Abstract
The ubiquitin ligase Mdm2 targets the p53 tumor suppressor protein for proteasomal degradation. Mutating phosphorylation sites in the central domain of Mdm2 prevents p53 degradation, although it is still ubiquitylated, indicating that Mdm2 has a post-ubiquitylation function for p53 degradation. We show that Mdm2 associates with several subunits of the 19S proteasome regulatory particle in a ubiquitylation-independent manner. Mdm2 furthermore promotes the formation of a ternary complex of itself, p53, and the proteasome. Replacing phosphorylation sites within the central domain with alanines reduced the formation of the ternary complex. The C-terminus of Mdm2 was sufficient for interaction with the proteasome despite an additional proteasome binding site in the Mdm2 N-terminus. In addition to binding to the proteasome, the C-terminus of Mdm2 bound to the central domain, possibly competing with, and therefore blocking, Mdm2/proteasome interaction. We propose that Mdm2 facilitates, or at least enhances, the association of p53 with the proteasome and that phosphorylation of the central domain of Mdm2 regulates this process.
Keywords: 19S subunit, protein degradation
P53 is one of the most important tumor suppressor proteins, as its gene is mutated or pathway inactivated, in nearly all human cancers. Because of its growth suppressing activities (1), tight control is imperative for maintaining normal cell growth. This control is largely provided by the Mdm2 protein (1). Mdm2 binds to the N-terminal transactivation domain of p53 (2), thereby suppressing p53-dependent transactivation. In addition, Mdm2 ubiquitylates p53 and promotes its rapid degradation (1).
Polyubiquitylated proteins are the classical target for degradation by 26 S proteasomes. Polyubiquitin chains target proteins to proteasomes (3, 4) and initiate the process of degradation (5). Polyubiquitylation of p53, however, is not sufficient to drive p53 degradation. When the central domain of Mdm2 was deleted, or when phosphorylatable central domain residues were replaced with nonphosphorylatable ones (6–8), p53 was not degraded although the protein was still polyubiquitylated. Thus Mdm2 encodes E3-independent activities that are obligatory for p53 degradation.
The proteasome is a cellular multisubunit protease that digests most of the short-lived proteins in a cell. It consists of a 20S catalytic subunit, and usually one or two 19S regulatory subunits (9). For specific activities of the proteasome, the 19S “cap” can be replaced by other protein complexes (10, 11). These regulatory subunits control the entry of the substrate proteins into the catalytic cavity of the 20S core. Accordingly, the major tasks of the regulatory subunit are considered to be recognition of polyubiquitin chains, deubiquitylation and denaturation of the proteins and translocation of substrates into the central cavity of the proteasome, where they are digested into oligopeptides. Nevertheless, it should be noted that certain nonubiquitylated proteins can also be degraded by proteasomes, provided that they are delivered to the catalytic core (9, 12, 13). The 19S subunit can be further divided into a base, which mainly harbors the six AAA-ATPases (S4, S6a, S6b, S7, S8, and S10b), and the lid. Base and lid are connected via the S5a protein.
In this study, we show that Mdm2 associates with the proteasome and this interaction strongly enhanced the association of p53 with the degradation machinery. This interaction of p53 with the proteasome was independent of ubiquitylation but strongly regulated by phosphorylation of the central domain of MDM2.
Results
Mdm2 Associates with the Proteasome.
Based on the result that ubiquitylated p53 is not degraded when serines in the central domain of Mdm2 have been replaced with alanine (6), we hypothesized that Mdm2 must be required not only for p53 ubiquitylation, but also for an additional activity that is required for p53 degradation. We speculated that this additional activity could be to directly link p53 to the proteasome.
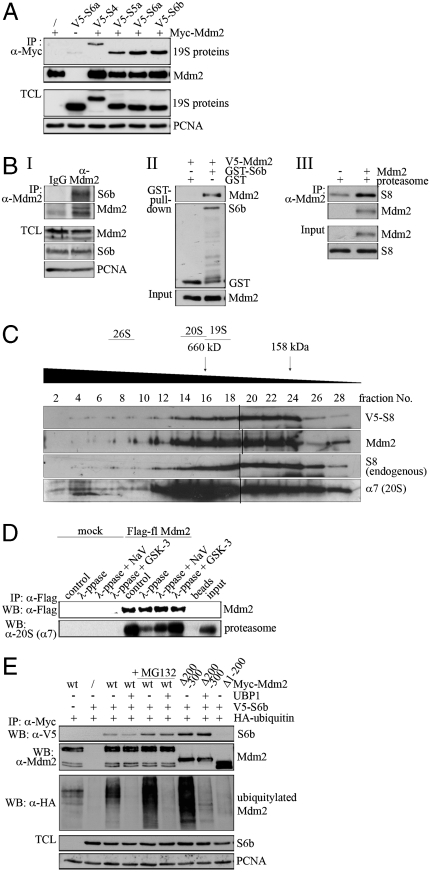
We reasoned that if Mdm2 indeed connects p53 directly with the proteasome, it should be found in a complex with the 19S proteasome. Indeed, exogenously expressed Mdm2 robustly coprecipitated with several of the proteins of the 19S regulatory subunit of the proteasome in vivo, when they were individually overexpressed including S2, S4a, S5a, S6a, and S6b (Fig. 1A and Fig. S1A). The association with S8 and S10b was detectable, but weaker. No interaction with Mdm2 was detected for S1, S5b, S12, and S15 (Fig. S1A).
Fig. 1.
Mdm2 associates with proteasomal proteins. (A) 293T cells were transfected with 7.5 μg of a plasmid encoding Myc-tagged Mdm2 and with 7.5 μg of a plasmid encoding the indicated V5-tagged proteins of the 19S complex. IP: Mdm2 was precipitated and associated proteasomal proteins were detected by Western blotting. TCL: Aliquots of cellular lysates were assayed by Western blotting for protein expression. (B) Section I: Mdm2 was precipitated from U2OS cells. Associated S6b protein was detected by Western blotting. Section II: Bacterially expressed and purified GST or S6b fused to GST were mixed with bacterially expressed V5-tagged Mdm2. GST was pulled down and associated Mdm2 was detected by Western blotting. Section III: Mdm2 expressed in insect cells was mixed with partially purified 26S proteasomes. Mdm2 was immunoprecipitated and associated S8 was detected by Western blotting. (C) H1299 cells were transfected with 7 μg of a plasmid encoding Mdm2 and 3 μg of a plasmid encoding V5-tagged S8. Cell extracts were separated by sucrose gradient. Mdm2, V5-S8, endogenous S8, and α7 (20S) were determined by Western blotting. The black lines indicate where two gels were spliced together. (D) Flag-tagged Mdm2 expressed in insect cells was dephosphorylated with λ-phosphatase in the presence and absence of sodium orthovanadate (NaV), phosphorylated with GSK-3 and mixed with 26S proteasomes. Mdm2 was immunoprecipitated and associated proteasomes were detected by Western blotting. (E) H1299 cells were transfected with 1 μg of a plasmid encoding HA-tagged ubiquitin, 7 μg of a plasmid encoding V5 tagged S6b and with 7 μg of a plasmid encoding wild type Myc-Mdm2 or the indicated mutants. Where indicated, MG132 was added 4 h prior to harvest. Mdm2 was precipitated and cell lysates were incubated with UBP1 or mock treated. Associating S6b was detected by Western blotting.
Whereas Mdm2/19S subunit associations were observed under conditions of overexpression, we also detected S6b in immunoprecipitates of endogenous Mdm2 (Fig. 1B, section I). A GST-pulldown with bacterially expressed GST-S6b and V5-Mdm2 proteins showed that the association between Mdm2 and at least one of the 19S proteins is direct and does not require additional proteins (Fig. 1B, section II). Likewise, Mdm2 also associated with the S8 protein when it was incorporated into the 26S proteasome (Fig. 1B, section III) and it associated with the S8 protein as part of the 26S proteasome in an in vitro pull down assay using insect cell-derived Mdm2 and purified proteasomes (Fig. 1B, section III).
To further confirm that Mdm2 associates with proteasomes in cells, we performed sucrose gradients. As shown in Fig. 1C and Fig. S1B, the largest amount of Mdm2 eluted in fractions sixteen to twenty-four corresponding to a molecular weight between 160 and 660 kDa. However, some Mdm2 protein also eluted with earlier fractions, where larger protein complexes are found. Importantly, Mdm2 showed an almost identical elution pattern as endogenous or transfected S8 protein (Fig. 1C and Fig. S1B), further supporting their physical interaction. It should be noted that the elution pattern of endogenous and ectopically expressed S8 is indistinguishable, proving that transfected proteasomal proteins are incorporated into larger complexes. Moreover, the elution profile of Mdm2 partially overlapped with the α7 subunit of the 20S core particle, showing that Mdm2 is incorporated into complexes which contain core proteasomal proteins. Other proteins of the 19S subunit (S1, S2, and S6a) showed a similar elution pattern, independently of whether they coprecipitated with Mdm2 (S2, S6a, S8) or not (S1; Fig. S1). It should, though, be noted that for S6a, we repeatedly saw an additional high molecular weight elution peak (Fig. S1B). Only a minority of proteasomal proteins (and Mdm2) eluted in fractions corresponding to the fully assembled 26S proteasome, despite the presence of ATP in the buffers. Probably the majority of proteasomal proteins are trapped in subcomplexes which may only assemble when full 26S proteasomes are required.
Regulation of Mdm2/Proteasome Association.
In previous experiments, we have shown that degradation of p53 depends strongly on phosphorylation of the central domain of Mdm2 (6, 14). Here we show that the association of Mdm2 with the proteasome also depended on its phosphorylation status. Treatment of insect cell-derived Mdm2 with λ-phosphatase reduced its association with the proteasome (Fig. 1D). This decrease was prevented, at least in part, by the presence of a phosphatase inhibitor (Fig. 1D). One of the kinases that regulate p53 abundance by phosphorylating Mdm2 is glycogen synthase kinase 3 (GSK-3) (14). Consistent with these data, phosphorylation of λ-phosphatase-treated Mdm2 with GSK-3 fully restored the association of Mdm2 with the proteasome (Fig. 1D).
Because Mdm2, like p53, is a substrate of the proteasome, one would expect that at some point (e.g., for its own degradation) Mdm2 must associate with the proteasome. This association shall, however, strongly depend on ubiquitylation of Mdm2. To differentiate between the association of Mdm2 in the course of its own destruction and in the course of p53 degradation, we deubiquitylated Mdm2 and compared the association of deubiquitylated Mdm2 and mock-treated Mdm2 with the proteasome. In Fig. 1E, we show that the association of Mdm2 with the proteasomal protein S6b was not affected by deubiquitylation of Mdm2, indicating that the association of Mdm2 with proteins of the 19S regulatory subunit was largely independent of ubiquitylation of Mdm2 (Fig. 1E). This result does, however, not exclude the possibility that a small minority of Mdm2 molecules that are beyond the detection level of this assay may associate with the proteasome in a ubiquitylation-dependent manner.
P53, Mdm2, and the Proteasome Form a Ternary Complex.
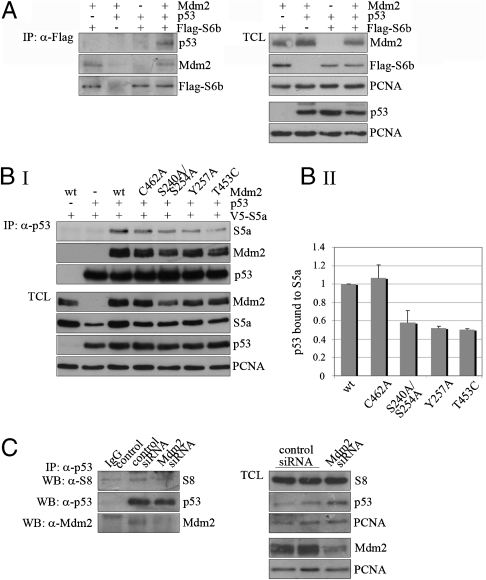
If our hypothesis that Mdm2 connects p53 physically with the proteasome is correct, then one would predict that the presence of Mdm2 should promote the association of p53 with the proteasome. This promotion of the association of p53 with the proteasome should furthermore be independent of ubiquitylation. Moreover, because the postubiquitylation function of Mdm2 is regulated by phosphorylation, the formation of a ternary complex of p53, Mdm2, and proteasomal proteins should require phosphorylation of the central domain of Mdm2. To investigate these possibilities, we cotransfected p53, Mdm2, and the proteasomal subunit S5a or S6b into p53-negative H1299 cells; immunoprecipitated p53 or S6b; and determined the amount of associated S5a or p53 in the presence and absence of wild type Mdm2 or different Mdm2 mutants. As is shown in Fig. 2A and Fig. S2A, the presence of Mdm2 strongly enhanced the association of p53 with the proteasome. Most interestingly, a RING (really interesting new gene) mutant of Mdm2 (C462A) that does not properly ubiquitylate p53 (15), facilitated the association of p53 with the proteasome to an extent comparable to wild type Mdm2 (Fig. 2B and Fig. S2A). In contrast, the association with the proteasome was strongly reduced when phosphorylatable residues in the central domain of Mdm2 were replaced with an alanine (S240A/S254A; S251A/S254A; Y257A). Moreover, an Mdm2 mutant (T453C), that is unable to target p53 for degradation (16), did not promote the formation of a ternary complex (Fig. 2B and Fig. S2A). Deletion of the central domain of Mdm2 also strongly reduced the formation of a ternary complex, possibly due to the absence of the second binding site for p53 (17). Consistent with a role for Mdm2 in the formation of a ternary complex, downregulation of Mdm2 by siRNA significantly reduced the association of endogenous p53 with the proteasome (Fig. 2C and Fig. S2B).
Fig. 2.
p53, Mdm2 and the proteasome form a ternary complex. (A) H1299 cells were transfected with 5 μg of a plasmid encoding p53 together with 5 μg of a plasmid encoding Mdm2 and with a plasmid encoding Flag-tagged S6b, where the amount of transfected plasmid was adjusted to receive equal levels. 4 h prior to harvest, 10 μM MG132 were added. IP:α-Flag: Flag-tagged S6b was precipitated and associated p53 and Mdm2 were determined by Western blotting. TCL: 50 μg of total cell lysate were separated on a SDS-PAGE gel. Mdm2, p53 and S5a were determined by Western blotting. (B) H1299 cells were transfected with 5 μg of a plasmid encoding p53 or with vector together with 5 μg of a plasmid encoding wild type Mdm2, the indicated mutants of Mdm2 or vector DNA and with 5 μg of a plasmid encoding V5-tagged S5a. 4 h prior to harvest, MG132 was added. (B) Section I: IP:α-p53: p53 was precipitated and associated S5a and Mdm2 were determined by Western blotting. TCL: 50 μg of total cell lysate were separated on a SDS-PAGE gel. Mdm2, p53 and S5a were determined by Western blotting. Section II: The signals for p53 and S5a from IP and TCL were quantified and the ratios between p53 and S5a were calculated. Mean values and standard deviations of the relative amount of S5a bound to p53 of three independent experiments were plotted. p53 bound to S5a in the presence of wt Mdm2 was set to 1. (C) U2OS cells were transfected with siRNA targeted against Mdm2 or with a control siRNA. 10 μM MG132 were added 4 h prior to harvest. P53 was precipitated and associated S8 was determined by Western blotting.
N and C-terminal Domains of Mdm2 Associate with the Proteasome.
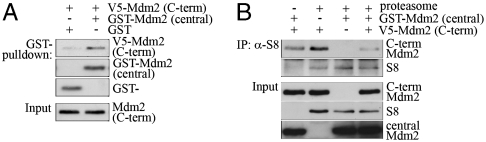
We next mapped the site of interaction for Mdm2 with S4, S5a, S6a, S6b, and S8. As before, full length Mdm2 associated with all tested proteasomal proteins whereas the Mdm2 mutant Δ1–200 interacted weakly or not at all with most proteasome subunits. The only reasonably robust association was with the S8 protein (Fig. S3B). In contrast, when we deleted the central domain (Δ200–300), an enhancement of the interaction was always observed. For some proteasomal proteins, we also found an enhancement of the association with Mdm2 by deleting amino acids 300–400 whereas deletion of the C-terminal region of Mdm2 (Δ460–493) reduced proteasome association (Fig. S3B). These results suggested that there are two independent binding sites for 19S proteasome subunit proteins in Mdm2 within the N- and C-termini. Indeed, the isolated N- and C-terminal domains of Mdm2 both associated equally well with S6b and S8 in vitro (Fig. 3A). At the same time, we failed to detect an association of the isolated central domain of Mdm2 (Fig. 3A).
Fig. 3.
The C-terminus of Mdm2 associates with the proteasome. (A) Bacterially expressed V5-tagged N-terminus (aa 1–200), V5-tagged central domain (aa 200–299) or V5-tagged C-terminal domain (aa 300–491) of Mdm2 were mixed with bacterially expressed and purified GST, S6b fused to GST or S8 fused to GST. GST was pulled down and associated Mdm2 was determined by Western blotting. (B) Schematic drawing of the C-terminal deletion mutants of Mdm2. Bacterially expressed V5-tagged C-terminal domain of Mdm2 and the indicated deletion mutants were mixed with bacterially expressed and purified GST or S6b fused to GST. GST was pulled down and associated C-terminal fragments of Mdm2 were determined by Western blotting. (C) Bacterially expressed C-terminal domain of Mdm2 harboring the indicated mutations were mixed with bacterially expressed and purified GST or S6b fused to GST. GST was pulled down and associated Mdm2 was determined by Western blotting.
When we mapped the proteasome binding site within the N-terminal domain of Mdm2, we found that the proteasome associates with the first 100 amino acids of the Mdm2 protein (Fig. S4A). Moreover, an active enantiomer of nutlin, a chemical that inserts into the p53 binding pocket of Mdm2 (18) reduced the association of Mdm2 with proteins of the 19S proteasome (Fig. S4B) and overexpression of p53 reduced the association of Mdm2 with the proteasome (Fig. S4C). Thus, the N-terminal interaction site on the Mdm2 proteins overlaps entirely with the p53 binding site (19). However, despite this overlap, Mdm2 strongly stimulated the formation of a ternary complex between p53 and the proteasome (Fig. 2 and Fig. S2). In light of these results, we assume that the association of the C-terminal domain of Mdm2 with the proteasome is the major determinant for the association of p53 with the proteasome.
By using Mdm2 mutants where individual parts of the C-terminal domain have been deleted (see Fig. 3B for deletion mutants), we mapped the binding site for the proteasome to amino acid 451–491 of Mdm2 (Fig. 3B). This result was further supported by the analysis of individual point mutations within this area: Replacement of histidine 457 with a serine (Fig. 3C) completely abrogated Mdm2 binding to S6b in vitro. Likewise, Mdm2-T453C also did not support the formation of a ternary complex in an in vivo coIP assay using overexpressed p53, Mdm2, and S5a (Fig. 2B). In contrast, replacing cysteine 462 with alanine was still competent for nucleating a ternary complex, suggesting that this amino acid is not directly involved in the association with proteasomal proteins (Figs. 3C and 2B and Fig. S2A). Because the C-terminal domain of Mdm2 contains an ATP-binding Walker motif (16), we investigated whether binding of ATP could influence the association of Mdm2 with the proteasome. However, neither the binding of the C-terminal domain nor of the full length Mdm2 protein to the proteasome, was altered by the presence of ATP (Fig. S5).
Central Domain of Mdm2 Regulates Binding of Mdm2 to the Proteasome.
Because deletion of the central domain of Mdm2 caused Mdm2 to associate more strongly with proteasomal proteins than full length Mdm2 (Fig. S3), we considered that the central part of Mdm2 might negatively regulate the association of Mdm2 with the proteasome, perhaps by interfering with the ability of the C-terminus of Mdm2 to interact with proteasomal subunits via intra- or intermolecular interactions of the central domain with the C-terminus of Mdm2. Consistent with this idea, GST-tagged central domain of Mdm2 pulled down V5-tagged C-terminus, indicating that these domains interact with each other (Fig. 4A). Moreover, the presence of an excess of the central domain of Mdm2 strongly reduced the association of the C-terminus, with the proteasome (Fig. 4B). The phosphorylated central domain of Mdm2 also strongly reduced the association of p53 with the proteasome (Fig. S6, section I), whereas p53 on its own was unable to associate with the proteasome (Fig. S6, section II) and the central domain of Mdm2 had no effect on the association of p53 with Mdm2 (Fig. S6, section III).
Fig. 4.
The C-terminus of Mdm2 binds to the central domain of Mdm2. (A) Bacterially expressed V5-tagged C-terminus of Mdm2 (aa 300–491) was mixed with bacterially expressed and purified GST or GST-fused to the central domain of Mdm2 (aa 200–299). GST was pulled down with Glutathione sepharose and associated Mdm2 was determined by Western blotting. (B) Bacterially expressed V5-tagged C-terminus of Mdm2 (aa 300–491) was mixed with bacterially expressed and purified GST or GST-fused to the central domain of Mdm2 (aa 200–299) and 26S proteasomes. The proteasomal protein S8 was precipitated and associated C-terminus of Mdm2 was detected by Western blotting.
Intriguingly, the central domain of Mdm2 harbors a sequence motif (EDY) that is also present on those 19S proteins (S2, S5a, S6a, S6b) that associate with Mdm2, whereas in others (S4, S8), the tyrosine of the EDY-motif is replaced with a phenylalanine (EDF; Fig. 5A). Consistent with the idea that the EDY-motif, probably together with surrounding amino acids, might provide the binding site for the C-terminal domain of Mdm2, a bacterially expressed and purified peptide derived from the central domain of Mdm2 (aa 245–264) that contained this “EDY motif” was sufficient to pull down the C-terminus of Mdm2 (Fig. 5B). Importantly, overexpression of an EDY peptide (fused to thioredoxin) in cells reduced p53 degradation, indicating that the association of the C-terminus with the EDY motif is required for p53 degradation. Moreover, the presence of such a peptide resulted in an accumulation of ubiquitylated p53 suggesting that overexpression of an EDY peptide blocks p53 degradation at a postubiquitylation step (Fig. 5C and Fig. S7A). Overexpression of thioredoxin alone or thioredoxin fused with a peptide where the tyrosine of the EDY had been replaced with an alanine (EDA) had no effect on p53 degradation (Fig. 5C and Fig. S7A). In addition to blocking p53 degradation, the EDY peptide also caused an accumulation of Mdm2 (Fig. 5C and Fig. S7A). In contrast, the degradation of c-Jun was not affected by the presence of an EDY-containing peptide, nor did we observe an accumulation of ubiquitylated c-Jun (Fig. S7A). Interestingly, when the aspartic acid and glutamic acid of the EDY motif were replaced with an alanine (AAY), we observed an even stronger binding of the C-terminus of Mdm2 to this peptide than to a peptide containing the wild-type EDY sequence (Fig. S7B).
Fig. 5.
An EDY motif is present on Mdm2 and proteasomal proteins. (A) Alignment of sequences containing the EDY motif from proteasomal proteins and Mdm2. (B) Bacterially expressed V5-tagged C-terminus of human Mdm2 (aa 300–491) was mixed with bacterially expressed and purified GST or GST fused to an EDY-containing peptide of Mdm2 (aa 245–264). (C) H1299 cells were transfected with 1 μg of a plasmid encoding His-tagged ubiquitin together with 0.4 μg of a plasmid encoding p53, 1.2 μg of a plasmid encoding Mdm2 and 30 μg of a plasmid encoding thioredoxin or 30 μg of a plasmid encoding an EDY (wt) or EDA (mu) containing peptide of Mdm2 (aa 245–264) fused to thioredoxin (TRX-EDY) or vector DNA for control. Where indicated, cells were treated with 10 μM MG132 for 4 h. Ubiquitylated proteins (ubi-p53) were purified by adsorption to Ni-agarose and p53 was detected by Western blotting. An aliquot of the cells was tested for expression of p53 and Mdm2. Detection of PCNA was used for loading control (TCL: total cell lysate).
Given the important role that Mdm2 has for the association of p53 with the proteasome, we wondered whether this interaction with 19S proteins might be a common principle for E3 ubiquitin ligases. We therefore tested two other ubiquitin ligases, c-Cbl and Siah-1, for their association with the proteasome. Notably, both c-Cbl and Siah-1 interacted with proteins of the 19S regulatory subunit. However, whereas Mdm2 interacted with S5a, S6a, and S6b, and to a lesser extent with S4 and S10b, c-Cbl and Siah-1 interacted only with S8 and S10b (Fig. S8).
Discussion
Mdm2 Promotes the Formation of a Ternary Complex of p53, Mdm2, and the Proteasome.
Mdm2 exhibits postubiquitylation functions in p53 regulation (6, 20). Here we show that Mdm2 promotes the formation of a ternary complex between p53, Mdm2, and the proteasome. Consistent with prior data linking Mdm2 central domain phosphorylation to degradation functions, the formation of a ternary complex depended on the presence of the same key phosphorylation sites. As a prerequisite for the formation of such a ternary complex for p53 degradation, we postulated an association of Mdm2 with the proteasome. Such an association has been reported previously for the ubiquitin ligases pVHL, RNF2, and Parkin (21–23), and for Mdm2 with the 20S core particle (12). We observed an association of Mdm2 with a variety of 19S subunit proteins, and also for two other ubiquitin ligases, c-Cbl and Siah-1.
The accumulating data that show an association of ubiquitin ligases with the proteasome raise the intriguing question whether it is an intrinsic property of all ubiquitin ligases to promote the formation of ternary complexes between themselves, their substrates, and the proteasome. Such a facilitation of the formation of complexes between the substrates and the proteasome would make ubiquitylation less important for this process. Nevertheless, polyubiquitylation is clearly crucial for p53 degradation and for degradation of most other substrates of the proteasome which leaves open the question as to the exact function of polyubiquitylation. Eventually ubiquitylation might be more important for steps that occur after a substrate has reached the proteasome, such as denaturation of the substrate, and/or regulation of substrate entry into the catalytic cavity.
C-terminus of Mdm2 Binds to Proteasomal Proteins.
The formation of a ternary complex between p53, Mdm2, and the proteasome is most likely promoted by simultaneous binding of p53 and the proteasome to Mdm2. The formation of a ternary complex implies that both interaction partners of the scaffold (Mdm2) bind at distinct sites. Surprisingly, we found a strong requirement of the N-terminus for efficient binding of Mdm2 to the proteasome. This binding site for proteasomal proteins is more or less identical to the p53 binding domain. However, in addition to this N-terminal binding site, we identified a second binding site for proteasomal proteins in the C-terminal domain of Mdm2, and it is most likely that this binding site is key for promoting the postubiquitylation degradation of p53. Consistent with this hypothesis, mutation of Mdm2 at the C-terminus (e.g., T453C) renders Mdm2 incapable of promoting an interaction between p53 and the proteasome. A contribution of the N-terminal proteasome binding site can, however, not be entirely ruled out, particularly because a second binding site exists for p53 within the central domain of Mdm2 (17). It is therefore possible that p53 might be captured by the very strong N-terminal binding site for p53 on the Mdm2 protein. Once bound, the weaker binding of p53 to the central domain of Mdm2 might be sufficient to keep the protein in place. This way, the N-terminus would be free for an association with the proteasome. Finally, despite the evidence presented for the Mdm2-mediated formation of a ternary p53–Mdm2–proteasome complex, we cannot completely exclude the possibility that upon binding of p53 to Mdm2, Mdm2 could promote a conformational change in the p53 molecule which would enhance its affinity for direct binding of the proteasome.
The Central Domain of Mdm2 Regulates the Association of the C-terminal Domain of Mdm2 with the Proteasome.
The C-terminal proteasome binding site of Mdm2 is possibly sequestered by the central domain due to an intra/intermolecular interaction, and therefore plays only a minor role for the association with the proteasome in the context of an isolated full length Mdm2 protein. Evidence for this scheme is deduced from the negative impact that the central domain has on the association of Mdm2 with the proteasome. Moreover, we observed a strong binding of the C-terminal domain of Mdm2 to its central domain and an isolated central domain of Mdm2 blocked binding of the C-terminus of Mdm2 to the proteasome in vitro. Whether the binding between the C-terminus and central domain of Mdm2 occurs intra- or intermolecularly remains to be determined.
Within the central domain of Mdm2, we have identified a three amino acid motif (EDY). This motif is surrounded by those phosphorylation sites that are most critical for p53 degradation (6). A peptide of 20 amino acids containing this motif in its center is sufficient to bind the C-terminal domain of Mdm2. Moreover, this motif is present on those 19S proteins that associated with Mdm2. Thus, the C-terminus of Mdm2 most likely associates with the proteasomal proteins via this motif. Further support for this idea is given by the result that overexpression of a peptide containing this motif reduced p53 degradation and led to the accumulation of ubiquitylated p53.
Taken together, these results lead to the following model for the regulation of the association of (presumably ubiquitylated) p53 with the proteasome: The C-terminus of Mdm2 appears to be the most critical proteasome binding site for p53 degradation. When its activity is not required, (when it does not need to promote p53 degradation) it is masked by the central domain by direct interaction. In the course of p53 degradation, it is released from the central domain of Mdm2 so that it can then associate with the proteasome and deliver p53 that is bound to the N-terminus and central domain of Mdm2 in the correct manner to the proteasome, resulting in p53 degradation.
How the C-terminus of Mdm2 is released from the central domain is unclear. One possibility is that phosphorylation of the central domain releases the C-terminus from the interaction. Most interestingly, we and others have shown previously that p53 binds to the central domain of Mdm2 (1). This interaction is, moreover, regulated by phosphorylation of the central domain of Mdm2 (17). An attractive possibility is therefore that p53 displaces the C-terminus of Mdm2 from its sequestration by the central domain, thus giving the signal for the association of Mdm2 with the proteasome, when the tumor suppressor protein is ready for destruction.
This model not only explains the postubiquitiylation function of Mdm2 but also other unresolved phenomena. One of these mysteries is the stabilization and accumulation of ubiquitylated p53 mutants (24). These p53 mutants associate with Mdm2 not via the classical N-terminal domain, but via the RING-finger in the C-terminus of Mdm2. Because this part of the protein is also required for the interaction with the proteasome, these p53 mutants cannot be degraded despite their ubiquitylation and association with Mdm2. Another example is the accumulation of ubiquitylated p53 after ionizing radiation (25). We have recently shown that p53 accumulates in response to ionizing radiation via a signaling cascade that involves DNA-PK, Akt/PKB, and GSK-3 (26). In resting cells, GSK-3 phosphorylates the central domain of Mdm2 (14). IR leads to the inactivation of GSK-3 (14). In consequence, the central domain of Mdm2 is hypophosphorylated. According to our model, this should lead to the sequestration of the C-terminus of Mdm2 by the central domain and inhibition of the association of p53 with the proteasome, resulting in its accumulation.
Material and Methods
Plasmids.
The plasmids Myc-Mdm2 (pDWM659), pcDNA3-p53, pcDNA3-Mdm2, pcDNA3-Mdm2-C464A, and His-ubiquitin have been described previously (6). The plasmid for UBP-1 has been provided by M. Scheffner and the plasmid for HA-ubiquitin by M. Treier. All other constructs were cloned and mutated by PCR. Primer sequences and cloning strategies are available on request.
Antibodies.
The following antibodies were used in the study: 9E10 (antiMyc, Santa Cruz), HRP-conjugated antiV5 (Invitrogen), PC10 (antiproliferating nuclear cell antigen, Santa Cruz), 4B2 (antiMdm2, Merck), C-18 (antiMdm2, Santa Cruz), antiGST (Rockland), DO-1 (antip53, Santa Cruz), H89 (anticJun, Santa Cruz), antiS1, antiS5a, antiS6a, antiS6b, and antiS8 (Biomol), antiS2 (Calbiochem), M2 (antiFlag, Sigma), antiubiquitin (Sigma), antialpha7 (Biomol).
Cell Lines and their Treatments.
U2OS, 293T, and H1299 cells were cultured according to standard conditions (17) and transiently transfected by calcium-phosphate, jet-Pei (Biomol) or Lipofectamine™2000 (siRNA; Invitrogen) Sequences of siRNAs are available on request.
Immunoprecipitation and GST-Pulldowns.
Immunoprecipitations of cellular proteins were performed as described in ref. 17. For some experiments the concentration of NP40 was reduced to 0.5%, phospatase inhibitors (Phospho-Stop; Roche) were added and the beads were resuspended in 2× sample buffer.
For in vitro coimmunoprecipitation of Mdm2 and the proteasome, 2 μL of a partially purified proteasome preparation (USBio) were mixed with 500 ng of full length Mdm2 or the respective deletion mutants.
Protein expression, purification of bacterial proteins, GST-pulldowns, and Mdm2/proteasome interaction assays are described in detail in SI Text.
SDS-PAGE and Western blotting.
SDS-PAGE and Western blotting were performed as described in ref. 17.
Ubiquitylation Assay.
Ubiquitylation assays were performed as described in ref. 6.
Sucrose Gradient.
Cells were washed twice with ice-cold phosphate-buffered saline (PBS), scraped into PBS, collected by centrifugation, and lysed by mild sonication in 1 mL proteasome lysis buffer (25 mM Tris, pH 7.4, 10 mM NaCl, 5 mM MgCl2, 0.05% NP40, 2 mM ATP and 1mM PMSF). The protein extract was cleared by centrifugation at 16,000 g at 4 °C for 15 min, loaded onto a 10–40% sucrose gradient and centrifuged at 100,000 g at 4 °C for 18 h. Fractions were collected and analyzed by SDS-PAGE and Western blot.
Supplementary Material
Acknowledgments.
This work was supported by the DFG (Deutsche Forschungsgemeinschaft) Grant BL526/5-1. M.K. and S.R.G. were supported by National Institutes of Health Grant R01CA107532.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911716107/-/DCSupplemental.
References
- 1.Boehme KA, Blattner C. Regulation of p53—Insights into a complex process. Crit Rev Biochem Mol Biol. 2009;44:367–392. doi: 10.3109/10409230903401507. [DOI] [PubMed] [Google Scholar]
- 2.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 3.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 4.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 5.Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Int J Biochem Cell Biol. 2003;35:606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 6.Blattner C, Hay T, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170–6182. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argentini M, Barboule N, Wasylyk B. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene. 2001;20:1267–1275. doi: 10.1038/sj.onc.1204241. [DOI] [PubMed] [Google Scholar]
- 8.Grossman SR, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 10.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 19 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Deng XW. The COP9 signalosome: an alternative lid for the 26S proteasome? Trends Cell Biol. 2003;13:507–509. doi: 10.1016/j.tcb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Sdek P, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Orlowski M, Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch Biochem Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 14.Kulikov R, Boehme KA, Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol. 2005;25:7170–7180. doi: 10.1128/MCB.25.16.7170-7180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 16.Poyurovsky MV, et al. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol Cell. 2003;12:875–887. doi: 10.1016/s1097-2765(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 17.Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem. 2006;281:28575–28583. doi: 10.1074/jbc.M513311200. [DOI] [PubMed] [Google Scholar]
- 18.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 19.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 20.Brignone C, Bradley KE, Kisselev AF, Grossman SR. A post-ubiquitination role for MDM2 and hHR23A in the p53 degradation pathway. Oncogene. 2004;23:4121–4129. doi: 10.1038/sj.onc.1207540. [DOI] [PubMed] [Google Scholar]
- 21.Lee SJ, Choi D, Rhim H, Kang S. E3 ubiquitin ligase RNF2 interacts with the S6′ proteasomal ATPase subunit and increases the ATP hydrolysis activity of S6′. Biochem J. 2005;389:457–463. doi: 10.1042/BJ20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dachsel JC, et al. Parkin interacts with the proteasome subunit alpha4. FEBS Lett. 2005;579:3913–3919. doi: 10.1016/j.febslet.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Corn PG, McDonald ER, III, Herman JG, El-Deiry WS. Tat-binding protein-1, a component of the 26S proteasome, contributes to the E3 ubiquitin ligase function of the von Hippel–Lindau protein. Nat Genet. 2003;35:229–237. doi: 10.1038/ng1254. [DOI] [PubMed] [Google Scholar]
- 24.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maki CG, Howley PM. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehme KA, Kulikov R, Blattner C. p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc Natl Acad Sci USA. 2008;105:7785–7790. doi: 10.1073/pnas.0703423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.