Abstract
With a direct link to cancer, aging, and heritable diseases as well as a critical role in cancer treatment, the importance of DNA damage is well-established. The intense interest in DNA damage in applications ranging from epidemiology to drug development drives an urgent need for robust, high throughput, and inexpensive tools for objective, quantitative DNA damage analysis. We have developed a simple method for high throughput DNA damage measurements that provides information on multiple lesions and pathways. Our method utilizes single cells captured by gravity into a microwell array with DNA damage revealed morphologically by gel electrophoresis. Spatial encoding enables simultaneous assays of multiple experimental conditions performed in parallel with fully automated analysis. This method also enables novel functionalities, including multiplexed labeling for parallel single cell assays, as well as DNA damage measurement in cell aggregates. We have also developed 24- and 96-well versions, which are applicable to high throughput screening. Using this platform, we have quantified DNA repair capacities of individuals with different genetic backgrounds, and compared the efficacy of potential cancer chemotherapeutics as inhibitors of a critical DNA repair enzyme, human AP endonuclease. This platform enables high throughput assessment of multiple DNA repair pathways and subpathways in parallel, thus enabling new strategies for drug discovery, genotoxicity testing, and environmental health.
Keywords: base excision repair, comet assay, DNA repair
DNA damage is a critical risk factor for cancer, aging, and heritable diseases (1–3), and it is the underlying basis for most frontline cancer therapies (4). Increased knowledge about DNA damage and repair would facilitate disease prevention, identification of individuals at increased risk of cancer, discovery of novel therapeutics, and better drug safety testing. Despite their obvious translational impact, most DNA damage assays have not changed significantly in more than two decades, and thus are not compatible with modern high throughput screening technologies. To better assess the biological impact of damage, we need data that reflect the integrated effect of multiple DNA repair pathways and subpathways, in response to a range of DNA lesions, measured in an accurate and high throughput fashion.
The single cell gel electrophoresis or “comet” assay is based on the principle that relaxed loops (induced by single strand breaks) and DNA fragments migrate farther in an agarose gel than undamaged DNA (5–8). The alkali comet assay sensitively detects a range of DNA lesions, including strand breaks (single and double), as well as alkali sensitive sites. Although most base lesions are not directly detected, base adducts can be detected when converted to abasic sites or single strand breaks with the addition of purified DNA repair enzymes (7–11). The comet assay has been used in a variety of applications, including genotoxicity testing, human biomonitoring and epidemiology, environmental health, and basic research on DNA damage and repair (7, 8, 12–19). Compared with other DNA damage assays (20–22), the comet assay is relatively inexpensive, is very sensitive, and can assess the integrated cellular response to many kinds of DNA lesions simultaneously (7, 8, 19).
Wider acceptance of the comet assay has been limited, however, by low throughput; poor reproducibility between slides, users, and laboratories; and image processing and analysis methods that are laborious, time-consuming, and potentially biased. The need for improvement is widely recognized, and previous approaches include reducing space requirements for electrophoresis (CometAssay 96, Trevigen), using a multiwell format for treating samples (23), incorporating internal controls (24, 25), or improving imaging (26). These methods address individual issues, but do not provide a comprehensive solution for DNA damage analysis.
Here we present a high throughput platform for DNA damage analysis with the sensitivity and versatility of the comet assay. We spatially register cells by capturing them into a microwell array patterned directly into agarose (Fig. 1). This passive patterning method requires only gravity to capture cells and introduces minimal external stress. Array registration allows multiple experimental conditions to be spatially encoded on a single slide. Additionally, the microwells provide a unique method for assessing DNA damage in collections of cells or cell aggregates. The arrays also facilitate fully automated imaging and analysis, requiring no user input or special equipment.
Fig. 1.
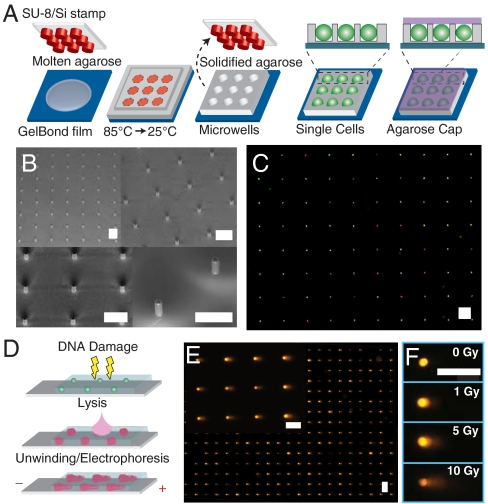
Single cell gel electrophoresis array. (A) Microwell fabrication. A microfabricated stamp is placed onto molten agarose. Agarose is cooled to set, and the stamp is removed. Cells are loaded into wells by gravitational settling, and an agarose overlay covers the cells. (B) Scanning electron micrograph of SU-8 posts patterned onto a silicon substrate. (C) Two populations stained red and green, loaded into wells concurrently, are shown by cytoplasmic staining. (D) Comet assay. Cells are treated with DNA damaging agent. Cells are lysed in the gel, exposing the DNA. The DNA is unwound and electrophoresed under alkaline conditions. Relaxed loops and low molecular weight fragments migrate out of the packed chromatin, forming a comet tail. (E) Arrayed microwell comets. (F) Microwell comets with varying doses of IR damage. Horizontal scale bars are 100 μm.
To facilitate the high throughput analysis of drugs, patient samples, or other experimental conditions, we have also developed 24- and 96-well implementations of the microwell array, which can be integrated with standard high throughput screening (HTS) techniques. We have validated this platform in two key applications. By incorporating multiple cell types and multiple DNA repair time points on a single plate, we directly compare DNA repair kinetics between human lymphocytes from individuals with different genetic backgrounds. We also demonstrate how this assay could be used in the context of drug screening by comparing the performance of potential inhibitors of base excision repair (BER).
Results
Micropatterned Cell Arrays.
The traditional comet assay uses cells randomly dispersed in agarose. Random cell placement leads to several problems, including difficulty in locating cells automatically and large numbers of unanalyzable cells due to overlap. Spatial registration of cells in a defined array obviates these problems and allows spatial encoding of multiple experimental conditions on the same slide, and cell spacing can be tuned to maximize the number of conditions per slide.
Several methods currently exist for patterning cells, including dielectrophoresis (27), hydrostatic trapping (28), and microwell cell traps (29–31). Of these methods, only microwells capture cells passively, eliminating the need for external energy sources and introducing minimal external stress on cells. Current microwell material systems, however, including polydimethyl siloxane and polyethylene glycol are not compatible with DNA electrophoresis. We have developed a method that uses a microfabricated stamp to pattern microscopic wells directly into hydrated agarose (Fig. 1A). The stamp consists of microposts (Fig. 1B), which are photolithographically defined and can be varied in width and depth to optimally accommodate specific cell types (see Materials and Methods). Molten agarose sets around the posts, and the stamp is removed to form arrayed microwells in the agarose gel. Cells are then trapped in the wells using gravity. Captured cells are protected from rinsing shear so that excess cells are easily rinsed away, leaving the captured cells patterned into a defined array. Fig. 1C shows patterned cells that have been labeled with a fluorescent cytoplasmic stain. The resulting arrays typically show at least 90% filling.
By directly patterning the agarose gel, we have created a system that is fully compatible with the comet assay (Fig. 1D). Fig. 1E shows that the morphology of the resulting comets is comparable to that seen in the traditional comet assay (8). There is a well-defined head consisting of tightly wound and high molecular weight DNA. The head is followed by a comet tail, which consists of relaxed loops and fragments. This is further demonstrated in Fig. 1F, which shows microwell comets from varying doses of ionizing radiation (IR). Typical comet morphology is also seen in the dose response, with heads growing dimmer and tails growing longer and brighter with increasing dose.
One potential application of the microwell array is multiplexing cell types or conditions using colors or other labels. Cell multiplexing is demonstrated in Fig. 1C, where two groups of cells, one stained red and the other green, have been loaded simultaneously into the microwell arrays. Fluorescent images of the cells are taken before lysis, and the color information can be recorded for each cell position and correlated with the final comet output.
Automated Imaging and Analysis.
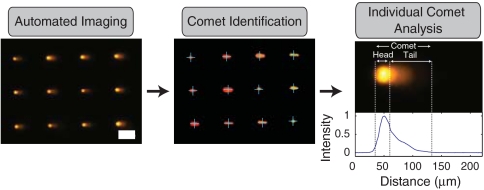
Imaging traditional comets is both laborious and tedious. Complications include sparse comet distribution and unanalyzable cell clumps, as well as random debris, which require users to distinguish analyzable objects. Automated imaging systems do exist, but they are expensive and either require manual comet selection or machine learning algorithms, which can be biased in their training. Comet analysis requires accurately discerning the transition from comet head to comet tail, which demands complex image analysis that can produce erroneous results. The microwell arrays obviate these problems and provide a simple route to automated imaging and analysis. We use a standard fluorescence microscope with automated stage (see Materials and Methods) combined with a suite of custom analysis software (Fig. 2), which utilizes simple algorithms to identify and analyze comets. Our software selects comets based on array registration, which eliminates problems with overlapping comets and debris. Further, because of the microwell fabrication method, all cells are located in a single focal plane, which eliminates the need for users to adjust the focus for each individual comet. Finally, the fixed head size, provided by the microwell geometry, simplifies the problem of identifying the head/tail transition, ensuring accurate determination of comet parameters (Fig. 2). The result is the capability to fully automate imaging and analysis with no user intervention and with no special equipment or complex software.
Fig. 2.
Comet analysis pipeline. Images of comets are acquired automatically. Identification software recognizes comets in a defined array. Image of arrayed comets before and after identification are shown. Identified comets are labeled with blue crosses. Scale bar is 100 μm. Finally comet analysis software identifies beginning and end of comet as well as head/tail division (dashed vertical lines) and calculates comet parameters. A comet is shown along with its corresponding line profile from which the comet parameters are calculated.
Spatially Encoded Microwell Comet Assay.
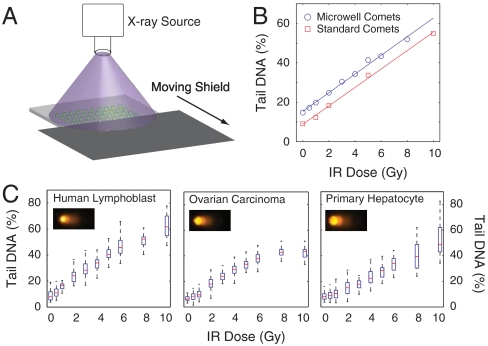
A major advantage of the micropatterned array is the ability to increase throughput by spatially encoding multiple dosing conditions on the same slide. As shown in Fig. 3A, using a moving shield to dynamically protect areas of the sample, different regions of the same comet slide can be treated with unique doses of IR, a key cancer therapeutic that has been well-characterized with the comet assay. To test the robustness of this platform for measuring DNA damage, we performed an IR dose response for human lymphoblast cells using either the traditional comet assay scored using commercial software or our spatially encoded microwell assay scored using our automated analysis tools. The results, shown in Fig. 3B, show consistent linear dose responses to IR in both implementations of the assay, demonstrating the efficacy of our platform for DNA damage analysis. Additionally, all IR doses for the microwell comets were incorporated on the same microscope slide. Due to differences in experimental protocols (see Materials and Methods), we did not compare absolute levels of damage between the two assays. Additionally, the microwell size can be tuned to accommodate virtually any cell type. For example, using different microwell sizes and spatial encoding of 10 IR doses, human lymphoblasts, ovarian cancer cells, and primary hepatocytes were analyzed (Fig. 3C). These data demonstrate efficacy for generating dose-response curves for multiple cell types.
Fig. 3.
Spatially encoded comet assay. (A) Method for spatially encoding IR doses on the same microscope slide. A moving lead shield exposes different regions of the slide to IR for varying lengths of time. (B) Comparison of IR dose response between traditional comet slides scored using commercial software and microwell comets scored using automated software. Each data point is the median of 50 individual comets. (C) IR dose responses for TK6 human lymphoblasts, OVCAR-8 human ovarian carcinoma cells, and freshly isolated primary rat hepatocytes. Representative images are shown from 10 Gy dose for each cell type. Box plots show median of 50 comets as a red line and the lower and upper quartiles as a blue box. Whiskers show extent of furthest data points within 150% of interquartile range.
Self-Calibrating Microwell Comets.
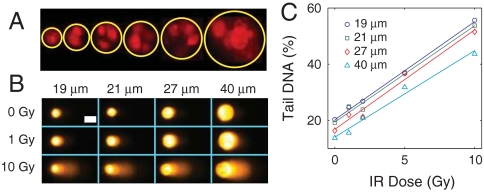
In the traditional comet assay, single cell suspensions are required. The microwell overcomes this limitation by physically restraining the DNA following lysis, giving rise to a uniform and analyzable comet head. The uniform head size, combined with an analysis that is normalized to the total amount of DNA, makes the microwells self-calibrating. This feature means that any number of cells can be combined in a microwell and analyzed as a single comet, providing a unique method for assessing DNA damage in microscopic clusters of cells. Fig. 4A shows human lymphoblast cells captured in wells with a range of diameters. The smallest wells (19 μm diameter) typically capture single cells, whereas the largest wells (54 μm diameter) can capture > 10 cells. Fig. 4B demonstrates that comets remain morphologically consistent over a range of IR damage and microwell diameters. The consistency with single cell comets is also reflected in the quantitative analysis (Fig. 4C). Further, all IR doses and well sizes were combined on the same microscope slide, demonstrating a 20-fold higher throughput than the traditional comet assay. Absolute levels of DNA damage appear to decrease with increasing microwell size, which could be the result of incomplete DNA migration from the larger wells. Longer electrophoresis times or a stronger field could lead to more complete DNA migration.
Fig. 4.
Self-calibrating microwells. (A) Fluorescence image of (from left to right) 19, 25, 29, 33, 40, and 54 μm diameter microwells filled with propidium iodide stained TK6 human lymphoblasts. (B) Morphological and (C) quantitative IR dose response of TK6 cells loaded into different sized microwells on the same slide. White scale bar in (B) is 25 μm. Each data point in (C) is the median of 50 comets.
Multiwell Comet Array.
To measure DNA damage induced by different chemicals, or among different samples of cells, we created a multiwell version of our micropatterned comet array (Fig. 5A). Fig. 5A illustrates the 24-well version of the assay, where the floor of each well is a patterned array of agarose microwells. Cells are loaded into the microwells and, once embedded in agarose, can be treated with chemical damaging agents, lysis solution, or repair media. After treatment, the multiwell structure can be removed, leaving only the cells embedded in agarose. The “comet plate” can then be carried through the standard comet assay protocol, enabling 24 or 96 samples to be run simultaneously. Importantly, this platform is fully compatible with our automated imaging and analysis tools.
Fig. 5.
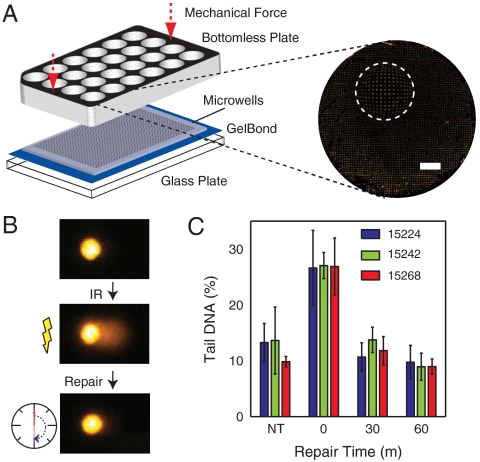
Multiwell comet array. (A) Assembly of multiwell comet array. Agarose gel with microwells is sandwiched between a glass substrate and a bottomless multiwell plate and sealed with mechanical force. Arrayed microwells comprise the bottom of the multiwell. A large field scan of one well of a 24-well plate is shown with a fish-eye magnification (2×) of a small region. White scale bar is 2 mm. (B) Schematic of repair study with representative comets. Cells are treated with IR and allowed to repair in media at 37 °C before lysis. (C) Repair kinetics of human B-lymphocyte lines GM15224, GM15242, and GM15268 after treatment with 7.5 Gy IR. Nontreated cells were not exposed to IR. Bars and error bars represent averages and standard deviations, respectively, of three independent experiments with at least 50 comets scored for each condition in each experiment.
There is significant interest in determining the extent of variation in DNA damage sensitivity and repair capacity between populations and individuals (24, 32). Although it is extremely sensitive, the traditional comet assay is impractical for most large-scale studies. Our multiwell comet plate, however, allows all samples and repair times to be analyzed simultaneously. Further, by including all replicates and conditions in the same agarose slab, this platform reduces the noise introduced by slide-to-slide variation. When comparing between wells of a 96-well plate, we observe an 8.8% coefficient of variation (CV) in the population medians (Fig. S1), which is less than the observed CV between slides in the traditional assay (25). As a proof of concept, we used the 96-well platform to evaluate the repair capacity in response to IR of three B-lymphocyte lines (Coriell Research Institute) from individuals with different genetic backgrounds. The experiment is shown schematically in Fig. 5B. All three cell lines, nontreated controls, and three repair time points (0, 30, and 60 m) were performed on a single plate by lysing cells in wells adjacent to cells that were allowed to repair at 37 °C. As shown in Fig. 5C, nearly all of the damage is repaired within 30 m of treatment with IR for all cell types. Additionally, it appears that no significant difference exists between the repair capacities of these cell lines for IR-induced damage. In contrast, these same cells were previously shown to have significantly different sensitivities to the methylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (33). Whereas the majority of IR-induced DNA damage is repaired by the BER pathway (4) (Fig. 6A), the cytotoxicity of MNNG is largely due to O6-methylguanine, which is primarily repaired by direct reversal. Thus, these cells show different repair capacities for different repair pathways.
Fig. 6.
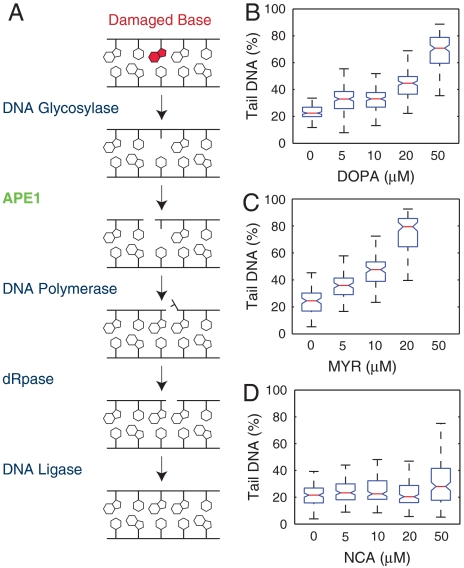
Potency of APE1 inhibitors. (A) Schematic of short patch BER for repairing methylated bases. The base is removed by a monofunctional glycosylase, and APE1 cleaves the backbone 5′ to the resulting abasic site. The new base is inserted by DNA polymerase, the 5′ flap is removed by a dRpase, and the nick is sealed by a DNA ligase. (B–D) Human lymphoblasts (TK6) treated with increasing doses of (B) DOPA, (C) MYR, or (D) NCA. Each dataset is a representative from three independent experiments. Box plots for NCA and DOPA include 150 individual comets per condition. 100 comets per condition are shown for MYR. Assay readout was saturated at 50 μm MYR. Notches represent 95% confidence interval of medians, determined using Kruskal–Wallis nonparametric analysis of variance. Boxes with nonoverlapping notches represent different populations with at least 95% confidence.
Small Molecule Inhibitors of Human AP Endonuclease.
DNA damaging agents are the frontline treatment for most cancers, but cells have numerous DNA repair mechanisms, which can limit therapeutic efficacy or contribute to resistance (4). Inhibiting DNA repair has the potential to sensitize cancer cells to treatment and is now a common theme in cancer therapy research (4). The BER pathway (Fig. 6A) has received particular interest because of its relevance to numerous types of cancer therapies, including IR and monofunctional alkylators (4). Following removal of a damaged base by a monofunctional DNA glycosylase, the major human apurinic/apyramidinic (AP) endonuclease, APE1, is responsible for cleavage of AP sites prior to processing by downstream BER machinery (Fig. 6A) (34). Inhibition of APE1 activity blocks the BER pathway and leads to accumulation of AP sites, which can be highly cytotoxic (35, 36). From a clinical perspective, APE1 overexpression has been observed in numerous cancer types (37). Additionally, reduced expression of APE1 using RNA interference has been shown to sensitize cells to treatment by numerous therapeutic agents (38–41). Small molecules that inhibit the AP endonuclease activity of APE1 could thus be extremely valuable in combination therapies for treating cancer.
Several small molecules have previously been identified as potential inhibitors of APE1’s AP endonuclease activity (42, 43, 44). Cytotoxicity of these molecules has been evaluated by clonogenic survival, which does not illuminate the mechanism by which the molecules are cytotoxic. The comet assay can directly reveal abasic sites that accumulate due to APE1 inhibition. As a proof of concept screen, we utilized the 96-well comet plate to test three potential small molecule inhibitors of APE1: 7-nitro-1H-indole-2-carboxylic acid (NCA) (42), myricetin (MYR), and 6-hydroxy-DL-DOPA (DOPA) (43). Cells treated with either DOPA (Fig. 6B) or MYR (Fig. 6C) show a dose-dependent increase in DNA damage levels, presumably as a result of the accumulation of BER intermediates that is expected from the high levels of spontaneous BER [estimated at > 10,000 BER events per day (10)]. All concentrations of these two molecules tested show a significant increase in damage over the controls. At 50 μm the damage induced by myricetin was so severe as to saturate the assay. Although there may be some direct damage resulting from these small molecules, the findings agree well with the observation of abasic site accumulation by Simeonov et al. (43). In contrast, cells treated with the candidate molecule NCA (Fig. 6D) do not show greatly increased damage, as compared to control. Only the highest concentration (50 μM) shows a statistically significant (21%) increase in damage over the control, whereas DOPA and MYR induce a 48% and 50% increase in damage, respectively, at 10-fold lower concentrations. Interestingly, two studies have reported no effect on cell survival when using NCA with DNA damaging agents (43, 45). Our results suggest that NCA may have some effect on cells, but both DOPA and MYR are far more potent. As potent inhibitors of APE1, these molecules could potentially be used in combination therapies to treat tumors more aggressively.
Discussion
We present a technique for measuring DNA damage that combines sensitivity, versatility, high throughput, and ease of use. This platform has potential to impact a broad range of applications in the laboratory and clinic. Having the capacity to handle dozens of conditions in parallel will shed light on how cells respond to multiple classes of DNA damaging agents, acted upon by a variety of DNA repair pathways and subpathways. With increased granularity in the human DNA repair landscape, it should become possible to reveal predictive biomarkers and prognostic indicators.
A major strength of the comet assay is its versatility. It has been used to study a variety of cell types and species from all three biological kingdoms (17). The microwell arrays further increase the versatility of this assay by enabling unique capabilities, including cell multiplexing and analysis of cell aggregates. Cell multiplexing could be used to analyze multiple readouts from the same cell. This could be useful in several contexts, including antibody labeling of mixed populations or determining viability of cryopreserved primary samples on a single cell basis. The microwells also facilitate DNA damage analysis in small cell aggregates (Fig. 4). Cell aggregates are common in research and the clinic, in the form of tissue samples, tumor spheroids or embryos, and embryonic stem cells. Maintaining cell–cell contact is often necessary to preserve important features of the sample, such as autocrine signaling and tight junctions that might influence cellular response to DNA damage. The microwell technique allows for DNA damage quantification in such samples, potentially without sacrificing important biological information.
We present here the first demonstration of a platform in which multiple cell types, repair time points, DNA damaging agents, DNA repair enzymes or inhibitors, and other conditions can be assayed simultaneously and in combination. Unlike other high throughput comet assay implementations [(23) and Trevigen], the multiwell platform is compatible with any cell type, whether cultured adherently or in suspension, and also incorporates significant improvements in imaging and analysis. Further, we provide the ability to assay repair kinetics on multiple cell types without introducing slide-to-slide variation, which could be critical for revealing subtle biological effects. The 8.8% CV observed between individual wells of a 96-well plate is less than the reported variation between individual comet slides using the traditional assay (25), and this could potentially be reduced further by including internal controls (25).
This platform has potential for both epidemiology and drug screening applications. Our findings on the repair kinetics of B-lymphocyte lines highlight the importance of measuring repair capacities through multiple pathways and by different agents, and they demonstrate the utility of a high throughput approach. For drug screening, the platform offers an integrated readout of multiple biological pathways. Specific subsets of DNA lesions can be detected by the application of DNA repair enzymes that convert otherwise undetectable damaged bases into detectable single strand breaks or AP sites. Modifications in the basic comet protocol provide increased specificity, thus enabling testing of therapeutics that damage DNA or that target DNA repair pathways. Additionally, the 96-well platform integrates with standard HTS techniques making it well-suited for drug development.
Conclusions
With the ability to measure acute DNA damage levels as well as repair kinetics, the comet assay is uniquely relevant to a variety of biological and clinical applications. The significant improvements afforded by this platform make the measurement of DNA damage easier and more readily applicable. Further, the approach is simple and scalable, offering a route to mass produce gels and distribute them in a manner similar to DNA and protein electrophoresis. This would bring critical information about DNA damage and repair to researchers and clinicians in a range of fields, and the use of this knowledge to facilitate disease prevention and treatment could be fully realized. Through the integration of traditional methods in biology and engineering, the platform described here represents a significant technological advance, providing high throughput, objective, and quantitative measurements that have the potential to become a new standard in DNA damage analysis.
Materials and Methods
Additional details in SI Text.
Cell Culture.
TK6 human lymphoblasts; Human B-lymphocyte lines GM15268, GM15242, and GM15224 (Coriell Institute); and OVCAR-8 human ovarian cancer cells were cultured in 1X RPMI medium 1640 with L-glutamine supplemented with 10% horse serum, 15% FBS, and 10% FBS, respectively. Primary rat hepatocytes were isolated and cultured as described elsewhere (46). Live cells in Fig. 1C were stained with CellTracker (Invitrogen).
Microwell Fabrication.
The microwell molds were fabricated by lithographically patterning SU-8 photoresist (SU-8 2025, MicroChem). Molten 1% normal melting point agarose (Omnipur, Invitrogen) was applied to a sheet of GelBond film (Lonza) and the mold was allowed to float until the agarose set. The mold was removed, leaving microwells.
Microwell Comet Preparation.
Cells were captured in microwells by gravity, and covered with low melting point agarose (Invitrogen). Traditional comet slides were prepared as previously described (8). The multiwell version of the comet platform was prepared by sealing a microwell gel between a glass plate and a bottomless 24- or 96-well plate (Greiner BioOne) (Fig. 5A).
Exposure to Ionizing Radiation.
After encapsulation in agarose, cells were irradiated at room temperature using 250 kVp X-rays at 1 Gy/m (Philips RT-250). After exposure cells were placed in lysis buffer. To evaluate repair kinetics, wells were synchronized during lysis after repairing in media for varying time intervals. Lysis at 37 °C for 0 m and 30 m repair times used 0.1% sodium dodecyl sulfate in the lysis buffer. After the final repair time, all samples were placed into standard lysis solution at 4 °C.
Potential APE1 Inhibitors.
7-Nitro-1H-indole-2-carboxylic acid (Gold Biotechnology) and Myricetin (Sigma–Aldrich) were dissolved in DMSO, and 6-Hydroxy-DL-DOPA was dissolved in 1 M HCl. Cells were loaded onto the 96-well comet plate and then incubated for 3 h in media with inhibitor. After treatment cells were placed in standard lysis solution at 4 °C. Three replicate wells were pooled for each condition.
Comet Assay.
The comet assay was performed using a modified version of the alkaline comet protocol as described by Singh et al. (6).
Fluorescence Imaging and Comet Analysis.
Slides were stained with SYBR Gold (Invitrogen). Images were captured automatically using an epifluorescent microscope and analyzed automatically using custom software written in MATLAB (The Mathworks). Traditional comet slides were scored manually using Komet 5.5 (Andor Technology).
Supplementary Material
Acknowledgments.
We thank Jacqueline Yanch and Dwight Chambers for IR facilities. We acknowledge Sukant Mittal’s and James Mutamba’s work in the project’s inception. We thank D. M. Wilson III for help with APE1 inhibitors. Yunji Wu, Drew Regitsky, Margaret Wangeline, and Jeffrey Quinn provided experimental and software development support. We also thank the Microtechnology Laboratory at the Massachusetts Institute of Technology (MIT) for microfabrication support. We acknowledge funding under the National Institutes of Health Ruth L. Kirchenstein postdoctoral fellowship, the Howard Hughes Medical Insititute, and the MIT Center for Environmental Health Sciences (Grant NIEHS P30-ES002109). This work was supported by the National Institute of Environmental Health Sciences (NIEHS) Genes, Environment, and Health Initiative Grant U01-ES016045, and D.M.W. was supported by the NIEHS Grant T32-ES07020-34.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004056107/-/DCSupplemental.
References
- 1.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2005. [Google Scholar]
- 2.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JHJ. DNA damage, aging, and cancer. New Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 4.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 5.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Bioph Res Co. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 6.Singh NP, McCoy MT, Tice RR, Scheider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 7.Collins AR. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 8.Olive PL, Banath JP. The comet assay: A method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 9.Dusinska M, Collins A. Detection of oxidised purines and uv-induced photoproducts in DNA of single cells, by inclusion of lesion-specific enzymes in the comet assay. Altern Lab Anim. 1996;24:405–411. [Google Scholar]
- 10.Fortini P, Raspaglio G, Falchi M, Dogliotti E. Analysis of DNA alkylation damage and repair in mammalian cells by the comet assay. Mutagenesis. 1996;11:169–175. doi: 10.1093/mutage/11.2.169. [DOI] [PubMed] [Google Scholar]
- 11.Collins AR, Dusinska M, Horska A. Detection of alkylation damage in human lymphocyte DNA with the comet assay. Acta Biochim Pol. 2001;48:611–614. [PubMed] [Google Scholar]
- 12.Brendler-Schwaab S, Hartmann A, Pfuhler S, Speit G. The in vivo comet assay: Use and status in genotoxicity testing. Mutagenesis. 2005;20:245–254. doi: 10.1093/mutage/gei033. [DOI] [PubMed] [Google Scholar]
- 13.Witte I, Plappert U, de Wall H, Hartmann A. Genetic toxicity assessment: Employing the best science for human safety evaluation part iii: The comet assay as an alternative to in vitro clastogenicity tests for early drug candidate selection. Toxicol Sci. 2007;97:21–26. doi: 10.1093/toxsci/kfl192. [DOI] [PubMed] [Google Scholar]
- 14.Valverde M, Rojas E. Environmental and occupational biomonitoring using the comet assay. Mutat Res. 2009;681:93–109. doi: 10.1016/j.mrrev.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Möller P. Genotoxicity of environmental agents assessed by the alkaline comet assay. Basic Clin Pharmacol. 2005;96:1–42. [PubMed] [Google Scholar]
- 16.Dusinska M, Collins AR. The comet assay in human biomonitoring: gene-environment interactions. Mutagenesis. 2008;23:191–205. doi: 10.1093/mutage/gen007. [DOI] [PubMed] [Google Scholar]
- 17.Dhawan A, Bajpayee M, Parmar D. Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol. 2009;25:5–32. doi: 10.1007/s10565-008-9072-z. [DOI] [PubMed] [Google Scholar]
- 18.McKenna DJ, McKeown SR, McKelvey-Martin VJ. Potential use of the comet assay in the clinical management of cancer. Mutagenesis. 2008;23:183–190. doi: 10.1093/mutage/gem054. [DOI] [PubMed] [Google Scholar]
- 19.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 20.Birnboim HC, Jevcak JJ. Fluorometric method for rapid detection of DNA strand breaks in human white blood cells produced by low doses of radiation. Cancer Res. 1981;41:1889–1892. [PubMed] [Google Scholar]
- 21.Ahnström G. Techniques to measure DNA single-strand breaks in cells: A review. Int J Radiat Biol. 1988;54:695–707. doi: 10.1080/09553008814552151. [DOI] [PubMed] [Google Scholar]
- 22.Kuo LJ, Yang LX. Gamma-H2AX—a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 23.Stang A, Witte I. Performance of the comet assay in a high-throughput version. Mutat Res. 2009;675:5–10. doi: 10.1016/j.mrgentox.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Trzeciak AR, Barnes J, Evans MK. A modified alkaline comet assay for measuring DNA repair capacity in human populations. Radiat Res. 2008;169:110–121. doi: 10.1667/RR1101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zainol M, et al. Introducing a true internal standard for the comet assay to minimize intra- and inter-experiment variability in measures of DNA damage and repair. Nucleic Acids Res. 2009:1–9. doi: 10.1093/nar/gkp826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McArt DG, et al. Systematic random sampling of the comet assay. Mutagenesis. 2009;24:373–378. doi: 10.1093/mutage/gep020. [DOI] [PubMed] [Google Scholar]
- 27.Gray DS, Tan JL, Voldman J, Chen CS. Dielectrophoretic registration of living cells to a microelectrode array. Biosens Bioelectron. 2004;15:1765–1774. doi: 10.1016/j.bios.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Di Carlo D, Lee LP. Dynamic single-cell analysis for quantitative biology. Anal Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 29.Rettig J, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77:5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 30.Chin VI, et al. Microfabricated platform for studying stem cell fates. Biotechnol Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 31.Rosenthal A, MacDonald A, Voldman J. Cell patterning chip for controlling the stem cell microenvironment. Biomaterials. 2007;28:3208–3216. doi: 10.1016/j.biomaterials.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcon F, et al. Assessment of individual sensitivity to ionizing radiation and DNA repair efficiency in a healthy population. Mutat Res. 2003;541:1–8. doi: 10.1016/s1383-5718(03)00171-2. [DOI] [PubMed] [Google Scholar]
- 33.Fry CF, et al. Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev. 2008;22:2621–2626. doi: 10.1101/gad.1688508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 36.Spek EJ, Vuong LN, Matsuguchi T, Marinus MG, Engelward BP. Nitric oxide induced homologous recombination in Escherichia coli is promoted by DNA glycosylases. J Bacteriol. 2002;184:3501–3507. doi: 10.1128/JB.184.13.3501-3507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res-DNA Repair. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 38.Lau JP, Weatherdon KL, Skalski V, Hedley DW. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. Brit J Cancer. 2004;91:1166–1173. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: Enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 40.Bobola MS, et al. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res. 2005;11:7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- 41.Fishel ML, Kelley MR. The DNA base excision repair protein APE1/ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Madhusudan S, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simeonov A, et al. Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS One. 2009;4:e5740. doi: 10.1371/journal.pone.0005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiple LA, Cardellina JH, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmacol. 2008;73:669–677. doi: 10.1124/mol.107.042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bapat A, Fishel ML, Kelley MR. Going APE as an approach to cancer therapeutics. Antioxid Redox Sign. 2009;11:571–574. doi: 10.1089/ars.2008.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hui EE, Bhatia SN. Micromechanical control of cell–cell interactions. Proc Natl Acad Sci USA. 2007;104:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.