Abstract
Zinc is an essential micronutrient for all living organisms. When facing a shortage in zinc supply, plants adapt by enhancing the zinc uptake capacity. The molecular regulators controlling this adaptation are not known. We present the identification of two closely related members of the Arabidopsis thaliana basic-region leucine-zipper (bZIP) transcription factor gene family, bZIP19 and bZIP23, that regulate the adaptation to low zinc supply. They were identified, in a yeast-one-hybrid screening, to associate to promoter regions of the zinc deficiency-induced ZIP4 gene of the Zrt- and Irt-related protein (ZIP) family of metal transporters. Although mutation of only one of the bZIP genes hardly affects plants, we show that the bzip19 bzip23 double mutant is hypersensitive to zinc deficiency. Unlike the wild type, the bzip19 bzip23 mutant is unable to induce the expression of a small set of genes that constitutes the primary response to zinc deficiency, comprising additional ZIP metal transporter genes. This set of target genes is characterized by the presence of one or more copies of a 10-bp imperfect palindrome in their promoter region, to which both bZIP proteins can bind. The bZIP19 and bZIP23 transcription factors, their target genes, and the characteristic cis zinc deficiency response elements they can bind to are conserved in higher plants. These findings are a significant step forward to unravel the molecular mechanism of zinc homeostasis in plants, allowing the improvement of zinc bio-fortification to alleviate human nutrition problems and phytoremediation strategies to clean contaminated soils.
Keywords: biofortification, zinc homeostasis regulation, plant nutrition, abiotic stress, adaptation
Zinc is an essential cofactor for many transcription factors, protein interaction domains, and enzymes in plants (1). Plants are thought to control zinc homeostasis by using a tightly regulated network of zinc status sensors and signal transducers controlling the coordinated expression of proteins involved in zinc acquisition from soil, mobilization between organs and tissues, and sequestration within cellular compartments (2). Although candidate genes for the required proteins such as zinc transporters and chelator biosynthesizing enzymes are found, no regulator of such network was ever identified in plants.
Zinc influx facilitators, members of the ZIP family of metal transporters, are thought to play a major role in zinc uptake in plants (3). In Arabidopsis there are 15 ZIP genes (4), with ZIP1, ZIP2, ZIP3, and IRT3 functionally characterized as zinc uptake transporters (3, 5). Gene expression analysis has shown that approximately half of the ZIP genes are induced in response to zinc deficiency (3, 5–8). The ZIP4 gene in particular is strongly induced upon shortage in zinc supply (3, 6–8).
We focused on the promoter of this zinc-deficiency-responsive gene as the starting point for unraveling the regulation of the zinc homeostasis network in plants. By using DNA fragments of the zinc-deficiency-responsive Arabidopsis ZIP4 gene promoter as bait in a yeast-one-hybrid assay, we have identified two transcription factors, which constitute an essential regulatory function for the adaptation of plants to zinc deficiency, and which are unique among known transcription factors of the plant zinc homeostasis network.
Results
Complementation Analysis and Zinc-Deficiency Response of the Arabidopsis ZIP4 Gene.
The ZIP4 gene is induced upon zinc deficiency and resembles known zinc transporters. To show its function as a zinc transporter we used it to complement the Saccharomyces cerevisiae zrt1zrt2 mutant, which is defective in high- and low-affinity zinc uptake transporters causing an increased zinc requirement (9). Complementation was effective in a drop spotting assay and in liquid culture (Fig. 1 A and B), supporting the conclusions that ZIP4 mediates zinc uptake. The analysis of ZIP4 transcript levels by quantitative RT-PCR (qPCR) on 3-week-old Arabidopsis seedlings grown on agar plates with and without zinc supply (Fig. 1C) shows a very strong induction under zinc-deficient conditions, in agreement with previous reports (3, 6–8). We cloned the promoter of the ZIP4 gene upstream of the GUS reporter gene and transformed it to Arabidopsis. GUS expression was apparent only when transgenic plants were grown on zinc-deficient medium, in full analogy to endogenous ZIP4 gene expression (Fig. 1D).
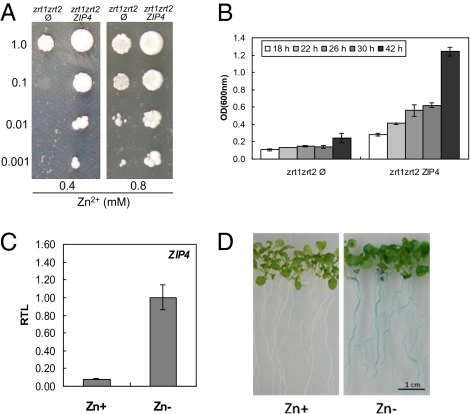
Fig. 1.
Gene expression and zinc transport of the Arabidopsis ZIP4 zinc transporter. (A) Growth of zrt1zrt2 S. cerevisiae cells carrying either the empty vector (zrt1zrt2 Ø) or expressing ZIP4 (zrt1zrt2 ZIP4) was assayed by spotting serial dilutions of cells (OD600 is shown on the left) on SD-URA selective medium with 0.4 or 0.8 mM ZnCl2. (B) OD measurements at the indicated time-intervals of zrt1zrt2 Ø and zrt1zrt2 ZIP4 on SD-URA selective liquid medium with 0.4 mM ZnCl2 (mean ± SEM). (C) Relative transcript levels (RTL) of ZIP4 in 3-week-old Arabidopsis seedlings grown in MS medium, with 30 μM ZnSO4 (Zn+) or without (Zn-) (mean ± SEM). (D) GUS expression analysis in roots of Arabidopsis 3-week-old seedlings, stably transformed with a ZIP4promoter::GUS construct, grown in MS medium, with 30 μM ZnSO4 (Zn+) or without (Zn-).
Yeast-One-Hybrid Screening and Identification of bZIP19 and bZIP23.
To identify transcription factors controlling ZIP4 expression, we used six overlapping ZIP4 promoter fragments as baits in a yeast-one-hybrid assay. In addition we used three tandem repeats of a 10-bp palindrome motif as bait. This motif is present in two copies close to the predicted transcription start in the ZIP4 promoter (Fig. 2A). Only when screening with baits containing two copies of the 10-bp palindrome or the three tandem repeats (Fig. 2A; fragments E, F, and G), several clones of bZIP19 (At4g35040) and bZIP23 (At2g16770) were identified (Table S1). These are two genes of the basic region/leucine zipper motif (bZIP) family of transcription factors. Arabidopsis contains 75 members of the bZIP family, divided over 10 groups based on sequence similarities (10). All bZIP proteins contain a characteristic and highly conserved basic domain, which binds DNA, and a leucine zipper dimerization motif (11, 12). bZIPs can form homo- and/or heterodimers, which bind DNA in a sequence-specific manner and are capable of binding short palindromic or pseudopalindromic target sequences (13). Plant bZIPs are important for the regulation of pathogen defense, environmental signaling, and development. However, so far no function is assigned to approximately two-thirds of the bZIP members (10). Among these are the bZIP19 and bZIP23 genes, which belong to bZIP group F (10). This group contains a third member, bZIP24 (At3g51960), not identified in the yeast-one-hybrid assay, which was recently found to be a regulator of salt response (14). The bZIP19 and bZIP23 predicted protein sequences share 69% of amino acid sequence identity, and each shares 28% and 32%, respectively, with bZIP24. All three members of the F group contain two characteristic histidine-rich motifs (Fig. S1A). Based on Genevestigator (www.genevestigator.com) assembled microarray expression data, bZIP19, bZIP23, and bZIP24 are expressed at low levels in all examined tissues (Fig. S2 A–C).
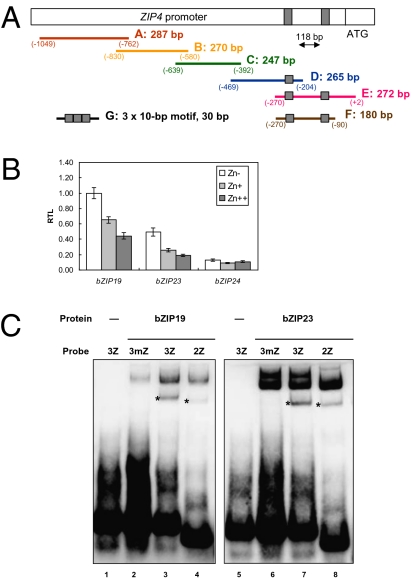
Fig. 2.
Yeast-one-hybrid baits, F group bZIP gene expression, and EMSA with bZIP19 and bZIP23. (A) Schematic diagram of the bait fragments (A-G) used to construct the reporter vectors in the yeast-one-hybrid assay. The gray box represents a 10-bp palindromic motif. (B) Relative transcript levels (RTL) of bZIP19, bZIP23, and bZIP24 in 3-week-old Arabidopsis seedlings grown in MS medium, without (Zn-), with 30 μM (Zn+) or with 300 μM ZnSO4 (Zn++) (mean ± SEM). (C) EMSA show that in vitro translated bZIP19 and bZIP23 protein can specifically bind to three tandem repeats of the 10-bp palindromic motif (3Z; 3 x ZDRE; lanes 3 and 7), corresponding to bait G in A, and to two tandem repeats (2Z; lanes 7 and 8), causing the bound fragments to migrate slower through the gel (*) than the labeled free probes found at the bottom of the gel. The observed shift (*) does not occur when using a three-tandem-repeat fragment of a mutated element (3mZ; lanes 2 and 6). A control assay, in which the empty vector was used for in vitro translation, does not show the band shift (lanes 1 and 5), indicating specific binding of bZIP19 and bZIP23 to the ZDRE probes.
To investigate the involvement of these three bZIP genes in controlling adaptation of plants to low zinc supply, we analyzed their transcript levels by qPCR in 3-week-old wild-type seedlings grown on agar plates at normal and high zinc concentrations and without zinc supply. Expression of both bZIP19 and bZIP23 was approximately two times higher when no zinc was supplied compared with normal zinc supply. bZIP19 was slightly higher expressed than bZIP23 (Fig. 2B). Expression of bZIP24 was lower than expression of the other two and not obviously affected by the zinc status of the medium. Therefore, we hypothesized that only bZIP19 and bZIP23 are involved in the regulation of the response to zinc deficiency in Arabidopsis. If so, single or double mutants for these genes should be impaired in a proper zinc deficiency response.
Double T-DNA Insertion Mutant m19m23 Is Hypersensitive to Zinc Deficiency.
To determine their mutant phenotypes, we obtained homozygous T-DNA insertion lines for the bZIP19 and bZIP23 genes (respectively named m19 and m23), which were devoid of full-length bZIP19 or bZIP23 transcript (Fig. S2 D and E). We crossed both single mutants to generate a homozygous double mutant line (m19m23). Wild-type plants and single or double mutants grown on soil did not show any obvious phenotypic differences (Fig. S3); however, when seedlings were grown on zinc-deficient agar media, only the double mutant line showed a zinc-deficiency hypersensitive phenotype (Fig. 3A), visible as very poor growth and strong chlorosis. To exclude in vitro effects during tissue culture, we grew plants for a longer time on hydroponics medium. Four-week-old double mutant plants growing at low zinc supply showed a strong growth reduction compared to wild-type plants, as determined by dry weight comparison (Fig. 3 B and C). These plants also had a decreased zinc uptake, with only 58% and 73% of the respective shoot and root zinc concentration of wild-type plants (Fig. 3C). Under these conditions, also the m19 single mutant showed reduction in growth and decreased zinc uptake when compared with the wild type (Fig. 3B). When growing in zinc-sufficient or zinc-excess media, we did not see differences between the mutants and the wild type (Fig. 3 A and B), neither for zinc concentration nor for dry weight (Fig. S4 A and B). Also when plants were grown on potting mix, containing Zn as trace element, there was no obvious morphological difference between wild-type and mutant plants, including plant fertility. To prove that mutations in bZIP19 and bZIP23 indeed caused the phenotype of the m19m23 double mutant we expressed either the bZIP19 or the bZIP23 cDNA, each under control of the constitutive CaMV 35S promoter, in the double mutant. This expression fully complemented its zinc deficiency hypersensitive phenotype (Fig. 4A).
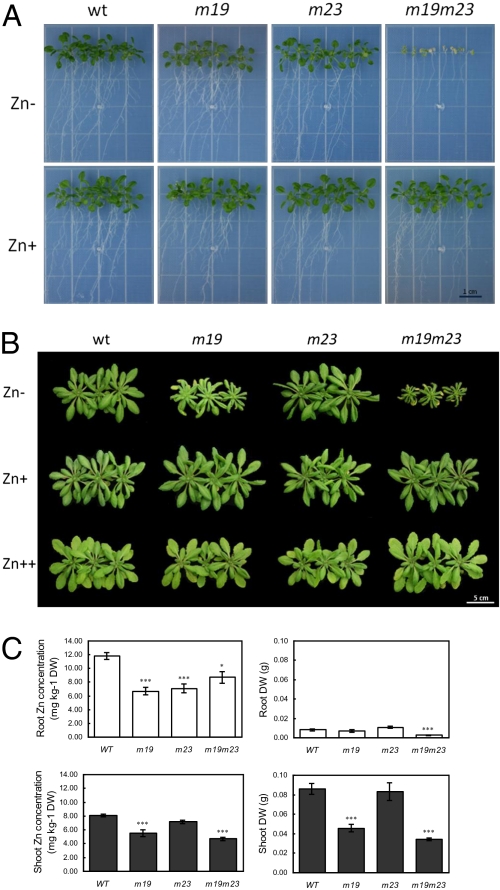
Fig. 3.
The double T-DNA insertion mutant m19m23 is hypersensitive to zinc deficiency. (A) Effect of zinc deficiency on 3-week-old seedlings of Arabidopsis (WT), bZIP19 T-DNA insertion mutants (m19), bZIP23 T-DNA insertion mutants (m23), and double T-DNA insertion mutants (m19m23) grown in MS medium without (Zn-) and with 30 μM ZnSO4 (Zn+). (B) Effect of zinc supply on 4-week-old plants of Arabidopsis (WT), m19, m23, and m19m23, grown in hydroponics at 0.05 μM (Zn-), 2 μM (Zn+), and 25 μM ZnSO4 (Zn++). (C) Zinc concentration, in mg·kg−1 of dry weight, and dry weight (DW), in g, of shoots (gray bars) and roots (white bars) of 4-week-old WT, m19, m23, and m19m23 plants, grown in hydroponics at 0.05 μM ZnSO4 (Zn-). *, P < 0.05; **, P < 0.01; ***, P < 0.001; representing significant differences of the mean in comparison with the WT mean (mean ± SEM).
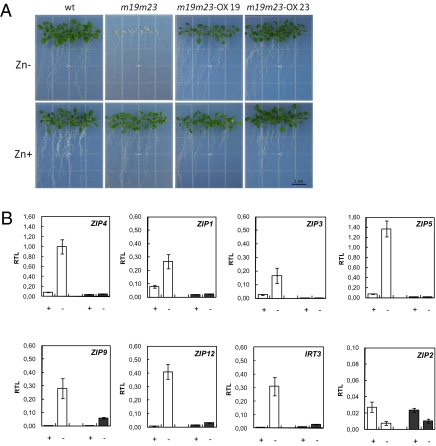
Fig. 4.
Complementation study and expression analysis of putative target genes. (A) Arabidopsis wild-type plants (WT), double T-DNA insertion mutants (m19m23), and double mutants constitutively expressing either bZIP19 (m19m23-OX19) or bZIP23 (m19m23-OX23), grown for 3 weeks in MS medium without (Zn-) and with 30 μM ZnSO4 (Zn+). (B) Relative transcript levels (RTL) of ZIP4, ZIP1, ZIP3, ZIP5, ZIP9, ZIP12, IRT3, and ZIP2 in 3-week-old wild-type seedlings of Arabidopsis (WT) (white bars) and m19m23 double mutants (gray bars) grown in MS medium with 30 μM ZnSO4 (Zn+) or without (Zn-) (mean ± SEM).
These findings show that bZIP19 and bZIP23 encode essential transcription factors that control the zinc deficiency response in Arabidopsis. They act redundantly, although bZIP19 is only partially redundant. bZIP transcription factors generally act as dimers. Because they are partially redundant, bZIP19 and bZIP23 are unlikely to act as strict hetero(di)mers, which is in line with previous predictions for bZIP proteins (15).
ZDRE Is Present in the Promoter of Arabidopsis Zinc-Deficiency-Responsive Genes.
The DNA target sequence for binding bZIP19 and bZIP23 should be within the 10-bp imperfect palindrome present as two copies in the ZIP4 promoter (Fig. 2A), because three tandem copies of this sequence were sufficient to identify both bZIP genes separately in the yeast-one-hybrid assay. To confirm that bZIP19 and bZIP23 specifically bind to this element, we performed an electrophoretic mobility shift assay (EMSA), using the three tandem copies of the 10-bp palindrome, as used in the yeast-one-hybrid assay. In addition we used two tandem copies of the palindrome and three tandem copies of a modified 10-bp palindrome in which the TCGA core has been mutated to TAGA. This analysis showed that both bZIP19 and bZIP23 can bind in vitro to three and two tandem copies of the 10-bp palindrome, but not to three tandem copies of a mutated version of the 10-bp palindrome (Fig. 2C). We therefore decided to refer to the consensus sequence of this unique cis element (RTGTCGACAY) as the Zinc Deficiency Response Element (ZDRE). The ZDRE does not have the typical ACGT core as found in the A-box (TACGTA), C-box (GACGTC) or G-box (CACGTG) (10, 16, 17) DNA elements, to which plant bZIPs are known to preferentially bind. Although other consensus bZIP binding sites have been reported (18, 19), the ZDRE element identified in this study is not among them.
To further confirm that ZDRE is indeed the characteristic cis element to target genes for transcriptional control by bZIP19/23, we screened the promoters of 14 other Arabidopsis ZIP transporter genes for ZDREs. ZIP1, ZIP3, ZIP4, ZIP5, ZIP9, ZIP12, and IRT3 contain one or two ZDRE copies in their promoters (allowing maximal two mismatches outside the core sequence), and these genes do not show the typical induction of expression in the m19m23 double mutant under zinc-deficient conditions as we saw in the wild-type (Fig. 4B). Expression of ZIP2, which has no ZDRE sequence in its promoter and is not zinc deficiency-induced (3), is not affected when comparing mutant and wild-type (Fig. 4B).
To determine the effect of loss of bZIP19/23 function on global gene expression we performed a microarray experiment comparing roots of 4-week-old m19m23 double mutant plants with those of wild-type plants. Plants were grown hydroponically and treated in their last week with low zinc supply. Only 23 genes showed a statistically significant alteration of transcript levels exceeding a 1.5-fold difference threshold (Table 1). Most of them (16 genes) were down-regulated in the double mutant, including the mutated bZIP19 gene. Among these, 11 genes are known to be induced in wild-type Arabidopsis roots upon zinc deficiency (3, 6–8) and nine genes contain one or more copies of the ZDRE (allowing one mismatch) in their promoter regions and are thus the likely direct targets of bZIP19 and bZIP23, important for the primary zinc deficiency response, whereas the other genes represent a secondary effect. These findings confirm the important role of the bZIP19/23 genes in controlling Arabidopsis zinc deficiency response. It also shows that the strong negative effect on growth and on zinc concentration of the m19m23 mutant when grown under zinc deficiency (Fig. 3 A–C) is largely explained by reduction in expression of a relatively small group of zinc homeostasis genes mainly involved in uptake and translocation of metals.
Table 1.
Differentially expressed genes detected by comparative microarray analysis of the root transcriptome of Arabidopsis wild-type and m19m23 double mutant plants
| Annotation | Gene model | FC (≥1.5) | A | Adjusted P value (BH) |
| Down-regulated | ||||
| bZIP transcription factor family protein | AT4G35040 | −35.87 | 7.31 | 9.27E-08 |
| ZIP3 (ZINC TRANSPORTER 3 PRECURSOR)* | AT2G32270 | −26.23 | 9.67 | 1.35E-09 |
| phosphatidylinositol 3- and 4-kinase family protein/ubiquitin family protein | AT5G24240 | −8.43 | 8.20 | 3.57E-08 |
| ZIP5 (ZINC TRANSPORTER 5 PRECURSOR)* | AT1G05300 | −8.34 | 6.90 | 4.15E-07 |
| ZIP4 (ZINC TRANSPORTER 4 PRECURSOR)* | AT1G10970 | −7.19 | 7.68 | 9.27E-08 |
| ZIP9 (ZINC TRANSPORTER 9 PRECURSOR)* | AT4G33020 | −3.23 | 6.90 | 4,41E-04 |
| ZIP1 (ZINC TRANSPORTER 1 PRECURSOR)* | AT3G12750 | −3.21 | 7.33 | 1.10E-05 |
| nicotianamine synthase, putative (NAS2)* | AT5G56080 | −3.02 | 9.24 | 1.28E-02 |
| ATPAP27/PAP27 (purple acid phosphatase 27) | AT5G50400 | −2.51 | 9.79 | 1.10E-05 |
| nicotianamine synthase, putative (NAS4)* | AT1G56430 | −2.46 | 7.68 | 1.98E-03 |
| ATARP9 (ACTIN-RELATED PROTEIN 9) | AT5G43500 | −2.33 | 7.28 | 3.16E-05 |
| FRD3 (FERRIC REDUCTASE DEFECTIVE 3) | AT3G08040 | −1.78 | 8.61 | 1.98E-03 |
| prolyl oligopeptidase, putative/prolyl endopeptidase, putative/postproline cleaving enzyme, putative* | AT1G20380 | −1.77 | 8.28 | 6.20E-03 |
| WR3 (WOUND-RESPONSIVE 3) | AT5G50200 | −1.60 | 11.63 | 2.48E-02 |
| ZIP10 (ZINC TRANSPORTER 10 PRECURSOR)* | AT1G31260 | −1.60 | 6.30 | 3.09E-02 |
| similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61260.1) | AT4G04990 | −1.59 | 8.41 | 4.22E-03 |
| Up-regulated | ||||
| LAC2 (laccase 2) | AT2G29130 | 1.82 | 7.01 | 1.82E-02 |
| ANR1; DNA binding / transcription factor | AT2G14210 | 1.71 | 8.90 | 2.85E-02 |
| similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21670.1) | AT1G21680 | 1.57 | 8.98 | 2.85E-02 |
| ATEBP/ERF72/RAP2.3 (RELATED TO AP2 3) | AT3G16770 | 1.55 | 9.68 | 4.18E-02 |
| COBL2 (COBRA-LIKE PROTEIN 2 PRECURSOR) | AT3G29810 | 1.54 | 7.40 | 3.42E-02 |
| kelch repeat-containing F-box family protein | AT1G80440 | 1.52 | 11.05 | 9.48E-03 |
| ATPSK2 (PHYTOSULFOKINE 2 PRECURSOR) | AT2G22860 | 1.52 | 9.20 | 4.24E-02 |
*Indicates genes that contain one or more copies of the ZDRE in their promoter region.
Roots of 4-week-old plants grown in hydroponics and exposed in their last week to low zinc supply (0.05 μM ZnSO4) were used. Fold change (FC) ≥ 1.5 and adjusted P values (Benjamini-Hochberg, BH) ≤ 0.05 were used as cutoffs. Average expression values (log2 scale) of the gene models in the data set are indicated as A.
bZIP19 and bZIP23 and the ZDRE Motifs Are Conserved in the Plant Kingdom.
bZIP genes that are closely related to Arabidopsis bZIP19 and bZIP23 are found in several species, including rice, poplar, and soybean (respectively OsbZIP48, PtrbZIP38/PtrbZIP39, and GmbZIP121/GmbZIP122), but also in gymnosperms like Picea glauca and Pinus taeda (PgbZIP4 and PtbZIP12, respectively) and lower plants like the Bryophyte Physcomitrella patens (PpbZIP18 and PpbZIP19) (20) (Fig. S1 B and C). In addition to conservation of the transcription factors, we checked for conservation of ZDREs in the promoter of target genes. In a bioinformatics analysis of the rice (O. sativa) and poplar (P. trichocarpa) genomes we determined: (i) the presence of the ZDRE motif in promoter regions; (ii) how many genes contain the motif; and (iii) whether the presence of the motif is conserved between apparent orthologs. As motif sequence we used ATGTCGACA[C/G/T], which is a simplification of RTGTCGACAY, assuming no bias for the orientation of the motif. The same motif search was used for Arabidopsis to verify the method. Promoter regions were defined as starting 1,000 bp upstream of the transcription start and including the 5′ untranslated leader or 1,500 bp upstream of the translation start when the transcription start was unknown. Using the search tool Vmatch (21), 141 genes were found for Arabidopsis, 219 for O. sativa and 115 for P. trichocarpa containing between one and three copies of the motif (without mismatches, except for the variation at the last nucleotide) (Dataset S1). Among this set of genes, the positions of the motif showed no preference relative to the transcription start. The number of genes with motifs identified in Arabidopsis is considerably larger than the number of genes we previously found to be differentially expressed in roots between the m19m23 double mutant and wild-type, because it will contain functional and nonfunctional cis elements. The screen did identify six of the nine genes involved in zinc deficiency that all contain one or more copies of the ZDRE (Table 1) and identified ZIP12 and IRT3, which we confirmed by qPCR to be nonresponding in the m19m23 double mutant seedlings (Fig. 4B). The ZDREs in the other three genes (ZIP1, ZIP3, and ZIP9) contained one more mismatch compared with the search motif than we allowed for detection.
We subsequently determined which genes from O. sativa and P. trichocarpa could be orthologs of any of the Arabidopsis genes containing at least one motif in its promoter. This way, 43 rice and 53 poplar orthologs of Arabidopsis genes were found (Dataset S2), among them the ZIP4 and ZIP5 orthologs for rice and the ZIP4, ZIP5, IRT3, NAS2, and NAS4 orthologs for poplar. Most of these genes contain 2 or 3 copies of the ZDRE in the promoter region, like their Arabidopsis orthologs. The conservation of the two bZIP transcription factors, the genes that respond to zinc deficiency, and the presence of the ZDRE transcription factor binding motif in their promoters strongly suggest that the regulatory functions of bZIP19 and bZIP23 have remained comparable over the course of evolution of higher plants before the separation of monocots and dicots and, perhaps, even in lower plants such as the moss Physcomitrella patens.
Discussion
We have shown that the bZIP19 and bZIP23 transcription factor function is essential for the response and adaptation of Arabidopsis to low zinc supply. They represent unique zinc homeostasis regulators identified in plants. The identification of these transcription factors, as well as the ZDRE cis element they bind to and the target genes they regulate, constitutes an important step forward toward unraveling the regulation of the zinc homeostasis network in plants. Up to now, the best studied plant micronutrient homeostasis regulatory network is the iron deficiency response involving bHLH transcription factors such as the Arabidopsis FIT, bHLH038, bHLH039, bHLH100, and bHLH101 genes (22–24), for which no transcription factor binding site has been found yet, and the rice IDEF1 and IDEF2 proteins—belonging to the ABI3/VP1 and NAC families of transcription factors—binding respectively to the IDE1 and IDE2 iron-deficiency-responsive elements (25, 26). The Arabidopsis bHLH transcription factors are strongly affected by iron status of the plant, whereas the rice transcription factors are not iron-regulated, more resembling the poor induction of bZIP19 and bZIP23 gene expression by zinc deficiency. There appears very little overlap between the iron and zinc deficiency responsive pathways. Only the ZIP9 gene, which we find to be nonresponsive in the m19m23 double mutant under zinc deficiency conditions (Table 1), is responsive to FIT under iron-deficient conditions (24).
The ZIP transporters are found in organisms at all phylogenetic levels including bacteria, fungi, plants, insects, and mammals and they play a major role in metal uptake in these organisms (27), but often their regulation is unknown. Only for the yeast Saccharomyces cerevisiae zinc responsive ZIP family, zinc transporters ZRT1 and ZRT2 are found to be regulated by the ZAP1 transcription activator, which binds to the zinc-responsive-element (ZRE) cis element (28). This way of transcriptional regulation appears to be specific for yeast, and ZAP1 orthologs have so far not been found in plants. The ZAP1/ZRE system of regulation does not resemble that of bZIP19/23 and the ZDRE. Instead it looks like plants have evolved their own zinc responsive regulation system controlling sufficient uptake of zinc in case of deficiency, depending on bZIP19- or bZIP23-like genes. Putative orthologs have been found in monocots and dicots, but also in gymnosperms and lower plants like Physcomitrella patens (20). These genes can be distinguished from putative paralogues resembling the bZIP24 gene of Arabidopsis (Fig. S1), which, like bZIP19 and bZIP23, belongs to group F of bZIP genes (10), and which has recently been identified as a regulator of salt stress response (14). Putative bZIP19/23 orthologs are markedly different from bZIP24 orthologs in their first 50 N-terminal amino acids, in the first of two conserved histidine-rich regions, which characterize the group F bZIP proteins, and in the region after the bZIP domain.
Such conservation of the zinc deficiency response regulatory network within the plant kingdom is interesting from an evolutionary perspective to understand the adaptation to extremely high zinc exposures that some plant species have evolved. It could also open up many new possibilities for the development of zinc biofortification strategies. Zinc deficiency afflicts up to 40% of the world's human population, mainly in developing countries where people depend on cereal-rich diets for sustenance (29, 30). Biofortification of crops with zinc, using plant breeding and other genetic technologies, offers a sustainable solution to this global problem (31, 32). Achieving a plant-based solution to alleviate zinc deficiency has thus far been hampered by insufficient knowledge on the mechanisms and regulation of the zinc homeostasis network in plants (32, 33). The identification of these transcription factors to regulate the zinc deficiency response may alter this for the better, as well as promote the development of zinc deficiency tolerant crops and of metal hyperaccumulator plants for phytoremediation of contaminated soil or water.
Methods
Plant Growth.
Arabidopsis ecotype Columbia (Col-0) was used in all experiments. Plants were grown in climate chambers with 16 h light at 22 °C, 8 h at 20 °C, 120 μmol photons m−2·s−1, and 50% relative humidity. Previous to germination, seeds had a 3-day stratification treatment in a cold room at 4 °C in the dark to promote uniform germination. For genetic analysis and transformation, plants were grown in pots with peat. For the plate-based assay, seeds were surface-sterilized by using chlorine vapor-phase seed sterilization and sown on plates with MS media (Duchefa Biochemie) supplemented with 1% sucrose and adjusted to pH 5.8. The MS media was prepared either without zinc (Zn-, zinc-deficient media), with 30 μM ZnSO4 (Zn+, zinc-sufficient media) or with 300 μM ZnSO4 (Zn++, zinc-excess media). For the hydroponically grown plants, seeds were sown on 0.55% agar-filled tubes and grown on a modified half-strength Hoagland's nutrient solution (34) prepared with either 0.05 μM (Zn-), 2 μM ZnSO4 (Zn+), or 25 μM ZnSO4 (Zn++). Zinc-sufficient (Zn+) and -excess (Zn++) media correspond to different zinc concentrations in agar-based media and hydroponics due to differences in zinc bioavailability. The hydroponic system consisted of 8-L-capacity containers (46 × 31 × 8 cm), with a nontranslucent 3-mm-thick plastic lid containing holes for placing agar-filled tubes in a 9 × 5 format. The nutrient solution was replaced once in the first week and twice in the weeks thereafter.
Construction of Reporter Vectors for the Yeast-One-Hybrid.
The bait sequence in six reporter vectors (named A to F) were PCR-amplified fragments of the ZIP4 promoter. These fragments covered the full promoter, starting 1,049 bp upstream of the start codon and had an overlap between fragments of 60–80 bp. The fragments were amplified from Arabidopsis genomic DNA by using proofreading polymerase (Pfu native; Stratagene), with PCR conditions as recommended by the manufacturer, and using primers with 5′-overhangs compatible with EcoRI/SacI (Table S2). The fragments were intermediately cloned into the pCR-Blunt II-TOPO vector (Invitrogen) according to the manufacturer's recommendations. The TOPO vector with each of the baits was digested with EcoRI/SacI, and the bait fragment was extracted from agarose gel (Qiagen Gel Extraction kit). The bait of reporter vector G consisted of a trimer of the following motif: ATGTCGACAT/C. Two antiparallel oligonucleotides, one representing the sense strand and the other its antisense complement strand, and containing 5′-overhangs compatible with EcoRI/SacI, were synthesized (Table S2). Each oligonucleotide strand (0.1 μg) were mixed in 10 μl of 50 mM NaCl, annealed by heating at 70 °C for 5 min, and slowly cooled down to room temperature. The digested, PCR-derived baits (A to F) and the annealed oligonucleotide (G) were cloned into pHISi, previously digested with EcoRI/SacI according to the manufacturer's recommendations. Each reporter vector was confirmed by digestion analysis and sequencing.
Yeast-One-Hybrid Screening.
The cDNA expression library was constructed with mRNA from Arabidopsis inflorescences and subsequently used to construct a Gateway compatible cDNA entry library by using of the CloneMiner cDNA library Construction Kit (Invitrogen). This cDNA entry library had a titer of 5 × 10E7 cfu·ml−1 and was cloned into the pDEST22 vector (Invitrogen) via an LR recombination reaction by following the protocol provided by the manufacturer, yielding an expression library with a titer of 2 × 10E6 cfu·ml−1. The reporter vectors were introduced into yeast strain PJ69-4A (35). For this purpose, yeast cells were transformed with digested (XhoI) linearized pHISi reporter vector by using a standard yeast transformation procedure (36). The empty pHISi vector, digested and undigested, and a nonintegrative reporter vector were used as controls. The cDNA expression library screening was performed by following the Large-Scale Yeast Transformation Protocol (PT3024-1; Clontech), which yielded a transformation efficiency of 5–9 × 10E5 cfu·μg−1 DNA. Screening with all of the reporter strains was performed on medium lacking His and in the presence of 20–40 mM 3-aminotriazole (3AT; the concentration was optimized for each reporter strain). Fifty-seven putative positive interactions were found (Table S3), and colony PCR was performed to amplify the cDNA inserts. Primers amplifying the regions adjacent to the attb recombination sites of pDEST22 vector were designed (5′-CGGTCCGAACCTCATAACAACTC-3′ and 5′-AGCCGACAACCTTGATTGGAGAC-3′). Amplification products of 47 putative positive colonies were sequenced, and the ones considered relevant are shown in Table S1. In total, 18 positive interactions (7 with bZIP19 and 11 with bZIP23) were found when using the reporter vectors E, F, and G (Fig. 2A and Table S1). Positive colonies were selected and the cDNA clone of the GAL4-AD library vector was isolated and sequenced.
Microarray Analysis.
Arabidopsis wild-type (wt) and T-DNA insertion double mutant m19m23 plants were grown in hydroponics medium as described above. They were grown for 3 weeks with normal zinc supply (Zn+, 2 μM ZnSO4) and 1 week with low zinc supply (Zn-, 0.05 μM ZnSO4) or normal zinc supply (Zn+). Roots of four plants per genotype and per treatment (Zn-/Zn+) were pooled in a two-biological-replica experiment, and RNA was extracted with the RNAeasy plant RNA kit (Qiagen). Transcriptomes were analyzed by using 1 μg of total RNA as starting material. Targets were prepared with the one-cycle cDNA synthesis kit followed by biotin-labeling with the IVT labeling kit (GeneChip One-cycle target labeling and control reagents; Affymetrix) and hybridized to the ATH1 gene chip for 16 h as recommended by the supplier (Gene expression analysis manual; Affymetrix). Raw data files were processed and quantile normalized in Bioconductor/R (37). Differential expression of each gene was tested for by applying an Empirical Bayes regularized t test (38). The P values were corrected for multiple testing by using the approach of Benjamini and Hochberg (39), providing control of the false discovery rate.
Supplementary Material
Acknowledgments
We thank R. Vooijs for assistance with the analysis of zinc concentration in plants, C. Hanhart for assistance with the hydroponics, M. Koornneef, T. Bisseling, and J. Keurentjes for comments on the manuscript, R. Joosen for assistance with figure preparation, K. Kaufmann for assistance with the EMSA, and D. Eide for providing the zrt1zrt2 yeast strain. We acknowledge the Kazusa DNA Research Institute (Kisarazu, Chiba, Japan) and the Centre National de Ressources Génomiques Végétales (Castanet Tolosan, France) for providing full length cDNA clones, and the Nottingham Arabidopsis Stock Centre (Loughborough, UK) for providing the T-DNA insertion lines. This work was funded by the Netherlands Organisation for Scientific Research, Innovational Research Incentives Scheme (Vernieuwingsimpuls), VENI scheme (A.G.L.A.) and was co-financed by the Centre for BioSystems Genomics, which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004788107/-/DCSupplemental.
References
- 1.Vallee BL, Auld DS. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 2.Clemens S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 2001;212:475–486. doi: 10.1007/s004250000458. [DOI] [PubMed] [Google Scholar]
- 3.Grotz N, et al. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mäser P, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y-F, et al. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009;182:392–404. doi: 10.1111/j.1469-8137.2009.02766.x. [DOI] [PubMed] [Google Scholar]
- 6.Wintz H, et al. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem. 2003;278:47644–47653. doi: 10.1074/jbc.M309338200. [DOI] [PubMed] [Google Scholar]
- 7.van de Mortel JE, et al. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2006;142:1127–1147. doi: 10.1104/pp.106.082073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talke IN, Hanikenne M, Krämer U. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 2006;142:148–167. doi: 10.1104/pp.105.076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakoby M, et al. bZIP Research Group. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 11.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 12.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 13.Fujii Y, et al. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- 14.Yang O, et al. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene. 2009;436:45–55. doi: 10.1016/j.gene.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Deppmann CD, Alvania RS, Taparowsky EJ. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- 16.Izawa T, Foster R, Chua N-H. Plant bZIP protein DNA binding specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- 17.Sibéril Y, Doireau P, Gantet P. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur J Biochem. 2001;268:5655–5666. doi: 10.1046/j.0014-2956.2001.02552.x. [DOI] [PubMed] [Google Scholar]
- 18.Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 19.Fukazawa J, et al. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell. 2000;12:901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corrêa LG, et al. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PloS ONE. 2008;3:e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurtz S. The Vmatch large scale sequence analysis software. 2008 (University of Hamburg, Hamburg, Germany). Available at www.vmatch.de. [Google Scholar]
- 22.Yuan Y, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008;18:385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- 23.Wang HY, et al. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta. 2007;226:897–908. doi: 10.1007/s00425-007-0535-x. [DOI] [PubMed] [Google Scholar]
- 24.Colangelo EP, Guerinot ML. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi T, et al. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc Natl Acad Sci USA. 2007;104:19150–19155. doi: 10.1073/pnas.0707010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogo Y, et al. A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J Biol Chem. 2008;283:13407–13417. doi: 10.1074/jbc.M708732200. [DOI] [PubMed] [Google Scholar]
- 27.Eide DJ. Zinc transporters and the cellular trafficking of zinc. Biochim Biophys Acta. 2006;1763:711–722. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, et al. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem. 1998;273:28713–28720. doi: 10.1074/jbc.273.44.28713. [DOI] [PubMed] [Google Scholar]
- 29.Welch RM. The impact of mineral nutrients in food crops on global human health. Plant Soil. 2002;247:83–90. [Google Scholar]
- 30.World Health Organization. The World Health Report. Reducing Risks, Promoting Healthy Life. 2002 (World Health Organization, Geneva) [Google Scholar]
- 31.Welch RM, Graham RD. Breeding for micronutrients in staple food crops from a human nutrition perspective. J Exp Bot. 2004;55:353–364. doi: 10.1093/jxb/erh064. [DOI] [PubMed] [Google Scholar]
- 32.Palmgren MG, et al. Zinc biofortification of cereals: Problems and solutions. Trends Plant Sci. 2008;13:464–473. doi: 10.1016/j.tplants.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Clemens S, Palmgren MG, Krämer U. A long way ahead: Understanding and engineering plant metal accumulation. Trends Plant Sci. 2002;7:309–315. doi: 10.1016/s1360-1385(02)02295-1. [DOI] [PubMed] [Google Scholar]
- 34.Assunção AGL, et al. Differential metal-specific tolerance and accumulation patterns among Thlaspi caerulescens populations originating from different soil types. New Phytol. 2003;159:411–419. doi: 10.1046/j.1469-8137.2003.00819.x. [DOI] [PubMed] [Google Scholar]
- 35.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gietz RD, Woods RA. Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 37.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 38.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.