Abstract
Human Cleavage Factor Im (CFIm) is an essential component of the pre-mRNA 3′ processing complex that functions in the regulation of poly(A) site selection through the recognition of UGUA sequences upstream of the poly(A) site. Although the highly conserved 25 kDa subunit (CFIm25) of the CFIm complex possesses a characteristic α/β/α Nudix fold, CFIm25 has no detectable hydrolase activity. Here we report the crystal structures of the human CFIm25 homodimer in complex with UGUAAA and UUGUAU RNA sequences. CFIm25 is the first Nudix protein to be reported to bind RNA in a sequence-specific manner. The UGUA sequence contributes to binding specificity through an intramolecular G:A Watson–Crick/sugar-edge base interaction, an unusual pairing previously found to be involved in the binding specificity of the SAM-III riboswitch. The structures, together with mutational data, suggest a novel mechanism for the simultaneous sequence-specific recognition of two UGUA elements within the pre-mRNA. Furthermore, the mutually exclusive binding of RNA and the signaling molecule Ap4A (diadenosine tetraphosphate) by CFIm25 suggests a potential role for small molecules in the regulation of mRNA 3′ processing.
Keywords: cleavage factor, CPSF5, mRNA processing, Protein-RNA complex, RNA recognition
The transcriptome complexity of higher eukaryotes requires the coordinate recognition of an array of alternative pre-mRNA processing signals in a developmental and tissue-specific manner (1, 2). The sequences that direct pre-mRNA splicing and 3′ processing are initially recognized within the nascent transcript in a process that is intimately coupled to transcription (3, 4). While the recognition of exons within the pre-mRNA is mediated by both RNA:RNA and protein:RNA interactions (5), the 3′ processing of polyadenylated mRNAs appears to rely solely on the interaction of protein factors (6) with unstructured RNA sequences (7) within the nascent transcript.
Vertebrate pre-mRNA 3′ processing signals are recognized by a tripartite mechanism through which a set of short RNA sequences direct the cooperative binding of three multimeric 3′ processing factors, cleavage factor Im (CFIm), cleavage and polyadenylation specificity factor (CPSF), and cleavage stimulation factor (CstF) (8). CPSF and CstF bind the AAUAAA hexamer and downstream GU-rich elements that flank the poly(A) site, respectively, whereas CFIm interacts with upstream sequences that may function in the regulation of alternative polyadenylation (9–11). SELEX and biochemical analyses have identified the sequence UGUAN (N = A > U > G, C) as the preferred binding site of CFIm (11). In this report we have taken a structural approach to determine the mechanism of sequence-specific RNA binding by CFIm.
CFIm is composed of a large subunit of 59, 68, or 72 kDa and a small subunit of 25 kDa (CFIm25, also referred to as CPSF5 or NUDT21) (12, 13), both of which contribute to RNA binding (14). The large subunit, encoded by either of two paralogs (CPSF6 and CPSF7), contains an N-terminal RNA Recognition Motif (RRM), an internal polyproline-rich region, and a C-terminal RS/RD alternating charge domain—a structure similar to that of the SR-protein family of splicing regulators. The small subunit (CFIm25) contains a Nudix domain, a protein domain that most often participates in the hydrolysis of substrates containing a nucleotide diphosphate linked to a variable moiety X (15). Found throughout all three kingdoms, Nudix proteins participate in a wide range of crucial housekeeping functions, including the hydrolysis of mutagenic nucleotides, the modulation of the levels of signaling molecules, and the monitoring of metabolic intermediates (15). CFIm25 possesses the characteristic α/β/α Nudix fold and is able to bind Ap4A (diadenosine tetraphosphate), but it has no hydrolase activity, due to the absence of two of the four glutamate residues that coordinate the divalent cations important for substrate hydrolysis (16).
While an array of different protein domains have been identified that bind RNA in a sequence-specific manner, only a limited subset functions in the sequence-specific recognition of single-stranded RNA (17). These domains include the ubiquitous RRM, hnRNP K homology domain (KH domain), zinc-binding domains, and the PUF domain. In this report, we present a previously undescribed mechanism for the sequence-specific binding of single-stranded RNA by the 25 kDa subunit of CFIm. Although the Nudix domains of the eukaryotic decapping enzymes (18), bacterial 5′ pyrophophohydrolase (19), and the trypanosome mitochondrial protein MERS1 (20) act on RNA, CFIm25 is unique among Nudix proteins in that it is capable of sequence-specific RNA binding. CFIm25 is highly conserved throughout the eukaryotic kingdom (Fig. S1), yet, interestingly, it has been lost in a subset of protists, including both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Fig. S2).
The structures of the CFIm25 homodimer in complex with RNA presented here not only reveal a unique mechanism for sequence-specific RNA binding, but also provide an insight into the coordinate recognition of multiple poly(A) site upstream elements, and the potential regulation of these interactions by small molecules.
Results
Overall Structure of CFIm25 Bound to UGUA Element.
Two 6-nucleotide RNA sequences containing a UGUA element: 5'-UGUAAA-3' and 5'-UUGUAU-3' were designed based on our previously published SELEX results (11). The second oligonucleotide with the extra uracil at the 5′-end was used to confirm that the UGUA core element was bound by CFIm25 specifically. The UGUAAA and UUGUAU complex structures were solved to a resolution of 2.1 and 2.2 Å, respectively (Table S1). The overall protein architecture of both CFIm25-RNA complexes is nearly identical to that of the previously published unliganded CFIm25 model [3BAP (16)] (RMSD 0.45 Å calculated on 194 Cα atoms). Briefly, CFIm25 is composed of a central domain encompassing residues 77–202, which adopts a α/β/α fold common to all Nudix proteins (21). In CFIm25, the central Nudix domain is sandwiched between N-terminal and C-terminal structural elements, which are major contributors to the dimer interface. The most notable difference between the apo and RNA-bound structures is the position of the N-terminal segment (residues 21–29), which swings backward instead of leaning toward the other monomer. Another interesting feature of CFIm25 is the loop connecting β2 and α1 (residues 51–60) (Fig. 1 and 4). This loop acts like a strap that occludes the canonical Nudix substrate-binding pocket. Contrary to earlier predictions (22), the loop does not move away upon RNA binding. Instead, it is an integral part of the RNA recognition pocket.
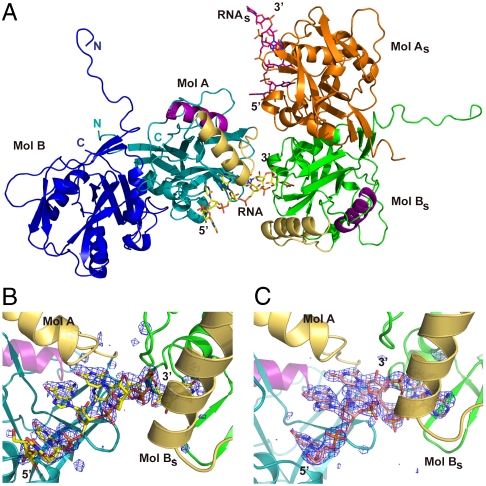
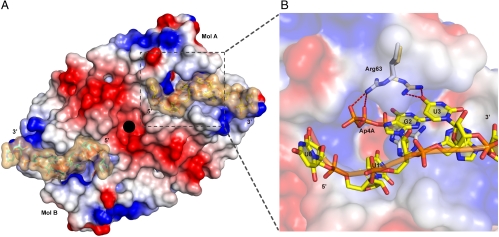
Fig. 1.
Overall structure of the CFIm25-RNA complex. (A) View of the crystal packing interactions of the CFIm25-UUGUAU complex. One asymmetric unit contains one CFIm25 homodimer (Molecule A in teal and Molecule B in dark blue) and one UUGUAA hexamer (yellow). The 5′-end of the RNA (UGUA element) binds to Mol A, while the 3′-end is bound by Mol B of an adjacent symmetry-related dimer (Mol Bs in green). Molecule A and the RNA of the adjacent dimer are shown in orange and pink, respectively. In Mol A and Mol Bs, the conserved Nudix box helix (residues 117–129) is highlighted in purple. Helix α1 and the loop connecting β2 and α1 (residues 51–74) are shown in gold. (B) Close up view of the CFIm25-UUGUAU interface between Mol A and Mol Bs. UUGUAU is shown as a stick model (yellow) with overlaid Fo-Fc electron density map (dark blue) contoured at 3σ. The difference map was calculated immediately after molecular replacement and prior to any refinement, in order to prevent model bias. Convincing density was observed for the entire RNA strand except for the base of the first U (U0). (C) Same view of the CFIm25-UGUAAA complex. UGUAAA is shown as a stick model (salmon), and the Fo-Fc map (3σ) (dark blue) was also calculated before any refinement. Strong density was observed for all six nucleotides.
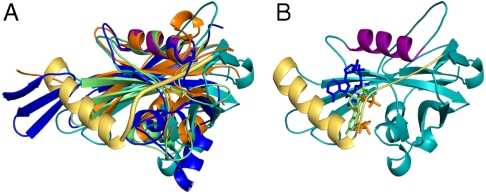
Fig. 4.
CFIm25 is the only Nudix protein of known structure in which the canonical Nudix substrate-binding pocket is occluded. (A) Superposition of the CFIm25-UUGUAU complex (Mol A) with three well-studied Nudix hydrolases (reviewed in ref. 21): MutT pyrophosphohydrolase (PDB ID: 1PPX) in lime, ADP-ribose pyrophosphatase (1V8L) in dark blue, and Ap4A hydrolase (1XSC) in orange. The CFIm25 color scheme is the same as in Fig. 1. The loop connecting β2 and α1 is shown as a thick yellow tube in the CFIm25-UUGUAU complex. (B) Superposition of the ligands from the three Nudix proteins in A onto CFIm25. The ligands (8-oxo-2’-deoxy-GMP, ADP-ribose, and ATP) are shown as stick models and colored as indicated in A.
Even though the asymmetric unit contains a dimer of CFIm25, we observe only one bound RNA molecule. The RNA hexamer is bound specifically by one molecule (designated as molecule A), and partially by molecule B of an adjacent dimer in the crystal (designated Bs, for symmetry equivalent) (Fig. 1A). In the UUGUAU-bound complex, we observed convincing density for all the bases except for the first U (referred to as U0) (Fig. 1B). The next four nucleotides, U1, G2, U3, and A4, are found in the RNA binding pocket of Mol A. Right after A4, the RNA backbone bends ∼105° toward Mol Bs of an adjacent dimer, leading U5 to insert into the RNA binding pocket of Mol Bs. In the UGUAAA-bound complex (Fig. 1C), the first three nucleotides, U1, G2, and U3, interact with Mol A in the same manner as in the UUGUAU complex. In contrast to the UUGUAU complex, however, the phosphate backbone of the RNA is twisted by ∼95° after the U3 nucleotide, flipping A4 and A5 into the RNA binding pocket of Mol Bs of the adjacent dimer. Interestingly, right after A5, the RNA strand twists back toward Mol A, enabling the interactions between G2 and A6, which are identical to the G2-A4 interactions observed in the UUGUAU-bound complex. All these observations support our earlier SELEX and biochemical analyses indicating that CFIm25 specifically recognizes the UGUA tetranucleotide sequence (11).
Sequence-Specific Recognition of UGUA by CFIm25.
CFIm25 binds to RNA through a variety of interactions, including hydrogen-bonding via both main-chain and side-chain atoms, aromatic stacking, and peptide bond stacking (17). Besides protein–RNA interactions, intramolecular interactions also play a substantial role in RNA recognition. A schematic representation of the interactions between CFIm25 and each of the RNAs is shown in Fig. S3. The interactions leading to sequence-specific recognition are common to the two complexes, unless otherwise noted.
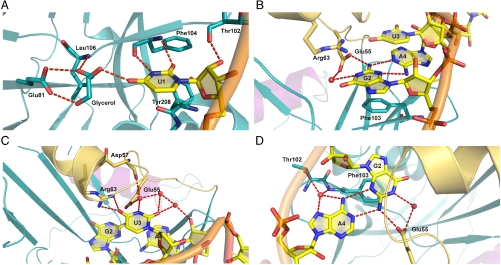
U1 forms three intermolecular hydrogen bonds through its Watson–Crick edge (Fig. 2A): O2 and N3 are recognized by the main-chain amide and carbonyl groups of Phe104, respectively, and O4 is stabilized by the side chain of Glu81 and the main-chain amide of Leu106 via a glycerol molecule, which was also found in the same location in the previously published unliganded CFIm25 structure (16). The glycerol molecule might mimic a small molecule or a network of ordered water molecules (23). Furthermore, a hydrogen bond is present between the O2’ hydroxyl of the ribose and the main-chain carbonyl of Thr102. In addition to these hydrogen bonds, U1 is further stabilized by stacking of the uracil base with the plane formed by the peptide bond between Tyr208 and Gly209 (17). This complex network of interactions indicates that uracil is the preferred base at the first position of the core UGUA recognition sequence.
Fig. 2.
Close-up views of the CFIm25-UUGUAU interactions. Close up views of CFIm25 interacting with each base within the UGUA element: (A) U1, (B) G2, (C) U3, and (D) A4. The protein color scheme is the same as in Fig. 1. The RNA backbone is shown in orange. Hydrogen bonds are represented by red dashed lines. Residues involved in RNA binding are shown and colored according to the domain they belong to. Water molecules involved in hydrogen bonding are shown as red spheres.
G2 participates in hydrogen bond interactions not only with the protein but also with A4 via an intramolecular contact (Fig. 2B). The N2 amino group of G2 hydrogen bonds with the side chain of Glu55, whereas N1 interacts with Glu55 via a water molecule. In addition to the recognition through its Watson–Crick edge, G2 forms two hydrogen bonds with A4 via its sugar edge. More specifically, N2 and N3 of G2 interact with N1 and N6 of A4, respectively. Steric considerations rule out the possibility of having a pyrimidine at the second and fourth positions of the tetranucleotide, because a smaller base at either position would not be able to establish complementary interactions with G2 or A4. A water molecule forms a four-way bridge between N6 and N7 of A4, the side chain hydroxyl of Thr102, and the main-chain carbonyl of Phe103, which provides another means to discriminate against pyrimidines at the fourth position (Fig. 2D). The interactions with Glu55 specify a G at the second position, which in turn determines the specific selection of the fourth base, namely adenine. In addition to the sequence-specific hydrogen bond interactions, the position of G2 is restricted by a stacking interaction with Phe103. Van der Waals contacts between A4 and both the main-chain carbonyl and side chain of Leu99 further strengthen the network of sequence-specific contacts holding G2, A4, and the protein together. These numerous interactions corroborate the observation that the substitution of G2 with C abolished CFIm25 RNA binding in vitro (Fig. 2 B and D).
All three polar atoms of the U3 bases are involved in hydrogen bonding with CFIm25 (Fig. 2C). O4 participates in two hydrogen bonds with the guanidinium group of Arg63. O2 and N3, on the other hand, are engaged in H bonds via two water molecules. One water molecule mediates the interactions between O2 and the O2’ hydroxyl of the A4 ribose. The other water molecule connects N3 to the side chain of Glu55 and the main-chain carbonyl of Asp57. The extensive interactions of U3 strongly support the results of the SELEX analysis (11) that indicated that a U is the preferred choice for the third position.
The nucleotides 3′ to the UGUA element are bound by the symmetry related molecule Bs (Fig. S3). U5 of UUGUAU is bound by Mol Bs at exactly the same position through identical hydrogen bonding and stacking interactions as U1 in Mol A (residues Glu81, Phe104, Tyr208, and Thr102). In the UGUAAA sequence, A4 and A5 are bound by Mol Bs nonspecifically: A4 is contacted by Phe103 and Glu55 whereas A5 interacts with Phe104 and Tyr208.
Mutational Analysis Supports the Structural Model for RNA Binding.
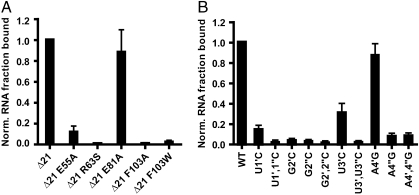
Of the five protein side chains involved in key protein–RNA interactions, four are highly conserved among those species that possess the CFIm25 protein (Fig. S1). Namely, Glu55, Arg63, and Glu81 are involved in specifying G2, U3, and U1, respectively. Phe103, even though it is not involved in specific recognition, provides strong stacking forces to stabilize the RNA strand. These four residues were substituted to validate the interactions we observed in the structure. All four single point mutation variants form crystals that have the same space group and similar cell parameters as full-length CFIm25, indicating that the protein variants are properly folded.
All the mutations tested reduced the affinity of CFIm25 for RNA, based on gel electrophoretic mobility shift analysis (EMSA) (Fig. 3A). The Glu55Ala and Arg63Ser mutations eliminate the hydrogen bonding to G2 and U3, respectively (Fig. 2 B and C), with a consequent reduction in RNA binding affinity of 88% for Glu55Ala and 99% for Arg63Ser. The Glu81Ala mutation reduced RNA binding by only 12%, which is not unexpected because Glu81 interacts with U1 only indirectly (Fig. 2A). The Phe103Ala variant lost 99% of its RNA binding affinity. Phe103 is involved in a three-layer stacking interaction with G2 and U3 (Fig. 2B), which is abrogated by the alanine mutation. Surprisingly, when Phe103 was replaced by Trp, the RNA binding affinity still decreased. Because tryptophan is more hydrophobic than phenylalanine, an increased binding affinity might have been expected. It is plausible that the larger tryptophan may displace other residues in the RNA binding pocket, leading to the reduced affinity. The varying degrees of RNA binding exhibited by the protein variants correlate well with the CFIm25-RNA interactions observed in the crystal structure and confirm the sequence-specific binding of CFIm25 to the UGUA sequence.
Fig. 3.
CFIm25 specifically recognizes two UGUA elements. (A) Bar graph representation of the electrophoretic mobility shift assay (EMSA) data of CFIm25 variants binding to a 21 nt PAPOLA poly(A) site RNA containing two UGUA elements. (B) EMSA data of wild type CFIm25ΔN21 binding to various RNA sequence variants. A single prime represents the mutation on the first UGUA element, and double prime represents the mutation on the second UGUA element. Experiments were done in triplicate and all the bound fractions were plotted relative to CFIm25ΔN21 and the wild type PAPOLA RNA. The error bars represent the standard deviation.
The CFIm25 Homodimer Specifically Binds Two UGUA Elements.
CFIm25 forms a homodimer in solution (16, 22), and the same dimer conformation is retained upon RNA binding, as shown in our crystal structure. CFIm25 therefore has the potential to specifically bind two UGUA elements simultaneously. To test this hypothesis, we used a 21 nt RNA containing a sequence found upstream of the human PAPOLA poly(A) site that has previously been shown to function in mRNA 3′ processing (8). This sequence, located 39 nt upstream of the PAPOLA poly(A) cleavage site, contains two UGUA elements separated by 9 bases. The first UGUA element is designated U1’, G2’, U3’ and A4’, and the second U1”, G2”, U3” and A4”.
The binding profiles of RNA sequence variants were determined by EMSA (Fig. 3B). The U1C, G2C, U3C, and A4G mutations were designed to eliminate the hydrogen-bonding interactions observed in the crystal structures. Simultaneous changes in both UGUA elements at each of the four positions diminished the CFIm25 binding by more than 90% (Fig. 3B). These results confirm the RNA binding specificity of CFIm25 toward the UGUA sequence. In comparison, single mutations at each of the four positions decreased binding affinity but to a lesser extent than the double mutations (Fig. 3B). This observation indicates that both UGUA elements can engage in RNA binding. Among all the single mutations tested, the G2C single mutants are the most affected. This may be due to the fact that G2 is involved in both protein-RNA and intramolecular RNA interactions, while other nucleotides participate in one or the other. In addition, a G2C single mutation at either the first or second UGUA decreases the RNA binding affinity more than 95%, indicating that both UGUA are involved in RNA binding. In contrast, a notable difference between two of the A4G single mutations was observed, where the affinity was decreased by 90% for A4”G compared to only 10% for A4’G. This dramatic difference between the two elements might be caused by the nature of the nucleotide immediately succeeding A4. A4’ is followed by another A, which could interact with G2’. This could be achieved by looping out A4’, in a fashion similar to the RNA structure observed in the UGUAAA-bound crystal. A4”, on the other hand, is followed by a U, which would be unable to form a stable interaction with G2”. Taken together, the CFIm25-RNA complex structures and the RNA binding analysis not only confirm the RNA binding specificity of CFIm25 toward the UGUA sequence but also support the hypothesis that the CFIm25 homodimer specifically binds two UGUA elements.
Discussion
CFIm functions in poly(A) site recognition and the regulation of alternative 3′ processing through the binding of sequences upstream of the poly(A) site (8, 10, 11). In this report we have determined the mechanism by which the 25 kDa subunit of CFIm binds the poly(A) site upstream element UGUA. Structures of the CFIm25 homodimer bound to RNA reveal how a Nudix hydrolase domain has been transformed into a platform for the sequence-specific binding of single-stranded RNA.
CFIm25 is a highly conserved protein (Fig. S1) in which a unique N-terminal extension has been appended to a Nudix domain (residues 77–202). The importance of the CFIm25 Nudix domain is illustrated by the fact that 8 out of 12 residues involved in RNA binding are located within this domain (Fig. S3). The N-terminal extension, specifically residues 51–74, also plays an essential role in RNA binding, consistent with previous results (14). Two of the four key residues (Glu55 and Arg63) responsible for UGUA recognition are found within this region. Interestingly, the N-terminal extension occludes the canonical Nudix substrate-binding pocket (Fig. 4). The Nudix domain, along with β2 and α1 of the N-terminal extension (including Glu55 and Arg63), is highly conserved, supporting the conclusion that CFIm25 has coopted a Nudix hydrolase domain for sequence-specific RNA binding. Intriguingly, CFIm25 has been lost in several protists (Fig. S2), many of which are characterized by a paucity of alternative mRNA processing (24, 25).
Single-stranded RNA binding proteins have been found to achieve sequence specificity through a variety of mechanisms that involve the formation of hydrogen bonds with the polar atoms of RNA bases. While some proteins, such as the zinc finger (ZnF) proteins Tis11d (26) and the bacterial repressing clamp RsmA/CsrA (27), interact exclusively through protein main-chain atoms, others, such as Pumilio, interact through protein side chains (28). CFIm25 utilizes both binding modes, a characteristic it shares with the RRM and KH domains, and the zinc knuckle of the MMLV nucleocapsid (reviewed in ref. 17). CFIm25 recognizes U1 and U3 primarily through main-chain (Phe104) and side-chain (Arg63) interactions, respectively. Additional selective forces are provided by side-chain (Glu81) and main-chain (Asp57) contacts for U1 and U3, respectively. In addition to hydrogen bond interactions, stacking interactions contribute to the binding of the UGUA tetranucleotide, as previously observed in other RNA binding proteins (reviewed in ref. 17). These include π–π interactions between Phe103 and G2 (Fig. 2B) and stacking between U1 and the peptide bond plane of Tyr208-Gly209 (Fig. 2A).
Intramolecular sugar-edge/Watson–Crick base pair recognition between G2 and A4 distinguishes CFIm25 from other sequence-specific single-stranded RNA binding proteins. To date, only six examples of sugar-edge/Watson–Crick base pairs have been reported in the Noncanonical Base Pair Database, out of 1,860 base pairs (29). In each case, the base pair is located within a double-stranded segment of the ribosome or ribonuclease P (30–32). Canonical Watson–Crick base pairing, as in the RsmA/CsrA-RNA structure (27), or noncanonical sugar-edge/Hoogsteen G-A base pairs, as in the U4 snRNA-15.5 kDa spliceosomal protein–RNA structure (33), have been demonstrated to be essential for the formation of the protein–RNA complexes, but again these base pairs are located within double-stranded segments of structured RNAs. The CFIm25-RNA structures provide the second example of an intramolecular base pair playing a crucial role in the recognition of single-stranded RNA, first identified in the complex of the RRM domain of human alternative splicing factor Fox-1 with RNA, in which the same G-A base pair is observed (34). Interestingly, an identical G-A base pair provides the key recognition between G26 of the SAM-III riboswitch and A of S-adenosylmethionine (35, 36). We speculate that the array of recognition mechanisms that we observe provides strong selective pressure to maintain not only the integrity of the protein fold and the identity of key amino acids but also the specific RNA sequence required for binding. Indeed, the UGUA sequence has been found to be a component of the poly(A) signals of a wide range of organisms, from Chlamydomonas to humans (37, 38).
The CFIm25-RNA complex retains the same homodimer conformation previously observed in the apo structure (16, 22). Multimeric organization is a common feature of single-stranded RNA binding proteins and has been demonstrated to facilitate both higher affinity and specificity (39–43). Although only one RNA molecule was present in each of our structures, EMSA data suggest that both binding sites of the CFIm25 homodimer are occupied in solution. Specifically, a single G to C point mutation in either UGUA element of the PAPOLA poly(A) site sequence nearly eliminated CFIm25/RNA complex formation, while other single point mutations significantly reduced binding (Fig. 3B). Furthermore, the presence of two UGUA elements within the substrate enhances the RNA binding affinity dramatically (Fig. S4A). In a competition assay, unlabeled 21 nt PAPOLA RNA competes 100 fold more effectively than the 6 nt UUGUAU (75 nM and 7.5 μM, respectively), supporting the binding of two UGUA elements by the CFIm25 homodimer (Fig. S4B). This result is consistent with our earlier observation that multiple UGUA elements are often observed within poly(A) site upstream sequences (37). Thus the structure of the CFIm25 homodimer suggests a mechanism for the coordinate recognition of eight nucleotides: a set of two UGUA elements separated by a variable number of bases. We tested the optimal length between the two elements by sequentially shortening the 9 nt spacer of the PAPOLA sequence. EMSA experiments showed that RNA sequences with a spacer of 3 nt or less no longer bind CFIm25 (Fig S4C). The minimum length (5 nt spacer) between two UGUA elements is close to the estimated distance (∼30 Å) required to connect two CFIm25-bound UGUA elements (Fig S4C and E). It is likely that, in vivo, the large subunit of CFIm, which possesses a RRM domain, makes an essential contribution to the binding of two UGUA elements by the CFIm25 dimer, as suggested by previous in vitro experiments (14). Such a mechanism is supported by our observation that the two subunits of CFIm form a heterotetramer in solution. Fig. S4E illustrate how the binding of a 6-mer UUGUAU RNA in the binding pocket of Mol B dictates that the 5′-ends of the two RNAs face each other across the twofold axis.
The antiparallel orientation of the UGUA sequences bound to the CFIm25 homodimer is reminiscent of the polypyrimidine tract binding protein (PTB) (44), which organizes two RNA sequences in a similar fashion through the use of two RRM domains. PTB appears to function in the regulation of splicing through the sequestration of pre-mRNA sequences within an RNA loop formed by the juxtaposition of two pyrimidine tracts (45). In the case of the splicing regulator MBNL1 (46), two zinc finger domains (ZnF) form a chain-reversal RNA binding track for the target pre-mRNA. In a similar manner, by varying the length of RNA between two UGUA elements, the RNA loop formed by the binding of the CFIm complex may contribute to its role in the regulation of alternative mRNA 3′-end processing (8–11). A structure of CFIm 25/68 kDa complexed with RNA will be required to elucidate the path the RNA follows between the two CFIm25 binding sites.
Structures of CFIm25 bound to Ap4A and 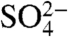 have previously been described. The
have previously been described. The 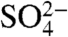 molecule was found to occupy the same location as the γ-phosphate of Ap4A (16, 22). Each of these small molecules binds CFIm25 in a manner that excludes the possibility of RNA binding. Arg63, a conserved residue which contacts
molecule was found to occupy the same location as the γ-phosphate of Ap4A (16, 22). Each of these small molecules binds CFIm25 in a manner that excludes the possibility of RNA binding. Arg63, a conserved residue which contacts 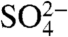 and the γ-phosphate of Ap4A, swings toward U3 upon RNA binding (Fig. 5B). The dramatic movement of Arg63 suggests it might act as a sensor for RNA. Another notable feature is that Ap4A makes the same stacking interaction with Phe103 as the guanine base of G2 (Fig. S5). The mutually exclusive binding of RNA and Ap4A, and possibly other small molecules, suggests a potential mechanism for the regulation of poly(A) site choice. Such a possibility is reminiscent of the allosteric regulation of the Nudix-related transcriptional regulator protein (NtrR) (47), in which a catalytically inactive Nudix domain serves a regulatory role through the binding of ADP-Ribose. As noted above, the Nudix domain of CFIm25 also appears to be catalytically inactive due to the absence of two key glutamate residues known to coordinate divalent cations (16). This feature is conserved among all known CFIm25 homologs. The potential for regulation of mRNA 3′ processing by small molecules is particularly intriguing in light of the observation that alternative 3′ processing can be modulated in response to synaptic activity in neurons (48). Future investigations of the interaction of CFIm25 with small molecules may provide an insight, not only into the biological function of CFIm25 but into the regulation of the mammalian mRNA processing machinery as well.
and the γ-phosphate of Ap4A, swings toward U3 upon RNA binding (Fig. 5B). The dramatic movement of Arg63 suggests it might act as a sensor for RNA. Another notable feature is that Ap4A makes the same stacking interaction with Phe103 as the guanine base of G2 (Fig. S5). The mutually exclusive binding of RNA and Ap4A, and possibly other small molecules, suggests a potential mechanism for the regulation of poly(A) site choice. Such a possibility is reminiscent of the allosteric regulation of the Nudix-related transcriptional regulator protein (NtrR) (47), in which a catalytically inactive Nudix domain serves a regulatory role through the binding of ADP-Ribose. As noted above, the Nudix domain of CFIm25 also appears to be catalytically inactive due to the absence of two key glutamate residues known to coordinate divalent cations (16). This feature is conserved among all known CFIm25 homologs. The potential for regulation of mRNA 3′ processing by small molecules is particularly intriguing in light of the observation that alternative 3′ processing can be modulated in response to synaptic activity in neurons (48). Future investigations of the interaction of CFIm25 with small molecules may provide an insight, not only into the biological function of CFIm25 but into the regulation of the mammalian mRNA processing machinery as well.
Fig. 5.
Surface presentation of the CFIm25-UUGUAU complex. (A) Electrostatic surface representation of the CFIm25 dimer, colored according to the electrostatic potential (blue, positive; red, negative). The UUGUAU RNA strand is shown as a stick model (yellow). A second UUGUAU molecule (shown in cyan) is modeled in Mol B in the same location as in Mol A. The surface of the RNA molecules is shown in beige. The crystallographic 2-fold axis is represented by a black circle. (B) A close-up view of the RNA binding pocket in Mol A. Superposition of the Ap4A-bound [PDB ID: 3BAP (16)] CFIm25 structure with the UUGUAU-bound structure. Ap4A and Arg63 from 3BAP are shown in white. Hydrogen bond interactions between Arg63 and its ligands are shown as red dashed lines.
Materials and Methods
Crystallization of the CFIm25-RNA Complexes.
The full-length CFIm25 was prepared as previously described (16). Two 6-nucleotide sequences containing one UGUA tetranucleotide were purchased from Dharmacon (Lafayette, CO): 5′-UGUAAA-3′ and 5′-UUGUAU-3′. The purified CFIm25 was mixed with the RNA in a 1∶1.2 molor ratio. The final concentration of CFIm25 was about 5 mg/ml. Crystals were grown in hanging drops and structures were determined as described in SI Text.
Gel Electrophoretic Mobility Shift Assays.
α32P-GTP-labeled RNAs containing the human PAPOLA upstream sequences (-56 to -39 relative to the poly(A) cleavage site were prepared as in (8). The CFIm25ΔN21 deletion construct and single amino acid substitution variants were made using a QuikChange II XL mutagenesis kit (Stratagene), expressed and purified using the same protocol as for the full-length CFIm25 (16). The RNA binding reactions were incubated at 30 °C for 5 min and the protein-RNA complexes were resolved by electrophoresis on a nondenaturing 5% (80∶1) polyacrylamide gel at 4 °C. After quantification, the percentage of bound RNA for the protein variants and RNA mutations were plotted relative to CFIm25ΔN21 and the wild type PAPOLA RNA. Details are in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Molly Coseno and Justin Meyette for help with protein expression, Dr. Joyce Heckman for help with RNA preparation, and Drs. Mark Rould and Frédérick Faucher for assistance with data collection and refinement. This research was supported by National Institutes of Health Grant GM62239 to S.D.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3MDG, 3MDI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000848107/-/DCSupplemental.
References
- 1.Licatalosi DD, Darnell RB. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet. 2010;11(1):75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4(12):e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Perales R, Bentley D. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36(2):178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahl MC, Will CL, Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33(3):365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graveley BR, Fleming ES, Gilmartin GM. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16(9):4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19(11):1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): Potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78(3):472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- 10.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34(21):6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12(6):1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 12.Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1(2):243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 13.Ruepp MD, et al. Mammalian pre-mRNA 3′ end processing factor CF I m 68 functions in mRNA export. Mol Biol Cell. 2009;20(24):5211–5223. doi: 10.1091/mbc.E09-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279(34):35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 15.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63(2):123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coseno M, et al. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 2008;36(10):3474–3483. doi: 10.1093/nar/gkn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34(17):4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA. 2002;99(20):12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451(7176):355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 20.Weng J, et al. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32(2):198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mildvan AS, et al. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys. 2005;433(1):129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Tresaugues L, et al. The crystal structure of human cleavage and polyadenylation specific factor-5 reveals a dimeric Nudix protein with a conserved catalytic site. Proteins. 2008;73(4):1047–1052. doi: 10.1002/prot.22198. [DOI] [PubMed] [Google Scholar]
- 23.Brown CJ, Verma CS, Walkinshaw MD, Lane DP. Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site. Cell Cycle. 2009;8(12):1905–1911. doi: 10.4161/cc.8.12.8742. [DOI] [PubMed] [Google Scholar]
- 24.McGuire AM, Pearson MD, Neafsey DE, Galagan JE. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol. 2008;9(3):R50. doi: 10.1186/gb-2008-9-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irimia M, Roy SW. Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 2008;4(8):e1000148. doi: 10.1371/journal.pgen.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11(3):257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 27.Schubert M, et al. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14(9):807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 29.Nagaswamy U, et al. NCIR: A database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30(1):395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 31.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407(6802):340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 32.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421(6924):760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 33.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6(6):1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 34.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25(1):163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo J, Westhof E. Base pairs and pseudo pairs observed in RNA-ligand complexes. J Mol Recognit. 2010;23(2):241–252. doi: 10.1002/jmr.978. [DOI] [PubMed] [Google Scholar]
- 36.Lu C, et al. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15(10):1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11(10):1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y, Liu Y, Liu L, Liang C, Li QQ. Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics. 2008;179(1):167–176. doi: 10.1534/genetics.108.088971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allain FH, Bouvet P, Dieckmann T, Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 2000;19(24):6870–6881. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98(6):835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 41.Handa N, et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398(6728):579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 42.Johansson C, et al. Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target. J Mol Biol. 2004;337(4):799–816. doi: 10.1016/j.jmb.2004.01.056. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8(2):141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 44.Oberstrass FC, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309(5743):2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 45.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15(12):1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang N, et al. Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism. Structure. 2009;17(7):939–951. doi: 10.1016/j.str.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.