Abstract
RNA virus polymerases must initiate replicative RNA synthesis with extremely high accuracy to maintain their genome termini and to avoid generating defective genomes. For the single-stranded negative-sense RNA viruses, it is not known how this accuracy is achieved. To investigate this question, mutations were introduced into the 3′ terminal base of a respiratory syncytial virus (RSV) template, and the RNA products were examined to determine the impact of the mutation. To perform the assay, RNA replication was reconstituted using a modified minireplicon system in which replication was limited to a single step. Importantly, this system allowed analysis of RSV RNA generated intracellularly, but from a defined template that was not subject to selection by replication. Sequence analysis of RNA products generated from templates containing 1U-C and 1U-A substitutions showed that, in both cases, replication products were initiated with a nontemplated, WT A residue, rather than a templated G or U residue, indicating that the polymerase selects the terminal NTP independently of the template. Examination of a template in which the position 1 nucleotide was deleted supported these findings. This mutant directed efficient replication at ∼60% of WT levels, and its product was found to be initiated at the WT position (−1 relative to the template) with a WT A residue. These findings show that the RSV replicase selects ATP and initiates at the correct position, independently of the first nucleotide of the template, suggesting a mechanism by which highly accurate replication initiation is achieved.
Keywords: Mononegavirales, paramyxovirus, RNA-dependent RNA polymerase, initiation
During RNA virus genome replication, the virus polymerase has to be able to initiate with a mechanism that will allow it to conserve the genome terminal sequences. RNA viruses have evolved several sophisticated mechanisms to achieve this (reviewed in ref. 1). For example, the picornaviruses use protein-primed initiation (2, 3). Hepatitis C virus polymerase initiates independently of a primer (de novo initiation), but with a mechanism that involves specific structural features in the polymerase and interactions between the template and initiating NTP that aid accurate positioning of the RNA 3′ terminus (4–9). The polymerases of double-stranded RNA viruses, ϕ6 and rotavirus, have highly specific interactions with the template also, and the rotavirus polymerase can generate a dinucleotide primer independently of the template, which might ensure correct positioning of the polymerase and faithful replication of the termini (10–13). Members of the segmented negative-strand viruses can initiate replication using a prime-realign mechanism, in which the polymerase initiates internally on the RNA to generate a short oligonucleotide primer, and then repositions so that the primer anneals to the 3′ terminus of the template (14, 15).
In contrast to the viruses described above, little is known regarding the mechanism of replication initiation in the nonsegmented negative strand RNA viruses, of which RSV, a member of the family Paramyxoviridae, is an example. At the 3′ and 5′ ends of the RSV genome are extragenic regions, known as leader (Le) and trailer (Tr), respectively (16). Encapsidated genome RNA acts as a template for replication. The polymerase initiates replication at a promoter within the Le, and continues along the length of the genome to produce a complementary antigenome RNA. The nascent RNA is encapsidated as it is synthesized, and there is evidence that concurrent encapsidation is necessary for replicase processivity (17–19). The Tr-complement (TrC) region at the 3′ end of the antigenome contains a promoter that in turn signals genome RNA synthesis and encapsidation (20). Previously, studies were performed to determine the template requirements for RSV replication. Nucleotides 1–13 of the Le promoter region are sufficient to signal RNA synthesis initiation (18, 21), and saturation mutagenesis analysis showed that nucleotides 3, 4, 5, 8, 9, 10, and 11 comprise a highly stringent promoter element that likely functions in polymerase recruitment and/or another aspect of RNA synthesis initiation (22, 23). Other sequences in the Le, up to position 36, are also required for RNA replication to occur, and evidence suggests that these sequences are required for encapsidation and replicase processivity (18). Studies that investigated the role of the 3′ terminus of the template RNA in directing replication revealed that the promoter must be at or near the 3′ end of the template to be active (21). However, the 3′ terminus of the RNA template does not determine the site of replication initiation, as the polymerase initiated correctly at the first position of the authentic Le sequence when there was an additional four nucleotides between residue 1 of the Le sequence and the 3′ end of template (21). These studies suggest the RSV polymerase binds to a sequence within bases 1–11 of the Le and is positioned such that it is constrained to initiate opposite position +1 of the promoter. Fewer studies have been performed on the TrC promoter region. This promoter is more active for replication than the Le, but its structural organization appears to be similar (24). Like the Le, the first 36 nucleotides of the TrC region are sufficient to signal replication initiation and the two promoter regions are very similar in sequence. Indeed, the first 11 bases of Le and TrC are identical, except at position 4; and mutation analysis of the first seven bases of the TrC promoter showed that the positions responded similarly to substitution, with the exception of positions 1 and 2, which showed slight differences (23, 25). Thus, it is believed that template requirements for replication initiation are similar in the Le and TrC promoters. Given that the RSV polymerase appears to be constrained to initiate opposite the first nucleotide of the Le, the aim of this study was to investigate the role of first position of the TrC promoter in replication initiation.
Results
Design of an Assay to Examine RSV Replication Initiation.
Experiments were performed using an intracellular plasmid-based mini-replicon system, which has been used extensively to study RSV transcription/replication mechanisms (e.g., refs. 18, 21, and 23–28). The minireplicon template is 900 nucleotides in length. The 3′ end consists of the first 36 nucleotides of the TrC region fused directly to chloramphenicol acetyl transferase (CAT)-specific sequence (Fig. 1). The 3′ end of the template is generated by the hepatitis delta virus antigenomic ribozyme (29). Studies of this ribozyme have shown that its cleavage site is not affected by the nature of the adjacent sequence (30, 31). Thus, it was possible to generate RSV templates with defined mutations within the first few nucleotides of the TrC promoter. For this study, it was important to ensure that the mutant templates being tested could not revert to a WT sequence. When reversion occurs, the repaired template rapidly replaces the mutant template population by amplification and selection, making it impossible to determine if a particular initiation event is occurring from a mutant or WT promoter. Therefore, the 5′ terminal 22 nucleotides of the Tr region of the minireplicon were deleted (Fig. 1). This deletion removed the promoter that would normally lie at the 3′ end of the primary replication product and so would be expected to restrict the minireplicon to a single step of RNA replication, thus avoiding the possibility of reversion. RSV RNA replication was reconstituted in cells by coexpressing minireplicon RNA together with the RSV proteins required for RNA synthesis in a transient transfection, and the RNA products generated from the minireplicon templates analyzed.
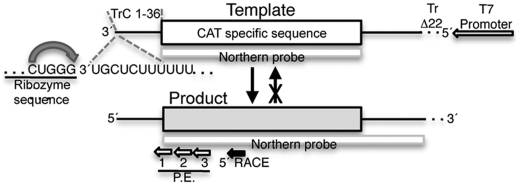
Fig. 1.
Schematic diagram (not to scale) illustrating the minireplicon template used in this study. The 3′ end of the minireplion is created by a ribozyme (curved arrow) and consists of the first 36 nucleotides of TrC sequence. Note that this promoter sequence lacks transcription-specific signals and so is optimized for detection of replication products. The 5′ end of the template contains the RSV Tr region, with a 22-nucleotide deletion at the 5′ terminus, indicated with a dotted line. This modification prevents the primary replication product from acting as a template for new minireplicons, as depicted by the arrow with a cross. Sites where the Northern blot riboprobes hybridize to the minireplicon template and replication product, and where reverse transcription primers for primer extension (P.E.) and 5′ rapid amplification of cDNA ends (5′ RACE) bind the replication product, are indicated.
RSV Polymerase Can Tolerate Mutations at First Position of Viral Promoter.
To determine the role of the first position of the RSV template in RNA replication, the effect of substituting residue 1U of the promoter to an A, C, or G was examined. RNA polymerases generally prefer to initiate RNA synthesis with a purine residue (32). Therefore, it was anticipated that a position 1 pyrimidine-to-pyrimidine mutation (1U-C) might support replication initiation, whereas the pyrimidine-to-purine mutants (1U-A and 1U-G) might not. In addition, deletion of position 1U (Δ1U) was examined. As the RSV polymerase is constrained to initiate RNA synthesis at the first position of the promoter, it was predicted that the Δ1U mutant would not support RNA replication.
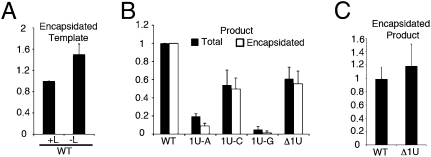
To assess the impact of the mutations on RNA replication, the levels of full-length replication products were determined by Northern blotting with strand specific probes (Fig. S1), followed by phosphorimage analysis (Fig. 2). Total intracellular RNA and RNA from micrococcal nuclease (MCN) treated cell lysates was analyzed. Encapsidated RNA is resistant to nuclease digestion, allowing confirmation of replicative template and product levels. Analysis of the input template RNA showed that addition of the plasmid encoding the major polymerase subunit L to transfections containing the WT minireplicon did not increase the amount of encapsidated template RNA present (Figs. 2A and S1A, comparing lanes 1 and 2). Thus, there was no amplification of the input minireplicon by the RSV polymerase, confirming that the minireplicon template was limited to a single step of replication. Analysis of the input RNA from transfections with the WT and mutant minireplicons indicated similar levels of encapsidated template were present in each case (Fig. S1A, lanes 2–6).
Fig. 2.
Impact of mutating the 3′ terminal nucleotide of the template on RSV RNA replication. (A) Quantitation of encapsidated template generated in transfections in which plasmid encoding L was present (+L) or absent (-L). (B) Quantitation of replication products generated from the mutant minireplicons. Black bars represent the replication product in the total RNA samples; white bars represent replication product measured in the MCN-treated RNA samples. (C) Direct comparison of the amounts of total and MCN-resistant replication product generated in the WT and Δ1U reactions. Each RNA value was determined based on Northern blot analysis, such as that shown in Fig. S1. Levels were calculated relative to the respective WT value for A and B, or relative to the WT or Δ1U total RNA values for C (set at 1.0 in each case). Each error bar represents the standard error of the mean from at least three independent experiments.
Examination of the products of RNA replication showed that, of the substitution mutants, the 1U-C mutant minireplicon supported the highest level of replication, as predicted, at ∼50% of WT levels. The 1U-A and 1U-G mutants directed lower levels at ∼20% and ∼2% of WT levels, respectively. Surprisingly, the Δ1U minireplicon generated relatively high levels of replication product, at ∼60% of the WT levels (Figs. 2B and S1B). These data show that the presence of the 3′ terminal uridylate is not critical for replication initiation, but that substitutions at this position are deleterious, and that different mutations affect replication to different extents. In each case, similar levels of replication product, relative to WT levels, were seen in both total and MCN-treated RNA samples (Figs. 2B and S1 B and C), and direct comparison of the amounts of MCN resistant compared with total replication product in the WT and Δ1 samples showed that essentially all of the full-length replication product was encapsidated (Fig. 2C). The impact of substituting positions 2 and 3 to each alternative NTP or deleting the first two residues of the minireplicon template was also examined (Figs. S2 and S3). Unlike the position 1 changes, all of these mutations ablated replication. These data are consistent with those of a previous study (25), and show that positions 2 and 3 play a key role in RNA replication.
RSV Polymerase Can Initiate at “Correct” Position on Mutant Templates.
The RNAs generated from the 1U-A, 1U-C, and Δ1U mutant templates were then examined by primer extension analysis to determine where the replication products were initiated (the 1U-G mutant was not analyzed further because of the very low levels of replication product that it generated). In the case of the 1U-C mutant, it was possible that the polymerase would be able to initiate opposite position 1, as this would still involve initiating with a purine residue. In contrast, initiation at position 1 of the 1U-A mutant would be expected to use a pyrimidine as the initiating residue, which the polymerase might not be able to accommodate, and so the 1U-A substitution might cause the polymerase to initiate elsewhere, for example, at position 4, the next uridylate in the template. Likewise, the Δ1U mutant might direct initiation from another site.
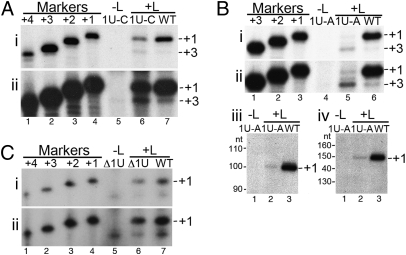
Total RNA rather than nuclease-treated RNA was analyzed, with the goal of examining as many initiation products as possible, including those that might not be efficiently encapsidated. Primer extension analysis was performed using a primer that hybridized close to the 5′ end of the product (bases 24–48 relative to the WT 5′ end; Fig. 1, primer 1), to detect initiations that might not be efficiently elongated and also to allow precise sizing of the cDNAs. This analysis showed that 1U-C RNA synthesis was frequently initiated at the same position as the RNA from the WT minireplicon, at position 1 of the template (Fig. 3Ai, comparing lanes 4, 6, and 7). Longer exposures also showed evidence for initiation at position 3C, the next pyrimidine residue (Fig. 3Aii). There was evidence for +3 initiations from the WT template also, demonstrating that the site is used to a minor extent, even with optimal template sequence. Primer extension analysis of the 1U-A product detected initiation from both positions +1 and +3 of the template, with initiations from position 3 being dominant by approximately 9-fold (Fig. 3 Bi and Bii). This suggested that although the polymerase could initiate at position 1 from this mutant template, this initiation site was less favored, as might be expected. Surprisingly, primer extension products from the Δ1U and WT reactions aligned with each other, demonstrating that the Δ1U mutant could direct initiation from the −1 position relative to the template (Fig. 3C, lanes 4, 6, and 7). There was no evidence of initiations at any other position on this template. These data show that the RSV polymerase has a strong preference to initiate at the correct position relative to the promoter, even in a situation that involves initiating RNA synthesis at the –1 position relative to the input template.
Fig. 3.
Effect of the 3′ terminal mutations on initiation site selection. Primer extension analysis of the RNA generated from the 1U-C, 1U-A, and Δ1U mutant minireplicons (A, B, and C, respectively). In each case, the mutant RNA was compared with RNA generated from a WT minireplicon. In i and ii, the primer hybridized at positions 24–48 relative to the +1 initiation product. Lane 4 or 5 (as indicated) is a negative control of RNA from cells transfected with plasmid encoding the relevant mutant minireplicon but no L polymerase plasmid. Molecular weight markers present in lanes 1–3 or 1–4 are end-labeled primers, representing products initiated from positions +3 to +1 or +4 to +1 of the template, respectively. In each case, ii is a longer exposure of the gel shown in i to show the +3 initiations. (B iii and iv) Primer extension analysis of the same RNA samples as shown in i and ii, using primers that hybridized at positions 74–100 (iii) or 124–148 (iv) relative to the 5′ end of the +1 initiation product.
RNAs Initiated at Position 3 Are Not Efficiently Extended.
There is accumulating evidence for the Mononegavirales that sequences at the 5′ end of an RNA product can affect polymerase processivity on the RNA template (reviewed in ref. 22). Therefore, it was of interest to determine whether the RNA initiated at position 3 (and lacking the 5′ terminal two nucleotides) was efficiently extended to the end of the template. To examine this, RNA from the 1U-A mutant was analyzed by primer extension with primers that hybridized at increased distances from the 5′ end of the product (Fig. 1, primers 2 and 3). This analysis showed that whereas the primer that hybridized within 50 bases from the 5′ end of the product predominantly detected RNA initiated at position 3 on the 1U-A minigenome, primers that hybridized 74–100 and 124–148 bases from the 5′ end of the product only detected the RNA that was initiated at position 1 (Fig. 3 Biii and Biv). Thus, it appears that, whereas RNA initiated at position 1 was efficiently extended, RNA initiated at position 3 was elongated only a distance of between 50 and 100 nucleotides.
RSV Polymerase Selects 5′ Terminal NTP Independently of Template.
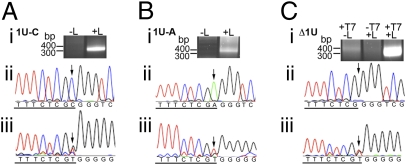
As it was found that the RSV polymerase can initiate RNA replication at the correct position, even if the template contains an unfavorable residue or is lacking a nucleotide at this site, the 5′ ends of the replication products were sequenced to determine what nucleotide was incorporated at the first position. 5′ RACE was performed using a primer that hybridized 290 bases from the 5′ end of the RNA product (Fig. 1), such that +1 initiation products were selectively amplified. Comparison of 5′ RACE products from transfection samples lacking or containing L plasmid confirmed that in each case the 5′ RACE reaction was specific for RSV replication products (Fig. 4 A–C, i). The 5′ RACE products were sequenced directly to identify the dominant sequence(s) in the population. Surprisingly, analysis of the RNA generated from the 1U-C template showed that most products contained an ATP at position 1 (indicated by a T residue in the sequence trace; Fig. 4Aiii). Thus, the 5′ of the product contained the WT but nontemplated residue. A smaller C peak was also observed at this position, suggesting that a smaller fraction of the population contained templated G residue at the 5′ terminus. The sequence of the transfected DNA plasmid is shown for comparison in Fig. 4Aii, and the result was confirmed in experiments performed with a C tailing reaction (Fig. S4 Ai and Aii). To gain quantitative information on the frequency with which the RSV polymerase could initiate with a nontemplated ATP, 5′ RACE products were cloned and individually sequenced (Table S1 and Fig. S4B). This analysis showed that a significant majority of clones (37/46) were initiated at the correct position with a nontemplated A residue, whereas a much smaller fraction (4/46) were initiated with a templated G residue.
Fig. 4.
Sequence analysis of RNA produced from mutant templates. 5′ RACE analysis of the RNA generated from the 1U-C, 1U-A, and Δ1U mutants (A, B, and C, respectively). In each case, i shows agarose gel electrophoresis of the 5′ RACE product generated from RNA from cells transfected with plasmid encoding the relevant mutant minireplicon and either lacking or containing the L polymerase plasmid or lacking MVA-T7, as indicated; ii shows sequence analysis of the input minireplicon plasmid DNA isolated from the transfected cells, and iii shows the corresponding sequence trace derived from the bulk 5′ RACE product (tailed with dGTP). The sequence traces are presented as template sense DNA; TrC sequence is underlined, with position 1 relative to the WT template indicated by an arrow. Sequencing of 5′ RACE reactions tailed with dCTP and of clonal isolates (Fig. S4) showed that the G peak at position 1 in iii is a sequencing artifact.
The same analyses were undertaken with the 1U-A replication products. Similarly to the results observed with the 1U-C mutant template, sequence analysis revealed that the replication products were frequently initiated with a nontemplated WT ATP, with 31/36 clones showing this assignment (Fig. 4B, Table S1, and Fig. S4). Thus, the data from the reactions involving the 1U-C and 1U-A mutants demonstrate that during initiation opposite position 1 of the promoter, the RSV polymerase favors initiating replication with a nontemplated WT ATP over Watson–Crick base pairing between the template and daughter strands.
Sequence analysis of the RNA generated from the Δ1U mutant showed that the RNA was initiated at the WT (−1) position relative to the template, confirming the primer extension data. Similarly to the substitution mutants, this template generated replication products that contained nontemplated ATP at their 5′ termini. In this case, ATP was found with 100% frequency (58/58 clones; Fig. 4C, Table S1, and Fig. S4). This result shows that the RSV polymerase initiated replication at the –1 position relative to the 3′ end of the Δ1U mutant template and preferentially inserted a WT ATP at the first position of the product, despite having no template base present with which the ATP could interact.
Discussion
The sequences required for RSV RNA replication are circumscribed to the 3′ terminus of the template and do not involve repetitive sequences or require terminal complementarity (16, 20, 24). For this reason, RSV RNA replication is believed to initiate opposite the 3′ terminal nucleotide of the template by a de novo mechanism in which the initiating NTP essentially acts as a primer for creation of the first phosphodiester bond. The results presented here show that when the 3′ terminal nucleotide of the TrC promoter was substituted or deleted, replication still frequently initiated at position +1 relative to the WT template. In contrast, nucleotides 2G and 3C were essential for initiation and full-length replication (Figs. S2 and S3). The ability to initiate with limited reference to the 3′ end of the template is consistent with our previously published data that showed that the RSV polymerase can initiate RNA replication at an internal site, as has also been shown for the paramyxovirus Sendai virus (21, 33), and suggests that the promoter sequence is the major feature anchoring the polymerase and directing start site selection. The data also show that the RSV polymerase could select the position 1 ATP independently of the first base of the template, and importantly, could use the ATP (presumably to prime initiation) even when there was no standard Watson–Crick bond between the ATP and the template. It should be noted that the residue at the 5′ end of the nascent replicative RNA might be important for polymerase processivity and RNA stability, such that 5′ A containing RNAs are extended and stabilized, potentially biasing the results somewhat. Nonetheless, the nonamplifying minireplicon system that was used for these studies provides evidence that these nontemplated initiation events occur at high frequency: the Δ1U mutant generated nontemplated replication products at ∼60% of WT levels, suggesting that the nontemplated mechanism of initiation identified in this study is relevant for understanding what occurs during normal replication initiation. Other virus polymerases have been shown to have a strong preference for the initiating NTP (8, 9, 13, 34, 35). However, the 3′ nucleotide of the template of other viruses plays a key role in initiation, with mutations at this position significantly hindering replication (34, 36, 37), suggesting that the 3′ terminal nucleotide is usually important for stabilizing the interactions between the polymerase and the initiating NTP. The findings described here differ, suggesting that RSV uses a distinct mechanism of initiation.
The data presented here, together with our earlier studies of RSV promoter function, suggest a model for RSV RNA replication initiation, illustrated in Fig. 5A. According to this model, the RSV polymerase complex binds to nucleotides 3, 5, 8, 9, 10, and 11 in the RNA promoter region (18, 21, 23), but is positioned such that its active site is located opposite nucleotides 1 and 2 of the template. ATP and CTP are recruited into the priming and incoming NTP sites of the polymerase, respectively. One possibility is that ATP is engaged first, solely by virtue of specific interactions of the adenine base with the polymerase complex (this could potentially occur before template binding). Hydrogen bond pairing between the CTP and the template guanylate could stabilize the complex, allowing the polymerase to catalyze phosphodiester bond formation between the ATP and CTP residues. Alternatively CTP could be engaged first, followed by recruitment of ATP because of specific interactions with the polymerase and/or base stacking with CTP. Phosphodiester bond formation between ATP and CTP would then initiate RNA synthesis. If either version of the model is correct, it represents a highly coordinated mechanism that would enable RSV to ensure accurate initiation and maintenance of its genome termini. Currently, there is no in vitro assay or high-resolution structure available for the RSV polymerase to confirm this model. However, a related virus, vesicular stomatitis virus (which also has the sequence 5′ AC at the end of its replication products) has been shown to have an ATP binding site on the polymerase, providing support for this model (38). Furthermore, in the present study, the levels of replication product generated from the different mutants are consistent with the model. 1U-A and 1U-G substitutions would result in a larger base being present in template binding site of the polymerase, which would be predicted to be incompatible with the recruited ATP, resulting in a decrease in replication levels. Conversely the model would predict that the 1U-C substitution would be tolerated to some extent, as the space occupancy of a C residue is similar to that of a U, allowing accommodation of ATP. Likewise, the lack of a template base at position 1 of the Δ1U minireplicon would be expected to result in the highest levels of replication, as it would enable the viral polymerase to select an ATP without constraint. This is the pattern that was observed (Fig. 2).
Fig. 5.
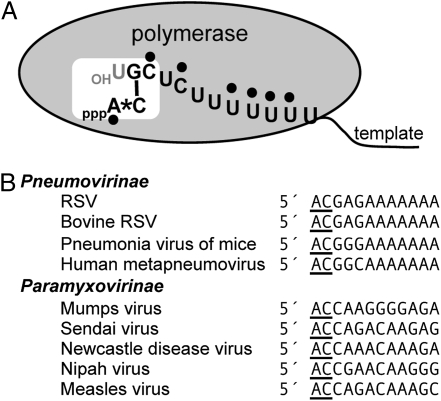
Conserved mechanism for replication initiation. (A) Schematic diagram depicting a proposed model for RSV replication initiation (detailed in text). White box depicts active site of polymerase. Black dots represent putative interactions between the polymerase and the 5′ terminal ATP and the template. Asterisk represents a putative stacking interaction between the ATP and CTP residues. Black bar represents hydrogen bonding. The 3′ terminal position of the template (1U) is shown in gray to illustrate that it is not required for initiation. (B) Alignment of the 5′ terminal trailer sequences of representatives of the family Paramyxoviridae. Alternative terminal sequences were found in the databank but appear to be incomplete.
Because of the similarity of the genomic and antigenomic promoter regions, it seems likely that initiation of plus strand replicative RNA from the Le promoter uses a mechanism similar to that which we have observed here for minus strand synthesis, although it should be noted that influenza virus plus and minus strand synthesis can occur through different mechanisms (14). Unfortunately, the WT RSV Le replication promoter is approximately 4-fold less efficient than the TrC promoter, and so a similar analysis to that described here is currently not technically possible. A third type of initiation also occurs in RSV and related viruses, namely subgenomic mRNA initiation from internal gene start signals. In contrast to the data shown here, the RSV polymerase has been shown to have a strong preference for initiating mRNAs with a G residue (39). It is intriguing that the base preferences should be different for RNA replication and mRNA transcription initiation; and this finding suggests that the polymerase might have different conformations that cause it to select different NTPs for the two processes.
Initiations from position +3C were observed in addition to initiations at position 1, with some mutants favoring initiations at position +3 versus +1 (Fig. 3). There was also evidence for some initiations from +3C of the WT template. It is possible that occasionally the replicase fails to initiate at position 1 and initiates at the next available pyrimidine instead, as has been described for HCV and rotavirus (9, 13, 35). However, it is also possible that position +3C is recognized as an initiation site by the viral transcriptase complex, as bases 3–12 of the TrC promoter region share identity with 7/10 bases of the RSV L gene start signal. If this is the case, the significance of such an initiation signal is currently unclear. The data show that whereas RNA initiated at position 1 was elongated, RNA initiated at position 3 was not efficiently extended beyond 50 nucleotides (Fig. 3B). In the case of the 1U-A mutant, most initiations were from position 3, and only a relatively small fraction was from position 1. Therefore, although this mutant could direct initiation from the replication promoter, most of the RNA it generated was not extended, explaining why this mutant yielded so little full-length replication product (Fig. 2 and Fig. S1). The length of the prematurely terminated RNA is consistent with a previous study that suggested that the RSV polymerase has an elongation checkpoint at ≈50 nucleotides (40). Data suggest that replication products must be encapsidated concurrently with synthesis for the replicase to be processive; therefore, a plausible explanation for why this RNA was not extended could be that it was not encapsidated, either because it was lacking an essential cis-acting encapsidation sequence or because it was generated by a transcriptase rather than a replicase.
Although the replication initiation model shown in Fig. 5A is consistent with the data, there are other possible explanations for the results. First, it is possible the 5′ RACE technique is inherently biased toward sequences containing ATP at the 5′ end. However, the same approach has been used for analyzing RSV mRNAs, and these studies detected many sequences containing a GTP and UTP at the 5′ end, depending on the template base at the first position of the gene start signal (39), indicating that the technique is unbiased. Second, a number of RNA viruses have been shown to possess terminal transferase activity (41–44). It is possible that the RSV polymerase shares this activity, allowing it to repair terminal truncations (such as the Δ1U mutation) by adding nontemplated nucleotides to the 3′ end of an RNA. However, the 3′ termini of the templates were generated by the hepatitis delta virus ribozyme, which would create a 2′, 3′ cyclic phosphate group at the end of the RNA, rather than a 3′ hydroxyl group (45). Therefore, these templates are highly unlikely to act as substrates for viral (or cellular) terminal transferase, particularly with the efficiency required to yield the levels of replication product that we observed. Another possibility is that the 2′, 3′ cyclic phosphate itself affects polymerase nucleotide selection; but if this is the case, it is unclear why the substitution and deletion mutants yield similar results. A fourth alternative is that the RSV polymerase recruits a primer from the cytoplasmic milieu to initiate RNA synthesis. The turnip crinkle carmovirus polymerase has been shown to repair truncations at the 3′ end of a subviral satellite RNA C using abortive transcripts derived from the intact turnip crinkle virus genome (46). However, in this study, there was no helper RSV or terminal redundancy in the minireplicon template. Finally, it is possible that RSV uses a prime-realign mechanism for initiation, initiating on an internal UG pair to generate a primer, and then realigning to the 3′ end; however, the only other UG in the first 36 bases of TrC sequence is at positions 24 and 25, and this dinucleotide is not conserved in the TrC promoters of close relatives of RSV, which can signal replication by the RSV polymerase (28). Thus, although other explanations of the data exist, the model shown in Fig. 5A seems the most plausible.
Inspection of paramyxovirus promoter sequences suggests that the results described here may be relevant across the family. There are significant differences between the paramyxovirus promoters in terms of whether they must lie in the correct phase relative to nucleoprotein (47–49), are bi- or monopartite (20, 50, 51), and in their 3′ terminal sequences (Fig. 5B). However, despite these differences, it is striking that the genomes and antigenomes of all paramyxoviruses sequenced to date contain 5′ AC (Fig. 5B). These observations suggest the mechanism by which the polymerase selects the terminal ATP and initiates RNA replication is conserved throughout the Paramyxoviridae.
Materials and Methods
Plasmid Construction and Mutagenesis.
The plasmid encoding the minireplicon templates is based on plasmid MP-28, which has been described previously (23). Briefly, MP-28 carries a dicistronic CAT-containing minireplicon, flanked with a T7 promoter and a hepatitis delta virus ribozyme to generate the RNA template 3′ end. For this study, plasmid MP-28 was modified as follows. First, 22 nucleotides from the 5′ terminus of the Tr region were deleted, and a hammerhead ribozyme was inserted between the remaining Tr region and the T7 promoter. Second, the 3′ end of the minireplicon was replaced with nucleotides 1–36 of the TrC promoter region, such that the promoter sequence directly abutted the nonspecific CAT sequence. This modification removed all known 3′ transcription signals, simplifying the RNA analysis and augmenting replication promoter activity. The minireplicon encoded by this plasmid thus has a WT TrC promoter at its 3′ terminus, and is referred to as the WT template. Substitutions and deletions were made at the 3′ terminus of the WT minireplicon using PCR mutagenesis. The 3′ and 5′ terminal sequences of each minireplicon were confirmed by sequence analysis.
Reconstituted Minigenome Replication and RNA Analysis.
Minireplicon replication was reconstituted in HEp-2 cells, as previously described (26). Cells were simultaneously infected with modified vaccinia virus Ankara-T7 (MVA-T7) to express the T7 polymerase (52). At 48 h RNA was either directly extracted to generate total intracellular RNA, or cells were lysed with nonionic detergent and incubated with MCN before RNA extraction, as described previously (26). RNA was isolated using an RNeasy kit (Qiagen) according to manufacturer's instructions. For Northern blot hybridization, RNA samples were subjected to electrophoresis on formaldehyde-agarose gels, transferred to nitrocellulose, and hybridized with specific riboprobes, as described previously (26, 27). Quantitative analysis was performed with a phosphorimager (Molecular Dynamics). To perform primer extension analysis, 0.2 pmol of 32P end-labeled primer was combined with total intracellular RNA representing one-tenth of a well of a six-well dish. The mixture was heated to 65 °C for 5 min and cooled on ice, and the resultant RNA-DNA hybrid was reverse transcribed using Thermoscript (Invitrogen) or Sensiscript (Qiagen) reverse transcriptase according to the manufacturer's instructions. It should be noted that the same reverse transcriptase was used within each experiment and that both enzymes gave similar results. End-labeled DNA oligonucleotides corresponding to template-sense nucleotides 1-, 2-, 3-, and 4–48 were used as molecular weight markers. Samples were subjected to electrophoresis on a 6% polyacrylamide gel containing 7 M urea and radiolabeled products were detected by autoradiography. For the primer extensions shown in Fig. 3 Biii and Biv, the reactions were migrated alongside a labeled molecular weight ladder. For 5′ RACE, total intracellular RNA representing one-tenth of a well of a six-well dish was annealed to a template-sense CAT-specific primer and reverse transcribed using Sensiscript reverse transcriptase (Qiagen) according to the manufacturer's instructions. Following purification, the cDNA was tailed with dGTP or dCTP using terminal transferase. The tailed cDNA was PCR amplified using a nested CAT-specific primer and a primer that annealed to either the dGTP or dCTP tail. The PCR reactions were carried out in triplicate, the products pooled and gel purified. Products were either sequenced directly using a nested CAT-specific primer, or were cloned into a pGEM vector for sequencing of individual cDNA clones. To eliminate clones in which the reverse transcriptase had terminated prematurely (i.e., that would be lacking the 5′ terminal sequence), the cDNA clones were PCR screened using a primer that hybridized at positions 13–33 of the TrC sequence, before sequence analysis. Minireplicon plasmid DNA present in total intracellular RNA samples was amplified using primers that annealed to CAT-specific and backbone plasmid sequence. The resultant PCR products were gel purified and sequenced using a CAT-specific primer.
Supplementary Material
Acknowledgments
We thank Dr. Peter Collins (National Institute of Allergy and Infectious Diseases, National Institutes of Health) for continued use of RSV minigenome and protein expression plasmids, Dr. Bernard Moss (National Institute of Allergy and Infectious Diseases, National Institutes of Health) for MVA-T7, Drs. E. Mühlberger and J. Connor for critical review of the manuscript, and the two anonymous reviewers for insightful comments. This work was supported by Biotechnology and Biological Sciences Research Council UK Grant 94/P17132 and by National Institutes of Health Grant AI079328 (to R.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913065107/-/DCSupplemental.
References
- 1.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 2.Herold J, Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli S, et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong Z, et al. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology. 2001;285:6–11. doi: 10.1006/viro.2001.0948. [DOI] [PubMed] [Google Scholar]
- 6.Lesburg CA, et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 7.Kao CC, et al. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2000;74:11121–11128. doi: 10.1128/jvi.74.23.11121-11128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo G, et al. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tortorici MA, Broering TJ, Nibert ML, Patton JT. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J Biol Chem. 2003;278:32673–32682. doi: 10.1074/jbc.M305358200. [DOI] [PubMed] [Google Scholar]
- 13.Chen D, Patton JT. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA. 2000;6:1455–1467. doi: 10.1017/s1355838200001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng T, Vreede FT, Brownlee GG. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol. 2006;80:2337–2348. doi: 10.1128/JVI.80.5.2337-2348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin D, et al. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mink MA, Stec DS, Collins PL. Nucleotide sequences of the 3′ leader and 5′ trailer regions of human respiratory syncytial virus genomic RNA. Virology. 1991;185:615–624. doi: 10.1016/0042-6822(91)90532-g. [DOI] [PubMed] [Google Scholar]
- 17.Gubbay O, Curran J, Kolakofsky D. Sendai virus genome synthesis and assembly are coupled: A possible mechanism to promote viral RNA polymerase processivity. J Gen Virol. 2001;82:2895–2903. doi: 10.1099/0022-1317-82-12-2895. [DOI] [PubMed] [Google Scholar]
- 18.McGivern DR, Collins PL, Fearns R. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J Virol. 2005;79:2449–2460. doi: 10.1128/JVI.79.4.2449-2460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidal S, Kolakofsky D. Modified model for the switch from Sendai virus transcription to replication. J Virol. 1989;63:1951–1958. doi: 10.1128/jvi.63.5.1951-1958.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins PL, Mink MA, Stec DS. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowton VM, Fearns R. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3′ end of the nucleocapsid and can scan to access internal signals. J Virol. 2005;79:11311–11322. doi: 10.1128/JVI.79.17.11311-11322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowton VM, McGivern DR, Fearns R. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J Gen Virol. 2006;87:1805–1821. doi: 10.1099/vir.0.81786-0. [DOI] [PubMed] [Google Scholar]
- 23.Fearns R, Peeples ME, Collins PL. Mapping the transcription and replication promoters of respiratory syncytial virus. J Virol. 2002;76:1663–1672. doi: 10.1128/JVI.76.4.1663-1672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fearns R, Collins PL, Peeples ME. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J Virol. 2000;74:6006–6014. doi: 10.1128/jvi.74.13.6006-6014.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peeples ME, Collins PL. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fearns R, Peeples ME, Collins PL. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 27.Grosfeld H, Hill MG, Collins PL. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marriott AC, Smith JM, Easton AJ. Fidelity of leader and trailer sequence usage by the respiratory syncytial virus and avian pneumovirus replication complexes. J Virol. 2001;75:6265–6272. doi: 10.1128/JVI.75.14.6265-6272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotta AT, Been MD. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa F, Fauzi H, Nishikawa S. Detailed analysis of base preferences at the cleavage site of a trans-acting HDV ribozyme: A mutation that changes cleavage site specificity. Nucleic Acids Res. 1997;25:1605–1610. doi: 10.1093/nar/25.8.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotta AT, Been MD. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao CC, Singh P, Ecker DJ. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 33.Vulliémoz D, Roux L. Given the opportunity, the Sendai virus RNA-dependent RNA polymerase could as well enter its template internally. J Virol. 2002;76:7987–7995. doi: 10.1128/JVI.76.16.7987-7995.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao CC, Del Vecchio AM, Zhong W. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 35.Shim JH, Larson G, Wu JZ, Hong Z. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J Virol. 2002;76:7030–7039. doi: 10.1128/JVI.76.14.7030-7039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai Z, Liang TJ, Luo G. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J Virol. 2004;78:3633–3643. doi: 10.1128/JVI.78.7.3633-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, Barros M, Spencer E, Patton JT. Features of the 3′-consensus sequence of rotavirus mRNAs critical to minus strand synthesis. Virology. 2001;282:221–229. doi: 10.1006/viro.2001.0825. [DOI] [PubMed] [Google Scholar]
- 38.Massey DM, Lenard J. Inactivation of the RNA polymerase of vesicular stomatitis virus by N-ethylmaleimide and protection by nucleoside triphosphates. Evidence for a second ATP binding site on L protein. J Biol Chem. 1987;262:8734–8737. [PubMed] [Google Scholar]
- 39.Kuo L, Fearns R, Collins PL. Analysis of the gene start and gene end signals of human respiratory syncytial virus: Quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liuzzi M, et al. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neufeld KL, Galarza JM, Richards OC, Summers DF, Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranjith-Kumar CT, et al. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: Implication for viral RNA synthesis. J Virol. 2001;75:8615–8623. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smallwood S, Moyer SA. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology. 1993;192:254–263. doi: 10.1006/viro.1993.1028. [DOI] [PubMed] [Google Scholar]
- 44.Tomar S, Hardy RW, Smith JL, Kuhn RJ. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol. 2006;80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu Rev Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 46.Nagy PD, Carpenter CD, Simon AE. A novel 3′-end repair mechanism in an RNA virus. Proc Natl Acad Sci USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samal SK, Collins PL. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology. 2004;318:463–473. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 49.Vulliémoz D, Roux L. “Rule of six”: How does the Sendai virus RNA polymerase keep count? J Virol. 2001;75:4506–4518. doi: 10.1128/JVI.75.10.4506-4518.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: A motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy SK, Ito Y, Parks GD. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyatt LS, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.