Abstract
The model plant Arabidopsis thaliana exhibits extensive natural variation in resistance to parasites. Immunity is often conferred by resistance (R) genes that permit recognition of specific races of a disease. The number of such R genes and their distribution are poorly understood. In this study, we investigated the basis for resistance to the downy mildew agent Hyaloperonospora arabidopsidis ex parasitica (Hpa) in a global sample of A. thaliana. We implemented a combined genome-wide mapping of resistance using populations of recombinant inbred lines and a collection of wild A. thaliana accessions. We tested the interaction between 96 host genotypes collected worldwide and five strains of Hpa. Then, a fraction of the species-wide resistance was genetically dissected using six recently constructed populations of recombinant inbred lines. We found that resistance is usually governed by single dominant R genes that are concentrated in four genomic regions only. We show that association genetics of resistance to diseases such as downy mildew enables increased mapping resolution from quantitative trait loci interval to candidate gene level. Association patterns in quantitative trait loci intervals indicate that the pool of A. thaliana resistance sources against the tested Hpa isolates may be predominantly confined to six RPP (Resistance to Hpa) loci isolated in previous studies. Our results suggest that combining association and linkage mapping could accelerate resistance gene discovery in plants.
Keywords: plant, quantitative trait loci, gene discovery
Only a minority of microbial species that interact with plants are parasitic. Host resistance to adapted parasites is frequently conferred by the presence of a functional allele of a resistance (R) locus (i.e., a locus contributing to host resistance that displays functional natural variation). In early genetic studies of immunity in major crop species and Arabidopsis thaliana (At), R loci specific for different parasites were often located on a limited number of genomic regions (1). This provoked the concept of major recognition complexes (MRCs) (2). Many R genes encode nucleotide-binding, leucine-rich repeat (NLR) proteins (3). Molecular mapping of At genes encoding NLR proteins revealed that regions enriched in NLR gene clusters and superclusters indeed correspond to MRCs (4). This circumstantial evidence and cloning of R genes helped establish that NLR genes are predominantly involved in parasite recognition in multiple organisms, including nonvascular plants, gymnosperms, monocots, and dicots (3). There are 149 NLR-homologous genes in the At accession Col-0 genome, of which ∼95% are expressed (5). Among them, 20 have been assigned a function. They include R genes mediating specific recognition of bacteria such as RPM1, RPS2, RPS4, RPS5, and RRS1 or oomycetes such as RAC1, WRR4, RPP1, RPP2, RPP5, RPP7, RPP8, and RPP13 (3, 6–12). RLM1 and RLM3 encode broad fungal resistance against ascomycete parasites (13, 14). Finally, HRT and RCY1 are alleles of RPP8 that confer virus resistance (15, 16).
RPP1, RPP2, RPP5, RPP7, RPP8, and RPP13 activate defense that culminates in the hypersensitive response (HR) on perception of the downy mildew parasite, Hyaloperonospora arabidopsidis ex parasitica (Hpa). Hpa is a highly specialized natural parasite of At. All Hpa are virulent on some accessions, but At frequently shows resistance (11, 17, 18). In A. thaliana, Hpa is the parasite species for which the most R loci and genes have been identified, consistent with an important role of Hpa in At evolution (19). Downy mildew infection on At provides a valuable molecular genetic model of gene-for-gene interactions. It can help to answer crucial questions regarding plant–parasite coevolution, important for both theoretical and applied purposes, including strategies to mine resistance in the germplasm or deploy it in crops. Previous genetic studies have identified at least 27 resistance to HyaloPeronospora Parasitica (RPP) genes, numbered RPP1 to RPP26 (excluding RPP15), RPP28, and RPP31 (2, 6–11, 20). After cloning of RPP1, RPP2, RPP5, RPP7, RPP8, and RPP13, another 8 loci within the 26 mapped were found to be allelic to one of the cloned genes (6, 10, 11, 21) (Table S1). Of 26 genetically mapped sources of resistance, 14 were found to be different alleles of only 6 R genes. However, all these studies were conducted in a restricted number of host genotypes, mostly Col-0, Ler, Nd-0, and Ws-0. It is unclear what fraction of species-wide Hpa recognition is accounted for by the six RPP genes cloned. The study of natural accessions is essential to comprehend ecological and evolutionary aspects of a species’ immunity (22).
Traditional approaches to dissecting natural variation in immunity involve the analysis of segregation of resistance in mapping populations. These can include recombinant inbred lines (RILs) to easily acquire low-resolution map positions of loci controlling the trait or so-called quantitative trait loci (QTLs) and map-based cloning of the genes using large F2 mapping populations (23). Nevertheless, carrying out F2 fine mapping to characterize the natural variation in any localized host population would be extremely laborious. Association or linkage disequilibrium (LD) mapping in Arabidopsis is emerging as a powerful alternative method to rapidly identify genome locations implicated in nearly every aspect of its biology (24). The current genotype datasets permit the mapping of putative causal loci with single-gene resolution. Yet, for gene discovery, it is important to avoid the major pitfall of association studies represented by false positives. To this end, it has been proposed that linkage and association mapping could be used in parallel (24, 25). Before embarking on candidate-gene validation, it is possible to validate association peaks by focusing on those that collocate with QTLs. In this work, we implemented this combined linkage and association mapping approach to dissect At resistance to Hpa at the population level.
Results
Natural Variation in Resistance to Five Isolates of H. arabidopsidis in a Set of 96 A. thaliana Accessions.
The Hpa isolates tested were Emco5, Emwa1, Hiks1, Noco2, and Emoy2. Of 441 host accession × parasite isolate interactions scored on leaves, 265 (60.1%) resulted in resistance, 156 (35.4%) resulted in susceptibility, and 20 (4.5%) were scored intermediate (I) (Tables S2 and S3). Thus, resistance was found to be a more probable outcome than susceptibility when challenging 1 of 96 At worldwide accessions with five isolates of Hpa originating from the United Kingdom. The resistance phenotype was not related to the geographic origin of accessions, although accessions from the United Kingdom were significantly more likely to be susceptible to Emco5 (P < 0.01) (Fig. S1). Ability to successfully infect a random host accession, or ratio of cases in the sample, was compared between different parasite isolates (Tables S2 and S3). Isolates were found to have a statistically distinct infection success rate [i.e., likelihood of causing full host susceptibility (P < 0.005) that ranged from 17% for Emco5 to 46.2% for Emoy2]. Thus, isolates of Hpa originating from the same location (e.g., Emco5 and Emoy2) can vary greatly in their virulence range on a subset of host accessions.
Analysis of Segregation of Resistance to Hpa in RIL Mapping Populations.
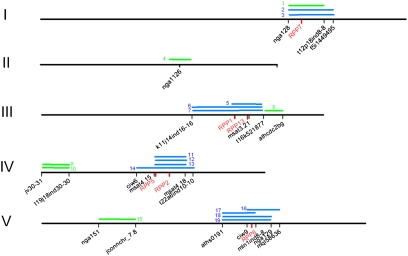
We studied six At RIL populations derived from crosses involving 12 parents included in the collection of 96 accessions selected for phenotypes unrelated to plant–parasite interactions. Five isolates of Hpa were used: Emco5, Emwa1, Hiks1, Noco2, and Cala2 to phenotype 17 RIL × Hpa isolate interactions for segregation of resistance. Segregation was observed in 11 interactions of 17 (Table 1). This included all four R × S interactions, as expected, and transgressive segregation in five of nine R × R interactions, one S × S interaction of three, and the only I × I interaction. We found that in the 13 cases where both parents have a similar disease-resistance phenotype, nearly one-half (six observed) showed no transgressive segregation, meaning the R genes are allelic or in the same genomic regions. This observation that R loci are often shared between accessions suggested that a small number of chromosomal (chr) regions are responsible for most of the observed natural variation in Hpa resistance in our sample. The mapping of 19 resistance specificities (i.e., recognition of a given parasite isolate in a given host accession) coming from 9 different accessions was consistent with this prediction (Table 1, Fig. 1, and Fig. S2 Top). Overall, the R loci that we mapped divided into two categories: 14 conferred single dominant resistance, whereas the remaining 5 conferred resistance with complex genetic basis (additive and epistatic). All 14 single dominant R loci (Table 1, blue, and Fig. 1) mapped to regions known as MRCs that contain cloned RPP genes (Fig. 1, red). The five complex R loci seemed to mostly locate in regions independent of MRCs (Table 1, green, and Fig. 1). As expected, our findings show that genome distribution of resistance sources is uneven and highly dependent on the type of resistance that they confer. Full resistance conferred by a single gene is the most common strategy of At seedlings for restricting Hpa infection, whereas resistance with complex genetic basis (additive and epistatic) plays a minor role in preventing Hpa. Therefore, in our sample of accessions, resistance at the seedling stage to the tested Hpa isolates is predominantly mediated by four MRC regions.
Table 1.
Identification of 19 A. thaliana resistance QTLs in a subset of interactions between six RIL populations and five isolates of H. arabidopsidis
| Hpa isolate | RIL population | Parental phenotypes | Inbred ratio | Predicted no. genes (ratio) | Est. theoretical proportions | χ2 | P | Detected loci | Predicted RPP locus location—candidate MRC cluster | |
| Parent A | Parent B | |||||||||
| Emco5 | Wt-5 × Ct-1 | R × R | 94:0 | NS | ||||||
| Cvi-0 × Ag-0 | R × R | 94:0 | NS | |||||||
| Nok-3 × Ga-0 | R × R | 74:18 | 2 (3:1) | 72.5:19.5 | 0.14 | 0.7 | 2 | Nok-3, ciw6 (IV-H)14 | Ga-0, k11j14ind16-16 (III-F)7 | |
| Bay-0 × Shahdara | I × R | 140:13 | 3 (7:1) | 133.8:19.1 | 2.23 | 0.13 | 1 | Shahdara, msat4.15 (IV-H)11 | ||
| Emwa1 | Wt-5 × Ct-1 | R × R | 94:0 | NS | ||||||
| Sorbo × Gy-0 | R × S | 25:67 | 2 (1:3) | 13.6:78.3 | 11.18 | <0.001 | 2 | Sorbo, t12p18ind8-8 (I-B)1, jconnchr5_7.8 (V-?)15 | ||
| Kondara × Br-0 | R × R | 80:14 | 3 (7:1) | 82.2–11.8 | 0.48 | 0.48 | 2 | Kondara, t12p18ind8-8 (I-B)3 | Br-0, ciw9 (V-J)17 | |
| Cvi-0 × Ag-0 | S × R | 62:32 | 1 (1:1) | 58.3:35.7 | 0.61 | 0.43 | 1 | Ag-0, ciw9 (V-J)18 | ||
| Nok-3 × Ga-0 | I × I | 11:81 | 3 (1:7) | 11.5–80.5 | 0.02 | 0.88 | 2 | Nok-3, nga1126 (II)4 | Ga-0, t19j18ind30-30 (IV-?)9 | |
| Bay-0 × Shahdara | R × R | 95:54 | 2 (3:1) | 122.1:26.8 | 33.6 | <0.001 | 2 | Bay-0, nga128 (I-B)2 | Shahdara, msat4.15 (IV-H)12 | |
| Hiks1 | Wt-5 × Ct-1 | R × R | 94:0 | NS | ||||||
| Nok-3 × Ga-0 | S × S | 18:74 | 2 (1:3) | 15.8:76.2 | 0.37 | 0.54 | 2 | Nok-3, athcdc2bg (III-?)8 | Ga-0, jv30-31 (IV-?)10 | |
| Noco2 | Sorbo × Gy-0 | R × R | 80:13 | 3 (7:1) | 81.3–11.7 | 0.16 | 0.69 | 2 | Sorbo, ciw9 (V-J)19 | Gy-0, k11j14ind16-16 (III-F)6 |
| Cvi-0 × Ag-0 | S × R | 59:34 | 1; (1:1) | 57.6:35.4 | 0.09 | 0.76 | 1 | Ag-0, ciw9 (V-J)16 | ||
| Nok-3 × Ga-0 | S × S | 0:92 | 0 | |||||||
| Cala2 | Nok-3 × Ga-0 | S × S | 0:92 | 0 | ||||||
| Bay-0 × Shahdara | R × R | 136:25 | 2 (3:1) | 132:29 | 0.67 | 0.41 | 2 | Bay-0, msat3.21 (III-F)5 | Shahdara, msat4.15 (IV-H)13 | |
The recognized Hpa isolate is reported at the left followed by the RIL population in which the QTL is segregated and the parental phenotypes are coded R (resistant), S (susceptible), or I (intermediate). Light and dark gray shades code for R × R and R × S or S × R crosses, respectively. Inbred ratio reports the observed segregation of the F8 RILs as number of R and S individuals. The predicted number of causal loci, expected R:S ratio, χ2, and P values of the observed segregation are reported together with the number of statistically significant QTLs (P < 0.05) for transgressively segregating RILs (NS indicates nonsegregating RILs). Estimated theoretical proportions were calculated using the segregation distortion observed in F9 at QTLs when known. The last two columns indicate the source and genome location of the QTLs, including single dominant R loci (blue) and additive, epistatic, and recessive R loci (green). The marker most linked to the QTL is indicated followed by the chromosome and the QTL interval or MRC when relevant. The reference number in superscript corresponds to the QTL mapping interval as depicted in Fig. 1.
Fig. 1.
Physical map locations of 19 resistance QTLs against H. arabidopsidis ex parasitica. Single dominant loci are depicted in blue bars, whereas additive, epistatic, and recessive loci are depicted in green bars. The markers flanking the mapping intervals are shown below in black. Cloned RPP genes are indicated in red. Chromosomes are pictured in scale and numbered I–V. The parental source and recognition specificity of the QTLs are reported in Tables S2 and S3, with the corresponding reference number.
Genome-Wide Association Mapping of Resistance to Downy Mildew in a Collection of At Accessions Reveals Signal at Known RPP Loci.
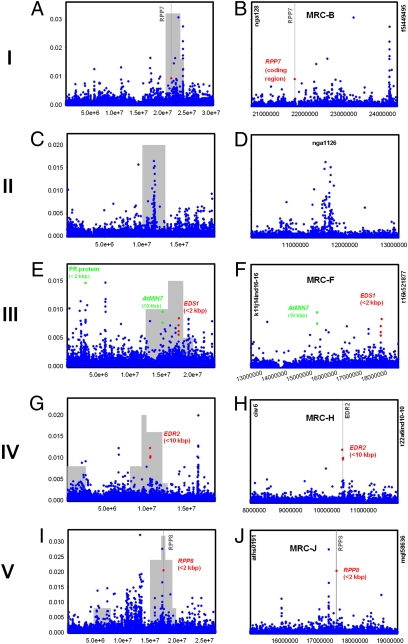
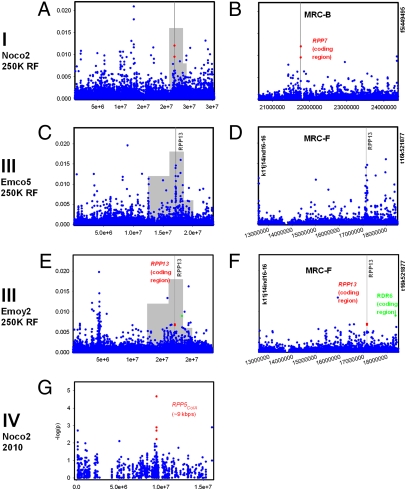
We conducted genome-wide association mapping (GWAM) of resistance in the collection of 96 accessions using the phenotypic data described in Tables S2 and S3, the 250K high-resolution SNP dataset, and random forest analysis method (Fig. S2) (http://arabidopsis.usc.edu/DisplayResults under the tab Defense). Then, focus was put on the QTL interval overlapping with MRCs where there is extensive linkage evidence of resistance. For example, GWAM of Hiks1 resistance is presented in Fig. 2. RPP7 is known to confer Hiks1 resistance in accession Ler and therefore, was used as a positive control (i.e., a gene required for Hpa resistance and known to exhibit natural variation). Accordingly, we found significant association to an SNP within RPP7 (At1g58602) (26). Although not within the top associations on chromosome I, when looking into the QTL interval overlapping with MRC-B (3.74 Mbp between nga128 and f5i1449495), the locus appeared as the fifth most associated genome region (Fig. 2B). Several top SNPs both on their respective chromosome and QTL interval located close to defense-related genes with a role in resistance to Hpa. These included enhanced disease susceptibility 1 (EDS1) (27) on chromosome III and enhanced disease resistance 2 (EDR2) (28) on chromosome IV (Fig. 2 F and H, respectively). Strikingly, on chromosome V, an SNP ∼2 kbp from RPP8 (At5g43470) came as the third highest association (Fig. 2I). Other noteworthy associations were observed in experiments with other Hpa isolates. The SNP most associated to resistance to Noco2 over the 3.74 Mbp of the QTL interval overlapping with MRC-B on chromosome I locates within the coding region of RPP7 (Fig. 3B). This evidence suggests that RPP7 is playing an important role in Noco2 resistance. SNPs within and surrounding RPP13 on chromosome III showed very high association with Emco5 resistance and a clear peak pattern centered around RPP13 (Fig. 3 C and D). Such high association for SNPs close to RPP genes was also observed in a GWAM experiment using the 2010 low-resolution genotype dataset (29). On chromosome IV, Noco2 resistance was most associated with a marker 9 kbp from RPP5ColA (At4g16860), which is known to confer resistance to Noco2 in accession Ler-0 (Fig. 3G). Finally, we found significant, yet relatively weak association in the RPP1 gene family or in RPP2a or RPP2b genes (involved in Cala2 recognition but not in recognition of the isolates tested here). Overall, this suggests that SNPs in RPP genes or neighboring regions, our positive controls in this study, usually stand out from other SNPs in terms of strength of association. They often correspond to the highest peaks within the QTL intervals, despite not being the highest chromosome-wide SNPs. A statistical test for enrichment in a priori candidate genes based on their putative role in plant immunity showed a clear enrichment in all GWAM experiments except Noco2 (Fig. S3), with enrichment being highest for the strongest associations. Overall, this shows that the GWAM retrieved primarily defense-related genes and that there is overrepresentation of SNPs close to positive-control genes among the top associated SNPs.
Fig. 2.
GWAM of resistance to Hiks1 in a collection of 96 A. thaliana accessions. GWAM of resistance was conducted using the 250K high-resolution dataset and random forest statistical method. Chromosomes are numbered I–V. Left corresponds to the whole chromosome. Right zooms in on the QTL intervals (flanking markers are on y axis) with prevalence to those overlapping with MRCs. (A) Significant association was found in the control gene RPP7 (At1g58602) on chromosome I. (B) When zooming in on the QTL interval on MRC-B between nga128 and f5i449495, it appeared as a moderately high SNP in the top 10 SNPs. (C and D) No defense-related candidate was found near the highest peaks on chromosome II. (E) Both SNPs show highly significant associations chromosome-wide that rank in the top 10 associated genome locations. (F) GWAM in a region of 5.9 Mbp corresponding to a QTL interval overlapping with MRC-F on chromosome III finds the top two SNPs to locate ∼10 kbp from AtMIN7 (At3g43300) and ∼1 kbp from the EDS1 gene (At3g48080). (G) On chromosome IV, the fourth highest peaks resides at ∼10 kbp from EDR2 (At4g19040). (H) When zooming in on the QTL interval between ciw6 and t22a6ind10-10, two more SNPs closer to EDR2 (<2 kbp) showed very high association. (I) On chromosome V, the third most associated SNP located ∼2 kbp from the coding region of RPP8 (At5g43470). (J) Over the 4.2 Mbp of the QTL interval overlapping with MRC-J (between aths0191 and mql58636), it was the second best SNP.
Fig. 3.
GWAM of downy mildew resistance in a collection of 96 A. thaliana accessions. GWAM of resistance was conducted on the 250K high-resolution and 2010 low-resolution genotype datasets, and it was calculated using random forest and unified mixed model K*+Q methods, respectively. Chromosomes are numbered I to IV. Left corresponds to the whole chromosome. Right shows a zoom in on the QTL interval (flanking markers are on y axis). (A) GWAM of Noco2 resistance using the 250K high-resolution dataset identified an SNP in RPP7 (At1g58602) coding region as the sixth most associated locus on chromosome I and (B) the most associated on the QTL interval overlapping with MRC-B between nga128 and f5i449495. (C) SNPs surrounding the positive control gene RPP13 appear as the third highest associations with Emco5 resistance on chromosome III and (D) present a very clear peak pattern centered on RPP13. (E, F) A similar observation was made when mapping Emoy2 resistance, although the peak pattern was not as clear. (G) GWAM of Noco2 resistance on chromosome IV using the 2010 low-resolution dataset identified the most associated SNP 9 kbp upstream of the RPP5ColA gene (RPP4 and At4g16860). Within the QTL interval overlapping with MRC-H, all four of the highest peaks of association reside 9 kbp north of RPP5 ColA.
Parallel GWAM Experiments Find Consistent Signal Predominantly in RPP Genes.
We surveyed in parallel associations with resistance to up to five isolates of Hpa. It is well-established that an RPP gene usually recognizes multiple Hpa isolates in an accession-dependent manner. Therefore, we partly expected to see association in the same genes in different experiments. In a preliminary 2010 low-resolution GWAM experiment, we observed that, in the QTL interval overlapping with MRC-F and more remarkably, on the whole of chromosome III, RPP13 was the only locus where SNPs showed significant association with resistance to all three strains tested, including Emco5 (Table S4). The 2010 low-resolution GWAM experiment also identified SNPs near RPP8 in very strong association with resistance to Emwa1/Emco5 (Table S4). In the 250K high-resolution GWAM experiment, we found that SNPs with importance > 0.004 (among the top 2% SNPs genome-wide; Materials and Methods) within or close to RPP13 and RPP5ColA were associated with resistance to Emco5/Emoy2 and Emco5/Noco2, respectively (SI RF and F candidates.xlsx). In addition to RPP13 and RPP5ColA, we found association with EDS1 and accelerated cell death 6 (ACD6) (30). This was not the case for RPP7 and RPP8 that only showed nearby signal in one experiment of five with the 250K high-resolution genotype data. Therefore, there was substantial consistency between and within the results from the 2010 low-resolution and 250K high-resolution experiments (Fig. 3 D and F and Table S4). Significant overlap was observed within the 250K high-resolution experiment using Random Forest and Fisher's exact test (http://arabidopsis.usc.edu/DisplayResults under the tab Defense). Altogether, we have performed different GWAM experiments with varying genotype datasets and multiple parasite strains, and we found that mainly, RPP genes and secondly, defense-related candidates tend to consistently harbor elevated association with resistance relative to the background.
Associations in RPP Genes Indicate Population-Wide Significance of Known Recognition Specificities and Putative Recognition Specificities.
We were able to observe very strong associations at the RPP13 and RPP7 loci with resistance to Emco5 and Hiks1, respectively, which is consistent with the known recognition specificities of RPP13Nd and RPP7Ler (Figs. 2B and 3D, respectively). We found very strong associations with SNPs near RPP5ColA in the Noco2 low- and high-resolution mapping experiments (Fig. 3G). This suggests that a moderate to large part of Emco5, Noco2, and Hiks1 resistance in our sample of accessions is conferred by RPP13, RPP5ColA, and RPP7, respectively. We were not able to find strong associations for known recognition specificities, such as RPP1/Noco2, RPP4/Emwa1, or RPP8/Emco5. Overall, highly associated SNPs were detected in three of six R locus/Hpa isolate combinations where a strong association was anticipated. Putative recognition specificities were also uncovered including RPP8/Hiks1 (Fig. 2 I and J) and RPP7/Noco2 (Fig. 3 A and B). The distribution of alleles within the collection of accessions for these two examples and EDS1/Hiks1 is shown in Fig. S4. Accessions carrying RPP13 haplotypes associated with resistance and susceptibility to Emco5 are shown in Fig. S5.
Identification of Candidate RPP Loci.
We used patterns of association found at positive controls and a priori candidates to select a threshold from which to determine new candidate RPP genes. The search focused on genomic regions corresponding to QTL intervals on four chromosomes for a total of ∼19.2 Mbp. We retained loci that showed consistent high signal (importance > 0.004) in at least two independent experiments (i.e., resistance with two or more isolates and SNPs less than 5 kbp apart). As mentioned previously, these criteria enabled the detection of the positive controls RPP13 and RPP5ColA and the defense-related genes EDS1 and ACD6 control genes. Using these criteria, we were able to select 56 candidate loci (Table S5). Enrichment for NLR encoding genes and defense-related genes accounting for 17 of 41 loci was observed (Table S5), consistent with genome-wide single-isolate resistance GWAM enrichment results (Fig. S3). These 17 loci included 9 genes with NLR signature, 3 defensin-like genes, an RbohD interactor (putatively involved in oxidative-stress response), and an avirulence-responsive gene (Table S5). The candidate genes occurred within ∼5.1 kbp average distance from the SNPs. The remaining 34 loci correspond to nonannotated genes or genes with a function not clearly related to parasite resistance. A large fraction of these SNPs with no obvious defense gene in their vicinity occurred near the borders of the QTL intervals, where resistance is not predicted to locate based on linkage evidence from the QTL analyses.
Discussion
Our study describes the implementation of single-gene resolution genome-wide association mapping of resistance to an obligate parasite in plants. The At–Hpa interaction was used as a model to develop GWAM genetics of resistance because of the straightforward phenotyping of large sets of accessions and prior knowledge of genes that control the interaction necessary to define selection criteria. As a proof of concept, we were able to identify four of six known RPP genes and two signaling genes that are known to show natural variation (EDS1 and ACD6) (31, 32). Several of these genes appeared as the top candidates in their respective QTL interval where the QTL is most likely to reside. In a gene-discovery pipeline, this means that we would have investigated these genes first. Results were consistent across experiments with different Hpa isolates and different genotype datasets of variable resolution.
Our results shed light on the species-wide basis of seedling resistance. Resistance is the most probable outcome of the interaction between At and the tested Hpa isolates. Single dominant resistance seems the most common mode of resistance, whereas additive resistance and epistatic resistance/susceptibility factors play a minor role. In addition to 28 previously identified loci reported in the literature, we have mapped 19 R loci from 9 accessions. In total, this comes to at least 47 RPP loci with assigned map location, including 38 distributed in 4 MRCs. To get an indication of how often the genetic basis for resistance or susceptibility is shared between accessions, we analyzed segregation in 13 RILs × Hpa isolate interactions. Nearly one-half showed no transgressive segregation, either because of tight linkage of different resistant sources or allelism (recognition of the same isolate by the same R locus). There is currently no straightforward way of verifying which condition prevails in all of the populations of interest. The frequent absence of segregation observed as well as the mapping of at least 38 loci to 4 MRCs prove that ∼80% of the R genes against the tested Hpa isolates locate in a very restricted portion of the genome. The GWAM results are consistent with this concentration of R loci, and they enable discrimination between allelism and tight linkage. Because of the nature of association studies, there needs to be sufficient occurrences of correlation of a phenotype with an allele (the RPP gene must play a significant enough role in the population in conferring resistance to the tested isolate) to implicate a locus. This suggests that the specificities known at RPP13 and RPP5ColA are likely to be spread among a large subset of the 96 accessions. The fact that other RPP genes such as RPP7 and RPP8 were top candidates suggests that they also play a significant role in this sample of accessions. This leads us to propose that the pool of A. thaliana resistance sources against the tested Hpa isolates consists predominantly of the six RPP loci revealed in previous studies and may also include some of the candidates identified in this work. For example, our data suggest that natural variation at EDS1 and ACD6 might also cause altered disease-resistance phenotypes.
There are several possible explanations for why no clear association was detected when a strong association was anticipated (three of six R locus/Hpa isolate combinations and two of six RPP loci with all five Hpa isolates). It could be that the specificity reported for an RPP gene does not represent a large part of what causes resistance to the tested isolate in the collection. This is in conjunction with shown epistasis at RPP genes; an allele at any of several loci can confer resistance to an Hpa isolate in an accession-dependent manner. Considering the small size of our collection, in these cases, the statistics may not be capable of detecting the faint signal. Increasing the sample size could solve this problem. In addition, accumulating evidence suggests that Hpa isolates are capable of suppressing the recognition of avirulence effectors on hosts that have functional R genes and subsequently, causing disease (33). Such hosts may have introduced a bias in the calculation of the association by behaving like susceptible plants while having similar haplotypes to resistant plants at some loci. It is possible that clearer associations and a reduction in false positives could have been observed by scoring the recognition of a given avirulence protein rather than resistance to an isolate. This was shown to work using bacterial avirulence effectors including AvrRpm1 and AvrRpt2 (24).
Several major peaks were observed in regions that do not harbor NLR genes in Col-0 reference genome. These could be false positives caused by population structure, they could correspond to R genes that are absent from Col-0 but present in other accessions, or they could be non-NLR genes with an important role in resistance, similar to EDS1.
We propose a general approach for R gene discovery from GWAM studies in plants that limits the major problem caused by false positives. The preliminary stage is the mapping of resistance to multiple parasite isolates using several segregating RIL or F2 populations. This serves to identify regions in the genome that are most likely to carry the causal genes. Then, GWAM of resistance to multiple parasite isolates is performed and analyzed using three criteria. First, SNPs in high association compared with background noise are selected. Second, the selected SNPs are examined with particular focus on those that are within regions where QTLs are predicted. Third, candidate SNPs from independent experiments are identified using different parasite isolates that colocate in the same regions. Ideally, loci fulfilling all three criteria may be genes that are likely to be involved in resistance, such as the nine NLRs that we identified. However, a locus fulfilling any two of these criteria would be worth looking at closely. To summarize, GWAM provides an insightful approach when used in concert with linkage analysis of resistance to increase mapping resolution from QTL down to candidate gene level and to reduce background. thus, we think that it has the potential to substantially accelerate gene-discovery pipelines.
Materials and Methods
A. thaliana Genotypes and Growth and H. arabidopsidis Materials and Infections.
Procedures described in SI Text.
Genetic Mapping Methods.
Linkage mapping of QTLs was performed using MAPQTL5 (34) following the multiple QTL mapping method after automatic cofactors selection. A permutation test was performed to calculate the 5% limit for statistical significance genome-wide. Binary scorings of RILs resistance phenotype were encoded in scores 95 and 5 for resistance and susceptibility, respectively. In association mapping experiments, binary phenotypes were used, excluding intermediate scores. The GWAM on the 2010 low-resolution genotype dataset scanned 1,214 fragments of 583 bp on average (35) and ∼800-bp fragments of 27 R genes (36). We scanned the genome using unified mixed model K*+Q on haplotypes (37) modified for binary inputs and selected SNPs with P < 0.05. For GWAM using the 250K high-resolution genotype dataset (216043–216121 SNPs after filtering), random forest (RF) tests (38) were performed using the random forest package in the R statistical program (R core development team, version 2.6.1). Essentially, this approach is a machine-learning based method of identifying which of the many SNPs in a genome best explain the phenotypes of interest. It assembles classification and regression trees for bootstrapped subsets of the SNPs in the data, resulting in a forest of 20,000 decision trees (38). Each tree had 464 random SNPs. The entire genome was evaluated for each phenotype with one RF. Then, the effect of an SNP on a phenotype was determined by evaluating each split of a node made on that SNP in the RF. The decision trees are designed such that the gini impurity criterion for subsequent nodes must be less than the parent node. The importance of an SNP to a phenotype is determined by evaluating the mean gini decreases for each individual SNP over all trees in the RF that included it; this yields higher scores for more important SNPs. In addition, Fisher's exact tests were conducted as per Atwell et al. (24). Fisher's exact test is expected to have false positives because of population structure, whereas RF may give false negatives. The overlap of RF and Fisher's exact results should, therefore, be more reliable. Finally, an in silico search was done in the Col-0 genome for candidate genes surrounding the selected associated loci.
Supplementary Material
Acknowledgments
This work was supported by the Gatsby Charitable Foundation (J.D.G.J.) and National Science Foundation Grant DEB-0519961 (to M.N.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913160107/-/DCSupplemental.
References
- 1.Crute IR, Pink DAC. Genetics and utilization of pathogen resistance in plants. Plant Cell. 1996;8:1747–1755. doi: 10.1105/tpc.8.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holub E, Beynon J. Andrews JH, Tommerup IC, editors. Symbiology of mouse-ear cress (Arabidopsis thaliana) and oomycetes. Advances in Botanical Research: Incorporating Advances in Plant Pathology. 1997;24:227–273. [Google Scholar]
- 3.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speulman E, Bouchez D, Holub EB, Beynon JL. Disease resistance gene homologs correlate with disease resistance loci of Arabidopsis thaliana. Plant J. 1998;14:467–474. doi: 10.1046/j.1365-313x.1998.00138.x. [DOI] [PubMed] [Google Scholar]
- 5.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botella MA, et al. Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell. 1998;10:1847–1860. doi: 10.1105/tpc.10.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinapidou E, et al. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- 8.Parker JE, et al. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDowell JM, et al. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000;21:177–188. doi: 10.1046/j.1365-313x.2000.00664.x. [DOI] [PubMed] [Google Scholar]
- 11.Slusarenko AJ, Schlaich NL. Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica) Mol Plant Pathol. 2003;4:159–170. doi: 10.1046/j.1364-3703.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- 12.Borhan MH, et al. WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Mol Plant Microbe Interact. 2008;21:757–768. doi: 10.1094/MPMI-21-6-0757. [DOI] [PubMed] [Google Scholar]
- 13.Staal J, Kaliff M, Bohman S, Dixelius C. Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 2006;46:218–230. doi: 10.1111/j.1365-313X.2006.02688.x. [DOI] [PubMed] [Google Scholar]
- 14.Staal J, Kaliff M, Dewaele E, Persson M, Dixelius C. RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 2008;55:188–200. doi: 10.1111/j.1365-313X.2008.03503.x. [DOI] [PubMed] [Google Scholar]
- 15.Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell. 2000;12:663–676. doi: 10.1105/tpc.12.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi H, et al. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 2002;32:655–667. doi: 10.1046/j.1365-313x.2002.01453.x. [DOI] [PubMed] [Google Scholar]
- 17.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holub EB, Williams PH, Crute IR. Natural infection of Arabidopsis thaliana by Albugo candida and Peronospora parasitica. Phytopathology. 1991;81:1226. [Google Scholar]
- 19.Salvaudon L, Héraudet V, Shykoff JA. Genotype-specific interactions and the trade-off between host and parasite fitness. BMC Evol Biol. 2007;7:189. doi: 10.1186/1471-2148-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holub EB, Beynon JL, Crute IR. Phenotypic and genotypic characterization of interactions between isolates of peronospora parasitica and accessions of Arabidopsis thaliana. Mol Plant Microbe Interact. 1994;7:223–239. [Google Scholar]
- 21.van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 2002;29:439–451. doi: 10.1046/j.0960-7412.2001.01229.x. [DOI] [PubMed] [Google Scholar]
- 22.Holub EB. Natural variation in innate immunity of a pioneer species. Curr Opin Plant Biol. 2007;10:415–424. doi: 10.1016/j.pbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Shindo C, Bernasconi G, Hardtke CS. Natural genetic variation in Arabidopsis: Tools, traits and prospects for evolutionary ecology. Ann Bot. 2007;99:1043–1054. doi: 10.1093/aob/mcl281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010 doi: 10.1038/nature08800. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- 26.Eulgem T, et al. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 2007;49:829–839. doi: 10.1111/j.1365-313X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 27.Parker JE, et al. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vorwerk S, et al. EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol. 2007;7:35. doi: 10.1186/1471-2229-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordborg M, et al. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2002;30:190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- 30.Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- 31.Caldwell KS, Michelmore RW. Arabidopsis thaliana genes encoding defense signaling and recognition proteins exhibit contrasting evolutionary dynamics. Genetics. 2009;181:671–684. doi: 10.1534/genetics.108.097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todesco M, et al. A fitness trade off between growth and disease resistance in Arabidopsis thaliana. Nature. 2010 in press. [Google Scholar]
- 33.Sohn KH, Lei R, Nemri A, Jones JDG. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell. 2007;19:4077–4090. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Ooijen JW. MAPQTL 5, Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Wageningen, The Netherlands: Kyazma B.V.; 2004. [Google Scholar]
- 35.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bureau A, et al. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.