Abstract
Purpose
To identify mutations in a Chinese family with congenital cataract and microcornea.
Methods
Detailed family history and clinical data were recorded. Genomic DNA was extracted from leukocytes of venous blood of the patients and noncarriers in this family along with 100 normal individuals. All six exons of crystallin, beta A4 gene (CRYBA4) were amplified by PCR methods and direct sequencing.
Results
We identified a c.225G>T sequence change that led to an amino acid substitution G64W in the CRYBA4-induced protein in two patients of this family; this nucleotide substitution was not detected in the other individuals.
Conclusions
A novel missense mutation in CRYBA4 was identified in our study. It expands the mutation spectrum of CRYBA4 and provides useful information to the study of molecular pathogenesis of cataract and microcornea.
Introduction
Congenital cataract can be defined as lens opacification presenting at birth or developing shortly thereafter. The lens alone may be involved, and this accounts for approximately 70% of congenital cataracts. Conversely, lens opacities may be associated with other ocular anomalies, such as microphthalmia, aniridia, other anterior chamber developmental anomalies, or retinal degenerations, seen in approximately 15% of case [1]. Congenital cataract is a leading cause of childhood blindness worldwide and results in about 10%–20% of children in developing countries to be blind [2]. Worldwide, 20 million children under the age of 16 suffer from cataract, and among these, 200,000 (10%) are severely visually impaired or blind. While this figure is relatively low compared to the 17 million (40%) adults who are blind caused by cataract [3-5].
Recently, more than 34 loci in the human genome have been reported to be associated with congenital cataract, and 22 specific genes have detected mutations, including encoding crystallins (crystallin, alpha A gene [CRYAA], crystallin, alpha B gene [CRYAB], crystallin, beta A1 gene [CRYBA1], crystallin, beta A4 gene [CRYBA4], crystallin, beta B1 gene [CRYBB1], crystallin, beta B2 gene [CRYBB2], crystallin, beta B3 gene [CRYBB3], crystallin, gamma C gene [CRYGC], crystallin, gamma D gene [CRYGD], and crystallin, gamma S gene [CRYGS] [6-14]), cytoskeletal proteins (beaded filament structural protein 1, filensin gene [BFSP1], and beaded filament structural protein 2, phakinin gene [BFSP2] [15,16]), membrane proteins gap junction protein, alpha 3 gene (GJA3) and gap junction protein, alpha 8 gene (GJA8), major intrinsic protein of lens fiber gene (MIP) and lens intrinsic membrane protein 2 gene (LIM2) [17-20]), transcription factors (heat shock transcription factor 4 gene [HSF4], paired-like homeodomain 3 gene [PITX3], and Maf-like protein gene [MAF] [21-23]), glucosaminyl (N-acetyl) transferase 2 gene (GCNT2) [24], chromatin modifying protein-4B gene (CHMP4B) [25], and transmembrane protein 114 gene (TMEM114) [26].
We report a novel missense mutation in CRYBA4 after analyzing a Chinese family with congenital cataract and microcornea. This mutation was not observed in any of the healthy family members.
Methods
Clinical evaluations
A three-generation Chinese pedigree that consists of 15 individuals, including two affected individuals, provided the basis for the study. Nine family members participated in the study (two affected and seven unaffected individuals; Figure 1). Two patients (both male) in this pedigree had congenital cataract and microcornea , and had shown symptoms of vision decrease before two years old. The proband was a 7-year-old boy who had a cataract extraction in another hospital, which provided us with post-operation photos (Figure 2). According to his medical records, this patient has congenital nuclear cataract with microcornea. The axial length of his eyes is 23.4 mm oculus dexter (OD) and 24.2 mm oculus sinister (OS); the corneal diameter is 9.5 mm. His father also has congenital nuclear cataract (post operation), and the axial length of his eyes is 24.6 mm (OD) and 25.2 mm (OS); the corneal diameter is also 9.5 mm. The corneal diameter and eye axial length of seven healthy members of this family were normal (Table 1). None of the family members had any other ocular or systemic abnormalities identified after a complete physical and ophthalmologic examination. One hundred normal controls (54 males, 46 females, age 2–42 years) were recruited from Physical Examination Center of Harbin Medical University the 2nd Affiliated Hospital, Harbin, Heilongjiang, China.
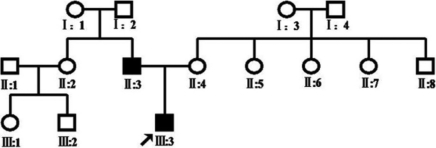
Figure 1.
Pedigree of a Chinese family with congenital cataract and microcornea. The proband (III:3) is indicated by an arrow. Two members (II:3 and III:3) in two generations were affected with congenital cataract and microcornea. Both of the two affected members had operations for bilateral congenital cataract.
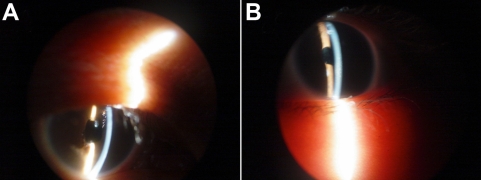
Figure 2.
Post-operation eye photographs of the two patients with congenital cataract and microcornea in a Chinese family. There were two patients in this family we studied with congenital cataract and microcornea. They had a cataract extraction in another hospital, which provided us with post-operation photos. A: Post-operation eye photographs of the proband’s father is shown. B: Post-operation eye photographs of the proband is displayed.
Table 1. Corneal diameter and eye axial length of nine members of the family.
| Member | Corneal diameter | Eye axial length (OD) | Eye axial length (OS) |
|---|---|---|---|
| III:3 |
9.5 mm |
23.4 mm |
24.2 mm |
| II:3 |
9.5 mm |
24.6 mm |
25.2 mm |
| I:1 |
11.6 mm |
24.3 mm |
24.7 mm |
| I:2 |
11.6 mm |
23.9 mm |
24.1 mm |
| II:2 |
11.8 mm |
24.2 mm |
25.1 mm |
| II:4 |
11.6 mm |
24.7 mm |
24.6 mm |
| II:5 |
11.5 mm |
24.5 mm |
24.1 mm |
| III:1 |
11.6 mm |
24.4 mm |
24.5 mm |
| III:2 | 11.5 mm | 24.2 mm | 24.7 mm |
Two members of this family (II:3 and III:3) in two generations were affected with congenital cataract and microcornea. Other seven members(I:1, I:2, II:2, II:4, II:5, III:1, and III:2) were not involved.
The family members were interviewed to obtain a detailed medical, ophthalmic, and family history after obtaining informed consent. This study was approved by the Institutional Review Board of Harbin Medical University, Harbin, China.
Molecular genetic studies
Peripheral blood samples (5 ml) were taken from nine members (two affected and seven unaffected individuals) of the family and 100 healthy controls, and were preserved at -20 °C in EDTA, and we then used the TIANamp Blood DNA kit (Tiangen Biltech Co. Ltd., Beijing, China) to extract genomic DNA. All six exons of CRYBA4 were amplified by PCR using the primers listed in Table 2. The PCR products were purified and sequenced by Shanghai Invitrogen Biotechnology Co. LTD (Shanghai, China). The data were compared with sequences from the NCBI GenBank (CRYBA4: NM_001886), and the modeled structures were built using Swiss-PdbViewer 4.0.1 (Torsten Schwede et al.,Basel, Switzerland) [27].
Table 2. Primers for mutational screening of CRYBA4.
| Exon | Forward primer | Reverse primer | Product length (bp) |
|---|---|---|---|
| 1 |
GTCCTTTCCCTCCCTGCTAA |
AGGATGAGGATGGCATTCAG |
316 |
| 2 |
TAGCCCAGTCACTCCTGGAC |
CCTAGGATTCATGGGGACCT |
238 |
| 3 |
TTTGCAATCCCTGCTTTACC |
CTTCAGGAGGGCACAACAGT |
350 |
| 4 |
ACCCCTGAATGGTTGTGACT |
CTTGAAGTGGCGACATGAGA |
350 |
| 5 |
CAAATGGCAAGGTTTCTGGT |
GTCCCTCAAATTCTGCCTGA |
465 |
| 6 | AGGGAATGGCATGATCAAAG | GGCCTGAAGTAAATAGAAGAAAGG | 633 |
Summary of the primers used for the amplification of CRYBA4 exons. Sequences are given in the 5′→3′direction.The primers were designed on line using primer 3.0.
Results
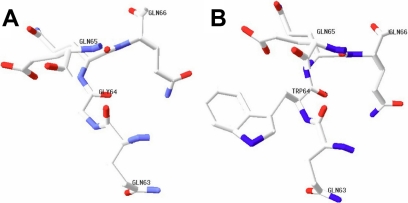
In this family, two patients showed the same clinical symptoms, congenital nuclear cataract and microcornea, and we identified a new mutation (c.225G>T, Figure 3) in exon 4 after direct sequencing of CRYBA4. This mutation was not detected in the healthy members of this family or in any of the normal control subjects. The mutation leads to an amino acid change (G64W). This substitution is located at a corner of the modeled structure of CRYBA4, as shown in the modeled structure (Figure 4 and Figure 5).
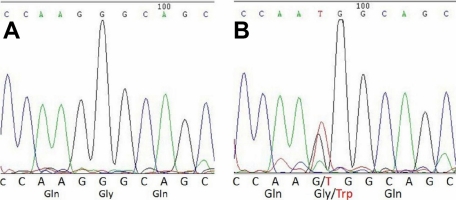
Figure 3.
Sequence analysis of the unaffected members and affected members in the family with congenital cataract and microcornea. A: The partial sequence of CRYBA4 in a normal individual is displayed. B: The corresponding nucleotide sequence of CRYBA4 in the proband is shown. Sequencing results showed a substitution of G→T in CRYBA4 (c.225G>T), which led to replacement of glycine by tryptophan at codon 64(G64W) in crystallin, beta A4 protein.
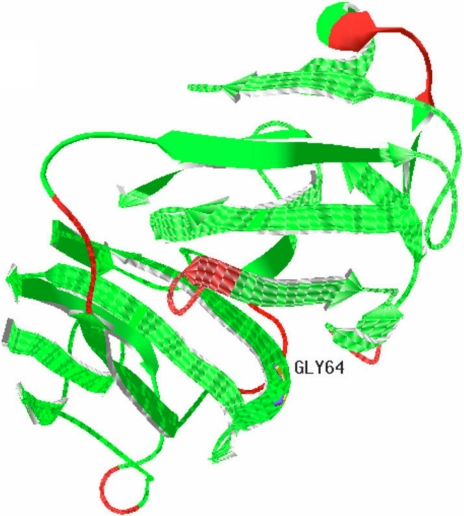
Figure 4.
The modeled structure of crystallin, beta A4 protein (CRYBA4). The modeled structure of CRYBA4 was built using special software (Swiss-PdbViewer 4.0.1 [39-41]). The mutation described in this study leads to the replacement of glycine by tryptophan at codon 64 (Gly64).The Gly64, located in the corner of the modeled structure, may form such secondary structures as β-sheet or β-turn.
Figure 5.
Portion of the crystallin, beta A4 protein (CRYBA4) modeled structure in the vicinity of residue 64. The portion modeled structures of the CRYBA4 in the vicinity of residue 64 were built using Swiss-PdbViewer 4.0.1 [39-41]). A: The normal modeled structure of CRYBA4 is displayed. B: The mutant modeled structure of CRYBA4 is shown. Glycine (Gly) is replaced by tryptophan (Trp) at codon 64. Modeled structures are shown by element type, using a default standard CPK (a popular color convention for distinguishing atoms of different chemical elements) scheme: n=blue, O=red, C=white.
Discussion
In this study, we identified a mutation (c.225G>T) in exon 4 of CRYBA4. This mutation segregates within the proband and his father (the two patients of this family) and was not detected in normal members of this family or in 100 healthy controls. We conclude that this sequence change results in the onset of congenital cataract and microcornea in this family.
Three major classes of crystallins are found in the mammalian lens [28]. They are α-crystallin (40% of total crystalline protein), β-crystallin (35%), and γ-crystallin (25%) [9]. The β- and γ-crystallins are members of a superfamily as they share a common two-domain structure composed of four “Greek key” motifs, two in the NH2- and two in the COOH-terminal domain [29].The β-crystallins are major constituents of the human lens and include three basic and four acidic protein forms. Each subgroup is encoded by three genes (CRYBA1, CRYBA2, and CRYBA4; CRYBB1, CRYBB2, and CRYBB3) [30].The protein encoded by CRYBA4 (belonging to the CRBA genes) contains 196 amino acids, and constitutes approximately 5% of the total soluble proteins in the young human lens [31].
β-Crystallins are expressed not only at the early developmental stages of the eye lens but also after birth. The temporal expression of crystallin genes vary in development. Different CRYB proteins can be found in both prenatal and postnatal developing lens; furthermore, they interact with each other [32,33]. Previous studies found homozygous changes in CRYBB2 that were associated with severe microphthalmia and cataract and found an interaction of CRYBB2–CRYBA4 monomers [34,35]. Human βA4-crystallin readily oligomerizes with human βB1-crystallin, a hetero-oligomer that can be purified [36].
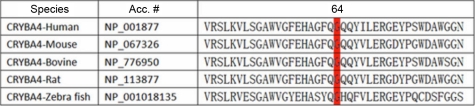
To date, Billingsley et al. [9] have reported two mutations (c.317T>C and c.242T>C) in exon 4 of CRYBA4 by genetic analysis of a large Indian family with an autosomal dominant cataract phenotype. It is worth noting that one of the two mutations (c.242T>C) and the mutation reported in this study (c.225G>T) are in a highly conserved area of CRYB exon 4 (Figure 6), which indicates that the sequence changes in this area play an important role in the onset of congenital cataract.
Figure 6.
Interspecies sequence alignment of a portion of the CRYBA4 (crystallin, beta A4) amino acid sequence. The alignment data indicate that glycine at position 64 (indicated by the red background) is highly conserved in CRYBA4 of different species.
The mutation described in this study leads to replacement of glycine by tryptophan at codon 64. From the modeled structure of CRYBA4 (Figure 4), which was built using special software (Swiss-PdbViewer 4.0.1; Torsten Schwede et al., Basel, Switzerland), we can report that this substitution takes place at a corner of the backbone structure. Moreover, previous studies support that glycine is often found in β-sheet secondary structures and is the amino acid appearing most frequently at position i+2 of β-turn [37,38]. Based on the above, we surmise that glycine at codon 64 probably forms similar secondary structures as β-sheet or β-turn. The substitution may result in damage to forming normal secondary structure during the CRYBA4 protein folding process so that the structure of the protein has reduced stability in the patient’s lens (Figure 4 and Figure 5). These series of changes may lead to disturbance of the lens transparency and functional integrity, resulting in cataract.
In the present study, we reported a novel missense mutation in two patients with congenital cataract and microcornea that come from the same Chinese family. This is the first report linking mutations in CRYBA4 to cataractogenesis and microcornea. Our findings expand the mutation spectrum of CRYBA4 and provide useful information in the study of molecular pathogenesis of congenital cataract.
Acknowledgments
We thank all members of the family and the normal volunteers for their cooperation in this study. The work was supported by Innovation Fund for Core Teachers of Heilongjiang Province Colleges and Universities, Project No. 1055G023.
References
- 1.Haargaard B, Wohlfahrt J, Fledelius HC, Rosenberg T, Melbye M. A nationwide Danish study of 1027 cases of congenital/infantile cataracts: etiological and clinical classifications. Ophthalmology. 2004;111:2292–8. doi: 10.1016/j.ophtha.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert CE, Rahi J, Quinn GE. visual impairment and blindness in children. In: Johnson GJ, Minassian DC, Weale R, West SK, editors. The Epidemiology of Eye Disease. 2nd ed. London: Arnold; 2003. p. 260–286. [Google Scholar]
- 3.Dawson CR, Schwab IR. Epidemiology of cataract - a major cause of preventable blindness. Bull World Health Organ. 1981;59:493–501. [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson GJ, Minassian DC, Weale R, West SK, eds. The epidemiology of eye disease. 2nd ed. London: Arnold; 2003. [Google Scholar]
- 5.Foster A, Gilbert C, Rahi J. Epidemiology of cataract in childhood: a global perspective. J Cataract Refract Surg. 1997;23:601–4. doi: 10.1016/s0886-3350(97)80040-5. [DOI] [PubMed] [Google Scholar]
- 6.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–4. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 7.Berry V, Francis P, Reddy MA, Collyer D, Vithana E, MacKay I, Dawson G, Carey AH, Moore A, Bhattacharya SS, Quinlan RA. Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet. 2001;69:1141–5. doi: 10.1086/324158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaA3/A1-crystallin gene. Mol Vis. 1998;4:21. [PubMed] [Google Scholar]
- 9.Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, Hosseini SM, Manisastry SM, Vijayalakshmi P, Gopinath PM, Graw J, Heon E. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79:702–9. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002;71:1216–21. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–8. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 12.Riazuddin SA, Yasmeen A, Yao W, Sergeev YV, Zhang Q, Zulfiqar F, Riaz A, Riazuddin S, Hejtmancik JF. Mutations in betaB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2100–6. doi: 10.1167/iovs.04-1481. [DOI] [PubMed] [Google Scholar]
- 13.Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, Lubsen N, Munier FL. The gamma-crystallins and human cataracts: a puzzle made clearer. Am J Hum Genet. 1999;65:1261–7. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, Gao Y, Ma W, Tang X, Li X, Shen Y. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–10. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran RD, Perumalsamy V, Hejtmancik JF. Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum Genet. 2007;121:475–82. doi: 10.1007/s00439-006-0319-6. [DOI] [PubMed] [Google Scholar]
- 16.Conley YP, Erturk D, Keverline A, Mah TS, Keravala A, Barnes LR, Bruchis A, Hess JF, FitzGerald PG, Weeks DE, Ferrell RE, Gorin MB. A juvenile-onset, progressive cataract locus on chromosome 3q21-q22 is associated with a missense mutation in the beaded filament structural protein-2. Am J Hum Genet. 2000;66:1426–31. doi: 10.1086/302871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rees MI, Watts P, Fenton I, Clarke A, Snell RG, Owen MJ, Gray J. Further evidence of autosomal dominant congenital zonular pulverulent cataracts linked to 13q11 (CZP3) and a novel mutation in connexin 46 (GJA3). Hum Genet. 2000;106:206–9. doi: 10.1007/s004390051029. [DOI] [PubMed] [Google Scholar]
- 18.Shiels A, Mackay D, Ionides A, Berry V, Moore A, Bhattacharya S. A missense mutation in the human connexin50 gene (GJA8) underlies autosomal dominant “zonular pulverulent” cataract, on chromosome 1q. Am J Hum Genet. 1998;62:526–32. doi: 10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry V, Francis P, Kaushal S, Moore A, Bhattacharya S. Missense mutations in MIP underlie autosomal dominant 'polymorphic' and lamellar cataracts linked to 12q. Nat Genet. 2000;25:15–7. doi: 10.1038/75538. [DOI] [PubMed] [Google Scholar]
- 20.Pras E, Levy-Nissenbaum E, Bakhan T, Lahat H, Assia E, Geffen-Carmi N, Frydman M, Goldman B, Pras E. A missense mutation in the LIM2 gene is associated with autosomal recessive presenile cataract in an inbred Iraqi Jewish family. Am J Hum Genet. 2002;70:1363–7. doi: 10.1086/340318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, Cui B, Xia Y, Liu J, Hu L, Zhao G, Hayden MR, Kong X. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31:276–8. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- 22.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 23.Vanita V, Singh D, Robinson PN, Sperling K, Singh JR. A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant “cerulean cataract” in an Indian family. Am J Med Genet A. 2006;140:558–66. doi: 10.1002/ajmg.a.31126. [DOI] [PubMed] [Google Scholar]
- 24.Pras E, Raz J, Yahalom V, Frydman M, Garzozi HJ, Pras E, Hejtmancik JF. A nonsense mutation in the glucosaminyl (N-acetyl) transferase 2 gene (GCNT2): association with autosomal recessive congenital cataracts. Invest Ophthalmol Vis Sci. 2004;45:1940–5. doi: 10.1167/iovs.03-1117. [DOI] [PubMed] [Google Scholar]
- 25.Shiels A, Bennett TM, Knopf HL, Yamada K, Yoshiura K, Niikawa N, Shim S, Hanson PI. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am J Hum Genet. 2007;81:596–606. doi: 10.1086/519980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamieson RV, Farrar N, Stewart K, Perveen R, Mihelec M, Carette M, Grigg JR, McAvoy JW, Lovicu FJ, Tam PP, Scambler P, Lloyd IC, Donnai D, Black GC. Characterization of a familial t(16;22) balanced translocation associated with congenital cataract leads to identification of a novel gene, TMEM114, expressed in the lens and disrupted by the translocation. Hum Mutat. 2007;28:968–77. doi: 10.1002/humu.20545. [DOI] [PubMed] [Google Scholar]
- 27.Schwede T, Diemand A, Guex N, Peitsch MC.Protein structure computing in the genomic era. Res Microbiol 2000151107–12.PP [DOI] [PubMed] [Google Scholar]
- 28.Bloemendal H, de Jong WW. Lens proteins and their genes. Prog Nucleic Acid Res Mol Biol. 1991;41:259–81. doi: 10.1016/s0079-6603(08)60012-4. [DOI] [PubMed] [Google Scholar]
- 29.Kumaraswamy VS, Lindley PF, Slingsby C, Glover ID. An eye lens protein-water structure: 1.2 A resolution structure of gammaB-crystallin at 150 K. Acta Crystallogr D Biol Crystallogr. 1996;52:611–22. doi: 10.1107/S0907444995014302. [DOI] [PubMed] [Google Scholar]
- 30.Piatigorsky J. Gene expression and genetic engineering in the lens. Friedenwald lecture. Invest Ophthalmol Vis Sci. 1987;28:9–28. [PubMed] [Google Scholar]
- 31.Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, David LL. Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J Biol Chem. 1997;272:2268–75. doi: 10.1074/jbc.272.4.2268. [DOI] [PubMed] [Google Scholar]
- 32.Bax B, Lapatto R, Nalini V, Driessen H, Lindley PF, Mahadevan D, Blundell TL. Slingsby C. X-ray analysis of beta B2-crystallin and evolution of oligomeric lens proteins. Nature. 1990;347:776–80. doi: 10.1038/347776a0. [DOI] [PubMed] [Google Scholar]
- 33.Norledge BV, Trinkl S, Jaenicke R, Slingsby C. The X-ray structure of a mutant eye lens beta B2-crystallin with truncated sequence extensions. Protein Sci. 1997;6:1612–20. doi: 10.1002/pro.5560060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer P, Yount J, Mitchell T, LaMorticella D, Carrero-Valenzuela R, Lovrien E, Maumenee I, Litt M. A second gene for cerulean cataracts maps to the beta crystallin region on chromosome 22. Genomics. 1996;35:539–42. doi: 10.1006/geno.1996.0395. [DOI] [PubMed] [Google Scholar]
- 35.Cooper PG, Carver JA, Truscott RJ. 1H-NMR spectroscopy of bovine lens beta-crystallin. The role of the beta B2-crystallin C-terminal extension in aggregation. Eur J Biochem. 1993;213:321–8. doi: 10.1111/j.1432-1033.1993.tb17765.x. [DOI] [PubMed] [Google Scholar]
- 36.Bateman OA, Sarra R, van Genesen ST, Kappe G, Lubsen NH, Slingsby C. The stability of human acidic beta-crystallin oligomers and hetero-oligomers. Exp Eye Res. 2003;77:409–22. doi: 10.1016/s0014-4835(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins JE, Creager MS, Lewis RV, Holland GP, Yarger JL. Quantitative Correlation between the protein primary sequences and secondary structures in spider dragline silks. Biomacromolecules. 2010;11:192–200. doi: 10.1021/bm9010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunasekaran K, Gomathi L, Ramakrishnan C, Chandrasekhar J, Balaram P. Conformational interconversions in peptide beta-turns: analysis of turns in proteins and computational estimates of barriers. J Mol Biol. 1998;284:1505–16. doi: 10.1006/jmbi.1998.2154. [DOI] [PubMed] [Google Scholar]
- 39.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 40.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]