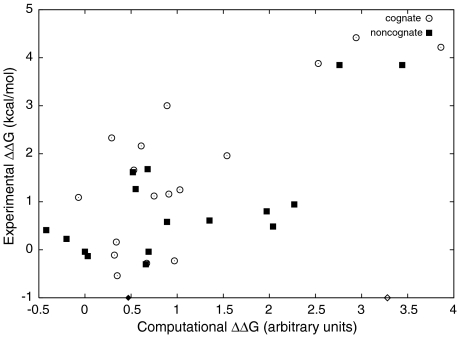
We used computational alanine scanning to further explore the interplay of hotspot and specificity contacts in the dual-recognition mechanism. RosettaDesign (35 and 38) was used to assess the effect of mutating key interface residues on complex stability (see SI Text). Computational alanine scanning within this framework has proven to be an efficient tool for predicting the locations of hotspot residues in protein-protein interfaces (38). This approach does not suffer, as does experimental alanine scanning, from the vagaries of expression and folding due to residue substitution. In the present analysis, we calculated the difference in (Rosetta binding) energy for single alanine mutations of E9 DNase and Im2 interface residues and compared these values to our previous computational analysis of the cognate E9 DNase-Im9 complex (see Fig. 4 and Table S4 and Table S5) (35). Moreover, we cross reference this analysis to our extensive experimental determinations of ΔΔG for both alanine mutations and specificity substitutions (26, 27, 35, 39). Although there are some discrepancies (notably N34A, D33A) on the whole, the computations match the experimental results in both the cognate and the noncognate interfaces, which span seven orders of magnitude of affinity.
Fig. 4.
Agreement between computational and experimental ΔΔG values. A scatter plot of per-residue calculated vs. experimental ΔΔG in cognate and noncognate complexes. The Pearson correlation coefficient for the data shown in this plot is r = 0.70. Open and closed diamonds mark the computational predictions for the effects of the mutation Y83A, in the cognate and noncognate complexes, correspondingly. See Table S4 and Table S5 for details, including computational data on other residues for which experimental mutation data have not been reported.
The key aspects of the dual-recognition mechanism are fully consistent with the new structure and are well supported by the computational and previous experimental work. Conserved hotspot residues (e.g. Im2 Y54, Y55 and E9 DNase F86) are highly stabilizing to cognate and noncognate complexes alike and key helix II sites (e.g., D33 and V37) reduce noncognate complex stability significantly (> -0.3 Rosetta energy units in Table S4 and Table S5). The importance of these sites in discrimination is further supported by experimental double mutant cycle analysis showing that positions 33 and 37 are slightly stabilizing in the cognate complex (35), but destabilizing in the noncognate complex, indicating that the major contributions to binding are correctly modeled. As discussed below, the computational analysis provides insight into contributions from residues, such Y83, that cannot be probed experimentally.
The destabilizing effect of charged-hydrophobic contacts at the heart of the E9 DNase-Im2 complex can be reversed through mutation. Mutating D33 in Im2 to alanine has a very significant impact on binding affinity in E9 DNase-Im2—KD decreases from 150 nM to ∼1 nM (Table S5), as revealed by isothermal titration calorimetry (ITC) measurements (29), consistent with the relief of frustration. Further recovery comes from mutation of D33 to leucine, the specificity residue in Im9, (KD ∼ 10-11 M) (27), emphasizing how well primed the system is for high-affinity binding from just a single substitution. Substituting three Im2 helix II residues with their Im9 counterparts (Im2 D33L/N34V/R38T) increased binding even further (KD ∼ 10-13 M) (27). No Im2 mutations have so far recovered full binding affinity to E9 DNase (10-14 M) suggesting that optimal binding specificity contains contributions from residues outside the interface core.
The Role of Im Protein Loop II in Binding Specificity.
Previous work on Im2/Im9 chimeras demonstrated that cross reactivity to E2 DNase was only abolished when loop II (between helices III and IV) from Im9 was incorporated into Im2 (40) but the basis for this effect was never established. Constructs lacking this loop retained binding to the parent colicin, E2 DNase. Our structure now sheds light on these observations. Im2, Im7, Im8, and Im9 all contain an aspartate at position 62 (D63 is the equivalent residue in Im7). The present structure shows that the orientation of this sidechain is sensitive to the nature of the binding partner. In E9 DNase-Im9, D62 hydrogen-bonds directly to N72 while in E7 DNase-Im7, D63 interacts with N74 via water-mediated hydrogen bonds. In E9 DNase-Im2, the interaction switches to water-mediated hydrogen bonding to S74 (Table S3) even though N72 is in close enough proximity (< 3.2 Å). Thus, in E9 DNase-Im2 D62 contacts are hybrid in character between those of the two cognate complexes. Our structure suggests that there will be a direct contact between these sites in the cognate E2 DNase-Im2 complex where N72 is now a lysine, implying D62 of the Im protein is required for development of E2 DNase- or E9 DNase-specific contacts. Correlation between the computational and experimental alanine-scanning data breaks down in the loop II region. Hence while the computational data suggest E9 DNase N72 forms a more stabilizing contact in the noncognate complex relative to that of Im9 (Table S4 and Table S5), experimental data show the mutation has essentially equivalent impact on the two complexes (Im2 D62 has yet to be mutated experimentally). Similarly, E9 DNase S74A has a bigger impact, experimentally, on noncognate Im2 relative to cognate Im9 binding yet the computational data suggest little difference in the two complexes. These discrepancies may be a consequence of Rosetta not accounting for water-mediated interactions, which clearly play a role in stabilizing loop II in the noncognate complex.
A Newly-Identified Putative Binding Hotspot Residue, E9 DNase Y83, Primes Colicin DNase-Im Protein Complexes for Maximal Affinity.
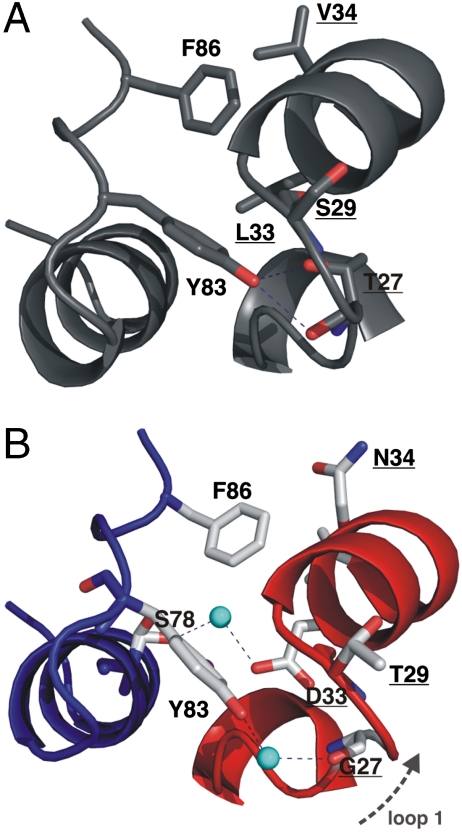
E9 DNase Y83 is on the edge of the interface, which in the cognate complex hydrogen bonds directly to T27 and S29 in loop I of Im9 (31), clamping the loop to the interface and ensuring optimal alignment of Im helix II and DNase specificity sites (Fig. 3A). Its aromatic moiety packs into a tight pocket formed by L33 and the aliphatic group of E30 on Im9 and is further buttressed by K81 of the E9 DNase. Our structure shows that in E9 DNase-Im2, where hydrogen-bonding capacity is removed by exchange of T27 for glycine, instead of direct interface contact a water molecule bridges G27 and Y83 (Fig. 3B). This intervening water molecule blocks direct hydrogen bonding between the tyrosine and loop I residues in Im2. The significance of this site in mediating specificity is again emphasized by computational alanine scanning (Fig. 4 and Table S4). On moving from cognate to noncognate complex, Y83A and G27A show the two largest decreases in the contribution to binding energy of any residue in the two proteins, with Y83 particularly affected (-2.8 energy units). Unfortunately, no experimental data for Y83A E9 DNase have been obtained to-date since the Y83A mutant fails to express in bacteria (35). Consequently, the identification of Y83 as a hotspot residue has yet to be verified experimentally.
Fig. 3.
Water molecules shape key E9 DNase-Im2 specificity interactions. A, Direct hydrogen bonds are formed between Im9 T27 and E9 DNase Y83 in the cognate complex with Im9. B, Bridging water molecules (cyan) interrupt Im2 G27-E9 DNase Y83 (loop I) and Im2 D33-E9 DNase S78 (helix II) hydrogen bonding; Im2 (red), E9 DNase (blue). See text for details.
Discussion
In using a disulfide-trapping strategy we have obtained a stable complex between Im2 and E9 DNase that does not result in a covalent link at the PPI. Instead, rotation about the engineered disulfide bond helps to crystallize the complex and capture the noncognate interface, formed between neighboring disulfided complexes. This was not an anticipated outcome of the disulfide strategy, but one that opens up the possibility of using a similar strategy for capturing other weakly binding PPIs that have resisted crystallisation.
Mechanisms by which families of essential high-affinity PPIs evolve new specificities are not well understood. Riley and coworkers have proposed that nuclease colicin-Im protein complexes evolve through diversifying selection (41) where a mutation must first occur in the Im protein conferring the ability to bind other colicins and increasing producer cell fitness. A subsequent (paired) mutation occurs in the colicin recognizing this new Im protein over the ancestral Im protein. Cells producing the new colicin are now able to kill cells producing the old colicin, promoting their survival over the ancestor and explaining why the binding site sequences of Im and colicin nuclease proteins are the most variable. But this hypothesis does not provide a mechanistic explanation as to how single mutations could affect specificity so profoundly within a complex that has fM affinity. By determining the structure of the noncognate complex E9 DNase-Im2 we are now able to cast light on the molecular features of specificity.
Loop Plasticity Provides a Template for Colicin DNase-Im Protein Interaction Specificity.
Two of the three loops in Im2 differ from Im9 in their contacts with E9 DNase. In loop I, hydrogen bonds are formed between Im9 T27 and E9 DNase Y83 which cannot be satisfied in the noncognate complex as a bridging water molecule blocks loop-interface alignment. In loop II, Im9 D62 forms a salt bridge with N72 of E9 DNase that is also interrupted by a bridging water molecule in E9 DNase-Im2. The importance of Im loop regions in Im-DNase recognition identified here has recently been highlighted by in vitro directed evolution studies where Im9 was evolved towards Im7 using both positive and negative selection methods (15 and 42). High-affinity, high-selectivity mutants exhibited multiple loop I mutations that generally appeared (in terms of the evolutionary trajectory) before mutations in direct contact with the interface (42). Loop mutations resulted in new (latent) contacts forming between the Im protein and the DNase due to a slightly altered binding configuration on the enzyme surface midway between that of Im9 and Im7. While the binding configuration of the Im protein in the E9 DNase-Im2 complex does not change the orientation of binding appreciably it nevertheless concurs with the directed evolution experiments; loop I substitutions affect binding specificity at the core of the interface by influencing key interactions with the predicted E9 DNase hotspot residue, Y83.
The Role of Bound Waters in Specificity.
Bound water molecules are at least as abundant at protein-protein interfaces as direct hydrogen bonds and are known to play a major stabilizing role through provision of bridging hydrogen bonds and by sculpting protein surfaces to improve interface complementarity (9, 43, 44). Here we find three roles for interface waters: First, where interface residues are conserved between cognate (E9 DNase-Im9) and noncognate (E9 DNase-Im2) complexes, bridging water molecules provide numerous interfacial hydrogen bonds and improve interface complementarity (Fig. 2A, Table 1). Second, where interface residues are variable, water molecules tend to offset unfavorable contacts (e.g., F86-D33; Fig. 2A). Third, water molecules also appear to act as spacers blocking correct loop alignment (e.g., E9 DNase Y83 to Im2 G27; Fig. 3B). Water molecules therefore play integral yet varied roles in shaping colicin DNase-Im protein specificity. This results in significantly more interface water being found in the noncognate than cognate interface (8 vs. 5 water molecules with ASA < 5 Å2), suggesting entropy gain on exclusion of interfacial water may be a means of heightening the affinity of cognate binding, which is borne out by calorimetric measurements (45). Greater interface hydration is likely a direct consequence of the conformational dynamics of noncognate vs. cognate complexes (29) and may be a contributing factor in the evolution of new colicin specificities.
Frustration at the Core of the DNase-Im Protein Interface.
The structure of E9 DNase-Im2 contains a relatively rare example of a highly unfavorable interaction between two amino acid sidechains, E9 DNase F86 and Im2 D33. While coplanar aromatic-carboxylate interactions are occasionally observed (46) deviations from such geometry are rare and thus likely to have a critical impact on affinity. In colicin complexes, binding within conserved parts of the interface, centring on the Im and DNase hotspots, is so highly optimized that the Phe and Asp are forced together and the resulting noncognate interface exhibits frustration. Hence optimal PPI specificity may follow similar principles to protein folding, as illustrated, for example, by frustration in the folding of Im7 (which exhibits an on-pathway intermediate) which is relieved on binding its cognate partner, E7 DNase (47). Frustration at the core of the E9 DNase-Im2 complex is given further credence by residue S78 of E9 DNase, where two equally populated conformers are observed in the electron density. Both conformers are within range of hydrogen bonding to D33 via bridging to a water molecule (Table S3), suggesting local instability in the F86-D33 region that is not observed in the cognate complex. In summary, the structure of the noncognate E9 DNase-Im2 complex suggests that frustration acts to tune the affinity of noncognate binding away from the cognate ideal in such a way as can be rescued by mutations in strategic positions (see below).
Dual Recognition—an Economical Strategy for Evolving Markedly Differing Specificities.
We have shown previously that the threshold colicin DNase-Im protein KD for cell survival is ∼10-10 M (26) (an effective concentration of less than one molecule per cell) even though fM affinity is encoded in a cognate interface. Previous studies suggest that the conserved hotspot of an Im protein provides a basal affinity of ∼10-6 M (26 and 29). How then does specificity develop in the first instance and mature towards cognate levels? Directed evolution studies and our observations above point towards Im loop mutations, which can modulate latent specificity contacts from preexisting residues in helix II (42), as the initial drivers of specificity. If these interactions improve binding to nanomolar levels—hence ensuring some surviving bacterial colonies—then colicin intoxication would represent a significant selective pressure for further mutations at key helix II specificity sites. Through the release of frustration at the core of the interface these later mutations greatly accentuate affinity maturation—a single mutation (e.g., position 33) can confer an increase in E9 vs. E2 DNase specificity of ∼104-fold (27). Cooperative interactions with conserved hotspot residues at the core of the interface likely explain the overcompensation in affinity, relative to that needed for cell survival, which may act as a buffer against deleterious mutations that reduce propagation of the new colicin specificity. The structure of E9 DNase-Im2 begins to explain how enormous differences in colicin DNase-Im binding affinity can be achieved with a minimal number of interface mutations, where affinity is modulated by the influence of unfavorable neighboring contacts. We predict this strategy of economy is essential to colicin-mediated competition as it allows new specificities to emerge and, perhaps more importantly, to persist where they would otherwise die out. The key to the success of this process is the interplay between a high-affinity, highly-optimized interface and a neighboring group of variable residues centered on the immunity specificity helix, but including contributions from loops and buried water molecules.
Materials and Methods
Construct Design and Protein Purification.
Im2 E31C and E9 DNase G95C were constructed using QuikChange mutagenesis (Stratagene). E. coli JM83 and BL21(DE3) pLysS strains were used for protein expression and proteins purified as previously described (24, 27, 48, 49).
Disulfide Complex Formation and Purification.
Im2 E31C (final concentration, 3.0 μM) and E9 DNase G95C (final concentration, 1.6 μM) in 25 mM Tris, 1 mM DTT, pH 8.0, were added to 1L 50 mM Tris-HCl, pH 8.0 (stirring, 20 °C). H2O2 was added to a final concentration of 10 μM and the mixture incubated (stirring, 20 °C, 1.5 h). The sample was concentrated by ultrafiltration, purified by reverse-phase HPLC (C8 matrix) and lyophilized from 20 mM ammonium acetate pH 7.0 (two rounds).
Crystallisation and Structure Determination.
Crystals were obtained using the hanging-drop vapor diffusion method (0.2 M NaNO3, 20% PEG-3350; 0.5 μL drops, 20 °C) and flash cooled using 20% glycerol as a cryoprotectant. Data were collected on a single crystal at 100 K at the European Synchrotron Radiation Facility Grenoble (beamline ID29) to 1.77 Å resolution. For further details of structure determination see SI Text.
Computational Alanine Scanning.
Computational alanine scanning of the E9 DNase-Im2 structure was carried out using the Rosetta software suite for macromolecular modeling (as in refs. 35, 38, 50). Details of the calculation are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Johan Turkenburg and Sam Hart for help with crystallographic data collection and Chris Spronk and Alexandre Bonvin for their advice on the use of the program HADDOCK. We also thank the referees of this paper for their helpful and constructive comments. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC). S.J.F. was supported by a long-term fellowship from the Human Frontier Science Program.
Footnotes
The authors declare no conflict of interest..
This article is a PNAS Direct Submission.
Data deposition: The coordinates of E9 DNase-Im2 have been deposited in the RCSB Protein Data Bank (2WPT).
References
- 1.Han JD, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 3.Humphris EL, Kortemme T. Design of multi-specificity in protein interfaces. PLoS Comput Biol. 2007;3:1591–1604. doi: 10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheridan C. TeGenero fiasco prompts regulatory rethink. Nat Biotechnol. 2006;24:475–476. doi: 10.1038/nbt0506-475. [DOI] [PubMed] [Google Scholar]
- 5.Reichmann D, et al. The molecular architecture of protein-protein binding sites. Curr Opin Struc Biol. 2007;17:67–76. doi: 10.1016/j.sbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Vajda S, Kozakov D. Covergence and combination of methods in protein-protein docking. Curr Opin Struc Biol. 2009;19:164–170. doi: 10.1016/j.sbi.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 8.Rühlmann A, et al. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. J Mol Biol. 1973;77:417–436. doi: 10.1016/0022-2836(73)90448-8. [DOI] [PubMed] [Google Scholar]
- 9.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 10.Chothia C, Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- 11.Janin J, Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 12.Laskowski M, Jr, Qasim MA, Yi Z. Additivity-based prediction of equilibrium constants for some protein-protein associations. Curr Opin Struc Biol. 2003;13:130–139. doi: 10.1016/s0959-440x(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 13.Kortemme T, Baker D. Computational design of protein-protein interactions. Curr Opin Chem Biol. 2004;8:91–97. doi: 10.1016/j.cbpa.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Karanicolas JKB. Computational design of affinity and specificity at protein-protein interfaces. Curr Opin Struc Biol. 2009;19:1–6. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernath K, Magdassi S, Tawfik DS. Directed evolution of protein inhibitors of DNA-nucleases by in vitro compartmentalization (IVC) and nano-droplet delivery. J Mol Biol. 2005;345:1015–1026. doi: 10.1016/j.jmb.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Thom G, et al. Probing a protein-protein interaction by in vitro evolution. Proc Natl Acad Sci USA. 2006;103:7619–7624. doi: 10.1073/pnas.0602341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortemme T, et al. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 18.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J Mol Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Potapov V, et al. Computational redesign of a protein-protein interface for high affinity and binding specificity using modular architecture and naturally occurring template fragments. J Mol Biol. 2008;384:109–119. doi: 10.1016/j.jmb.2008.08.078. [DOI] [PubMed] [Google Scholar]
- 20.Yosef E, Politi R, Choi MH, Shifman JM. Computational design of calmodulin mutants with up to 900-fold increase binding specificity. J Mol Biol. 2009;385:1470–1480. doi: 10.1016/j.jmb.2008.09.053. [DOI] [PubMed] [Google Scholar]
- 21.Kleanthous C. Protein-Protein Recognition. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 22.Kleanthous C, Walker D. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem Sci. 2001;26:624–631. doi: 10.1016/s0968-0004(01)01941-7. [DOI] [PubMed] [Google Scholar]
- 23.Kleanthous C, et al. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 24.Wallis R, Moore GR, James R, Kleanthous C. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion-controlled association and femtomolar binding for the cognate complex. Biochemistry. 1995;34:13743–13750. doi: 10.1021/bi00042a004. [DOI] [PubMed] [Google Scholar]
- 25.Wallis R, et al. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry. 1995;34:13751–13759. doi: 10.1021/bi00042a005. [DOI] [PubMed] [Google Scholar]
- 26.Li W, et al. Highly discriminating protein-protein interaction specificities in the context of a conserved binding energy hotspot. J Mol Biol. 2004;337:743–759. doi: 10.1016/j.jmb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Li W, et al. Dual recognition and the role of specificity-determining residues in colicin E9 DNase-immunity protein interactions. Biochemistry. 1998;37:11771–11779. doi: 10.1021/bi9808621. [DOI] [PubMed] [Google Scholar]
- 28.Wallis R, et al. Specificity in protein-protein recognition: conserved Im9 residues are the major determinants of stability in the colicin E9 DNase-Im9 complex. Biochemistry. 1998;37:476–485. doi: 10.1021/bi971884a. [DOI] [PubMed] [Google Scholar]
- 29.Keeble AH, Kleanthous C. The kinetic basis for dual recognition in colicin endonuclease-immunity protein complexes. J Mol Biol. 2005;352:656–671. doi: 10.1016/j.jmb.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Ko TP, et al. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7:91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 31.Kuhlmann UC, et al. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J Mol Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 32.Darby N, Creighton TE. Disulfide bonds in protein folding and stability. Methods in Molecular Biology. 1995;40:219–252. doi: 10.1385/0-89603-301-5:219. [DOI] [PubMed] [Google Scholar]
- 33.Greene NP, et al. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J Biol Chem. 2007;282:23937–23945. doi: 10.1074/jbc.M702972200. [DOI] [PubMed] [Google Scholar]
- 34.Buck E, Wells JA. Disulfide trapping to localize small-molecule agonists and antagonists for a G protein-coupled receptor. Proc Natl Acad Sci U S A. 2005;102:2719–2724. doi: 10.1073/pnas.0500016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keeble AH, et al. Experimental and computational analyses of the energetic basis for dual recognition of immunity proteins by colicin endonucleases. J Mol Biol. 2008;379:745–759. doi: 10.1016/j.jmb.2008.03.055. [DOI] [PubMed] [Google Scholar]
- 36.Glaser F, Steinberg DM, Vakser IA, Ben-Tal N. Residue frequencies and pairing preferences at protein-protein interfaces. Proteins. 2001;43:89–102. [PubMed] [Google Scholar]
- 37.Ueda Y, Taketomi H, Go N. Studies on protein folding, unfolding, and fluctuations by computer simulation. II. A. Three-dimensional lattice model of lysozyme. Biopolymers. 1978;17:1531–1548. [Google Scholar]
- 38.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Science Signaling - The Signal Transduction Knowledge Environment pI2. 2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 39.Wallis R, et al. Specificity in protein-protein recognition: conserved Im9 residues are the major determinants of stability in the colicin E9 DNase-Im9 complex. Biochemistry. 1998;37:476. doi: 10.1021/bi971884a. [DOI] [PubMed] [Google Scholar]
- 40.Li W, et al. Protein-protein interaction specificity of Im9 for the endonuclease toxin colicin E9 defined by homologue-scanning mutagenesis. J Biol Chem. 1997;272:22253–22258. doi: 10.1074/jbc.272.35.22253. [DOI] [PubMed] [Google Scholar]
- 41.Riley MA, Wertz JE. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84:357–364. doi: 10.1016/s0300-9084(02)01421-9. [DOI] [PubMed] [Google Scholar]
- 42.Levin KB, et al. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009;16:1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- 43.Rodier F, Bahadur RP, Chakrabarti P, Janin J. Hydration of protein-protein interfaces. Proteins. 2005;60:36–45. doi: 10.1002/prot.20478. [DOI] [PubMed] [Google Scholar]
- 44.Janin J. Wet and dry interfaces: the role of solvent in protein-protein and protein-DNA recognition. Structure. 1999;7:R277–279. doi: 10.1016/s0969-2126(00)88333-1. [DOI] [PubMed] [Google Scholar]
- 45.Keeble AH, Kirkpatrick N, Shimizu S, Kleanthous C. Calorimetric dissection of colicin DNase–immunity protein complex specificity. Biochemistry. 2006;45:3243–3254. doi: 10.1021/bi052373o. [DOI] [PubMed] [Google Scholar]
- 46.Jackson MR, et al. A preference for edgewise interactions between aromatic rings and carboxylate anions: the biological relevance of anion-quadrupole interactions. J Phys Chem B. 2007;111:8242–8249. doi: 10.1021/jp0661995. [DOI] [PubMed] [Google Scholar]
- 47.Sutto L, et al. Consequences of localized frustration for the folding mechanism of the IM7 protein. Proc Natl Acad Sci USA. 2007;104:19825–19830. doi: 10.1073/pnas.0709922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garinot-Schneider C, et al. Identification of putative active-site residues in the DNase domain of colicin E9 by random mutagenesis. J Mol Biol. 1996;260:731–742. doi: 10.1006/jmbi.1996.0433. [DOI] [PubMed] [Google Scholar]
- 49.Wallis R, et al. In vivo and in vitro characterization of overproduced colicin E9 immunity protein. Eur J Biochem. 1992;207:687–695. doi: 10.1111/j.1432-1033.1992.tb17096.x. [DOI] [PubMed] [Google Scholar]
- 50.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.