Abstract
Syndecans are a family of type-I transmembrane proteins that are involved in cell-matrix adhesion, migration, neuronal development, and inflammation. Previous quantitative genetic studies pinpointed Drosophila Syndecan (dSdc) as a positional candidate gene affecting variation in fat storage between two Drosophila melanogaster strains. Here, we first used quantitative complementation tests with dSdc mutants to confirm that natural variation in this gene affects variability in Drosophila fat storage. Next, we examined the effects of a viable dSdc mutant on Drosophila whole-body energy metabolism and associated traits. We observed that young flies homozygous for the dSdc mutation had reduced fat storage and slept longer than homozygous wild-type flies. They also displayed significantly reduced metabolic rate, lower expression of spargel (the Drosophila homologue of PGC-1), and reduced mitochondrial respiration. Compared to control flies, dSdc mutants had lower expression of brain insulin-like peptides, were less fecund, more sensitive to starvation, and had reduced life span. Finally, we tested for association between single nucleotide polymorphisms (SNPs) in the human SDC4 gene and variation in body composition, metabolism, glucose homeostasis, and sleep traits in a cohort of healthy early pubertal children. We found that SNP rs4599 was significantly associated with resting energy expenditure (P = 0.001 after Bonferroni correction) and nominally associated with fasting glucose levels (P = 0.01) and sleep duration (P = 0.044). On average, children homozygous for the minor allele had lower levels of glucose, higher resting energy expenditure, and slept shorter than children homozygous for the common allele. We also observed that SNP rs1981429 was nominally associated with lean tissue mass (P = 0.035) and intra-abdominal fat (P = 0.049), and SNP rs2267871 with insulin sensitivity (P = 0.037). Collectively, our results in Drosophila and humans argue that syndecan family members play a key role in the regulation of body metabolism.
Introduction
Obesity is a condition characterized by an excess of adipose tissue that adversely affects human health [1]. The clinical problem of excessive adipose tissue resides in its strong association with a number of chronic diseases, such as insulin resistance, type 2 diabetes mellitus (T2DM), coronary artery disease and stroke [1]. In 2007–2008, 33.8% of the adults in the United States were obese [2]. Another worrisome observation is the increase in percent of obese children and adolescents [3]. Obese children are more likely to become obese adults [4], and thus we may see profound public health consequences as a result of the appearance of associated co-morbidities in adulthood.
The role of genes in human obesity and related phenotypes is well established [5], [6]. The fruit fly D. melanogaster has emerged in recent years as a powerful model organism for studying the genetics of fat storage and obesity [7]–[9]. Previously, we used a recombinant mapping approach to identify chromosomal regions (quantitative trait loci or QTL) that contribute to variation in triacylglycerol (TAG) storage using two unrelated strains of D. melanogaster, Oregon R (Ore) and 2b [10]. Subsequent fine mapping of these QTL regions identified several candidate genes that contribute to variation in TAG storage, many of which have been verified to be important for TAG storage in both Drosophila and humans [7]. Fine-mapping localized one QTL affecting TAG to the 57E1;57E3 cytogenetic region on chromosome 2 [10]. Only five genes lie within this region, including the dSdc gene, which encodes a member of the syndecan gene family [11].
Syndecans are type-I transmembrane proteins that are present on the surface of all adherent cells [12]. While Drosophila appears to have only one syndecan protein, mammals have four syndecan proteins encoded by four separate genes. Three of them, SDC1, SDC2, and SDC3, are expressed in a tissue-specific manner, whereas the fourth, SDC4, is expressed in a variety of cell types [12]. All syndecan proteins are characterized by a core protein composed of an extracellular domain (ectodomain) followed by a single hydrophobic membrane-spanning domain and a short intracellular domain. The ectodomain contains attachment sites for heparan sulphate polysaccharide chains that mediate interactions with extracellular matrix (ECM) components [13], heparin-sulfate growth factors [14], cell adhesion molecules [15], lipases [16], chemokines, cytokines and their receptors [17], and pathogens [18]. As a result, syndecans function as co-receptors modulating signal transduction pathways initiated by growth factors and are involved in cell proliferation, adhesion and migration, lipid metabolism, and inflammation [12]. Via their intracellular domains, syndecans also interact with cytoplasmic proteins that control focal adhesion, cell spreading, and actin cytoskeletal organization [12], hence playing a direct role in transducing signals from the ECM to the cytoplasm. Previous studies confirmed the functional conservation of syndecan in Drosophila [19]. Furthermore, Drosophila syndecan has been reported to participate in normal axon guidance and neuronal development via regulation of the Slit/Roundabout signaling [20].
Based on these observations, the results of our genetic mapping experiment, and the similarity in structural characteristics between Drosophila and mammalian syndecans, we reasoned that dSdc was a candidate gene contributing to variation in TAG storage between Ore and 2b. To investigate this hypothesis, we performed quantitative complementation tests by crossing mutants of dSdc with flies of the Ore and 2b genotypes and found that allelic differences at dSdc produced differences in TAG storage between these two strains. We next examined the functional role played by dSdc in fat storage by testing whether flies homozygous for a hypomorphic mutation of dSdc (dSdcBG 02774) and flies homozygous for the corresponding wild-type allele differed in whole-body energy metabolism and associated traits in Drosophila. Collectively, our results in Drosophila showed that dSdc plays an important role in the regulation of body composition and metabolism in young flies.
In humans, the SDC4 gene includes 5 exons spanning 19.7 kb on chromosome 20q12 [21]. A number of whole-genome linkage studies have linked the chromosomal region 20q12-13 to T2DM and obesity [22]–[25]. Based on this observation and our results in Drosophila, we carried out an explorative study to examine the association between genetic variants in the human SDC4 gene and variation in traits associated with body composition, energy metabolism, and sleep duration in a cohort of 252 healthy early pubertal children. Although small in size, this human cohort was chosen for two main reasons. First, the phenotypic data available for each subject included robust measurements of body composition and glucose-insulin dynamics. Second, the cohort was characterized by genetic admixture, which allowed us to adjust for ancestry within ethnic groups and therefore limit false-positive results [26]. Consistent with our data in flies, association tests using SDC4 genotypes as independent variables revealed an impact of SDC4 variation on resting energy expenditure (REE), insulin dynamics, and sleep duration in our cohort of young children. These results motivate future genetic studies in independent human populations to verify the effects of this gene.
Results
Complementation tests to dSdc mutations
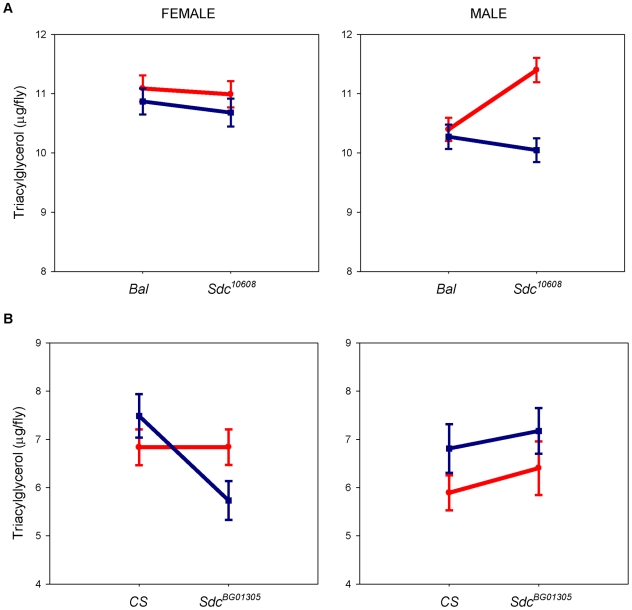
To investigate the relationship between dSdc and TAG storage, we first used complementation tests with two mutations of dSdc (Sdc10608 and SdcBG01305). Sdc10608 failed to complement the TAG phenotype of Ore and 2b QTL in males (Fig. 1A) and SdcBG01305 failed to complement the TAG phenotype of Ore and 2b QTL in females (Fig. 1B). The allelic effects of variation at this locus were consistent for both dSdc mutants, as flies with the Ore allele had higher levels of TAG than flies with the 2b allele. Thus, these results confirmed dSdc as a candidate gene affecting variation in TAG storage between these two strains of D. melanogaster.
Figure 1. Variation in dSdc affects variability in Drosophila fat storage.
(A and B) Quantitative complementation tests with two mutations of dSdc. TAG levels are assessed for four genotypes: M/2b, Bal or CS/2b, M/Ore, andz Bal or CS/Ore, where M denotes the dSdc mutation, Bal is the balancer chromosome, and CS is the Canton S strain. Values shown are the TAG least-squared means within genotype classes for n = 10 independent replicates. Details on the analysis are provided in the text. Ore and 2b heterozygote flies are color-coded red and dark blue, respectively. (A) In females, there is no evidence of quantitative failure of the Sdc10608 mutation to complement the Ore and 2b alleles because the relative difference in the average trait values between Ore and 2b over Sdc10608 mutation and the Bal is the same. In males, however, the difference in the average trait values between Ore and 2b alleles over the Sdc10608 mutation is greater than the difference in the average trait value between Ore and 2b alleles over the Bal (P value for Line x Genotype is 0.003), which indicates failure of the Sdc10608 to complement the Ore and 2b alleles. (B) Although there is no evidence of quantitative failure to complement in males, greater difference in female Ore and 2b lines when over SdcBG01305 mutation compared to female Ore and 2b over the CS strain indicates failure of the Sdc10608 to complement the Ore and 2b alleles (P value for Line x Genotype is 0.033). Collectively, these results indicate an interaction between the mutated allele (Sdc10608 or SdcBG01305) and the TAG QTL of Ore and 2b.
Effect of SdcBG02774 mutation on body weight mutation on body weight, total protein content, and metabolite storage
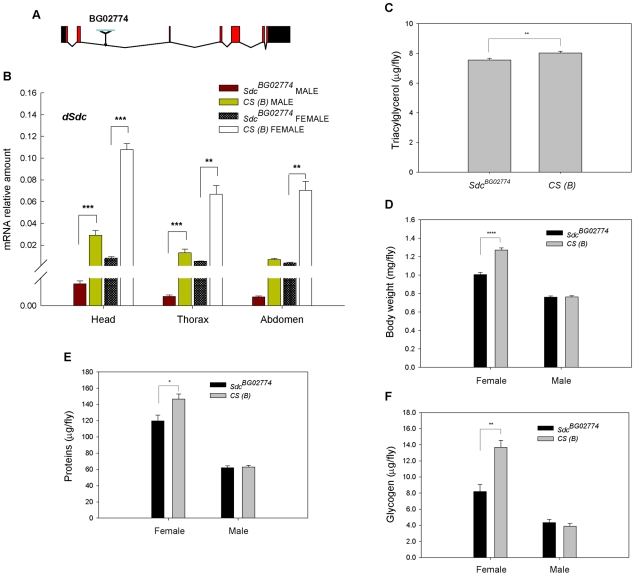
To examine the functional role played by dSdc in fat storage, we used a viable mutant allele of the gene, SdcBG02774. SdcBG02774 is a mutant generated in the w1118;Canton S (B) [CS (B)] strain by the insertion of a p[GT1]-element in the second intron of dSdc (Fig. 2A) [27]. We examined the effects of this P-element insertion on dSdc transcription in adult flies by performing Real Time-quantitative PCR (RT-qPCR) experiments using RNA isolated from three body parts (head, thorax, and abdomen). We found that the overall expression of dSdc is significantly reduced in the three body parts of SdcBG02774 flies (Fig. 2B).
Figure 2. Effect of SdcBG0277 4 mutation on body weight, total protein content, and metabolite storage.
(A) Schematic representation of dSdc gene region on the second chromosome at cytological position 57E1-57E6 (NCBI accession no. AE013599.4). Black and red boxes represent untranslated regions and exons, respectively. The location of the p[GT1] insertion site that creates the SdcBG0277 4 mutation is indicated with an arrowhead. (B) P-element insertion in the dSdc gene leads to significantly reduced dSdc transcript abundance. Messenger RNA levels analyzed by RT-qPCR (n = 6) on cDNA using primers that encompass a common region of alternative transcripts. Levels of dSdc mRNA were normalized to Drosophila ribosomal protein49 (rp49) mRNA levels. (C) SdcBG0277 4 mutation affects TAG levels. Because no differences were observed between male and female flies in TAG storage, we pooled male and female data for the analysis. Values represent least-square means from n = 20 independent replicates of homozygous SdcBG0277 4 and CS (B) flies. (D–F) SdcBG0277 4 mutation affects body weight, total protein content, and glycogen levels in females. Each value represents the mean body weight (panel D) and least-square means for protein (panel E) and glycogen (panel F) levels from n = 10 independent replicates. In all panels, error bars represent SEM. * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.0001 compared to control.
Next, we assessed the effect of the SdcBG02774 mutation on body weight and whole-body TAG, protein, and glycogen contents. After adjusting for body weight, we found a small but significant reduction in TAG storage in homozygous SdcBG02774 flies compared to controls (Fig. 2C). On average, TAG levels of SdcBG02774 flies were 6% lower than that of controls. However, sex-specific effects were observed for the other traits. Compared to controls, females homozygous for the SdcBG02774 mutation had significantly reduced body weight (21%) and body weight-adjusted protein (18%) and glycogen (40%) contents, while male SdcBG02774 did not differ from the control strain (Fig. 2D–F).
Homozygous SdcBG02774 flies have lower expression levels of dilps, are less fertile, and have reduced metabolic activity than controls
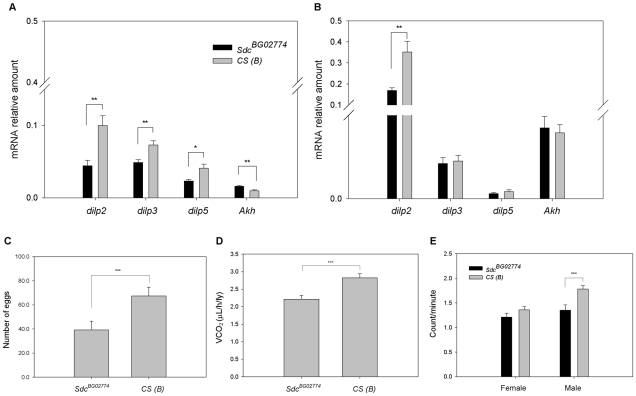
In insects, the insulin/insulin-like growth factor (IGF) signaling controls organ growth and final body size [28] as well as metabolism in adult flies [29], [30]. The Drosophila genome contains seven Drosophila insulin-like peptide (dilp) genes. Several of the dilps (dilp1-5) are expressed at high levels in insulin producing cells (IPC) of the pars intercerebralis in the brain of both larvae [31] and adult flies [29]. To investigate whether the SdcBG02774 had a defect in the production of DILPs, we measured mRNA levels of dilp2, dilp3, and dilp5, which are expressed in the IPC of adult flies [29], in dSdc mutants and controls. We also investigated the expression level of the gene that encodes the adipokinetic hormone (AKH), a putative glucagon homolog. Like pancreatic insulin and glucagon-producing cells in mammals, cross-regulatory interactions exist between DILP- and AKH-producing cells in Drosophila [32] and expression levels of Akh were previously shown to be increased in IPC-deficient larvae and adults [33]. In females, we observed a 55%, 33% and 43% reduction in expression of dilp2, dilp3 and dilp5, respectively, and a 38% increase in Akh expression in SdcBG0277 4 flies compared with controls (Fig. 3A). In males, we found no difference between mutants and controls for dilp3, dilp5, and Akh, but the SdcBG0277 4 flies showed a 52% reduction in expression of dilp2 (Fig. 3B).
Figure 3. Homozygous SdcBG0277 4 flies have lower expression levels of dilps, fecundity, and metabolic activity.
(A–B) Gene expression levels were measured by RT-qPCR using mRNA extracted from heads (dilps) and whole-body (Akh) of female (panel A) and male (panel B) flies (n = 6). All genes were normalized to rp49. (C) Values represent the average number of eggs laid by female flies over a five day period per n = 10 independent replicates. (D) CO2 production measured by indirect calorimetry. No differences were observed between male and female flies and the values represent VCO2 least-square means of n = 20 independent replicates. (E) Waking activity was measured counting the number of times a given fly crosses an infrared beam during a one-minute interval. Values represent the average number of activity counts per waking minute of two independent replicates of 16 flies. In all panels, error bars represent SEM. *P≤0.05, ** P≤0.01, *** P≤0.001 compared to control.
Recently, Zhang and collaborators [30] have shown that flies homozygous for a deletion of dilp1-5 were poorly fertile and had reduced metabolic activity. A significant reduction in egg-production was also observed by Gronke et al. [34] in dilp2-3,5 mutant females. Therefore, we tested the impact of the dSdc mutation on these phenotypes. First, we measured fecundity by counting the total number of eggs laid over a five day period. As expected, we found that dSdcBG02774 females laid significantly less eggs (42% less) than control flies (Fig. 3C). Next, we examined metabolic rate by measuring carbon dioxide (CO2) production. Because no differences were observed between male and female flies in CO2 production, we pooled male and female data for statistical analysis. We observed that dSdc mutants displayed a 22% reduction in metabolic rate as compared to controls (Fig. 3D). Finally, we monitored waking activity, the number of times flies crossed an infrared beam per minute spent awake [35]. We did not observe a difference in waking activity in SdcBG02774 female flies; however, SdcBG02774 male flies showed a 24% reduction in waking activity compared to controls (Fig. 3E).
Homozygous SdcBG02774 flies have lower transcript levels of Thor and spargel and reduced mitochondrial function compared to controls
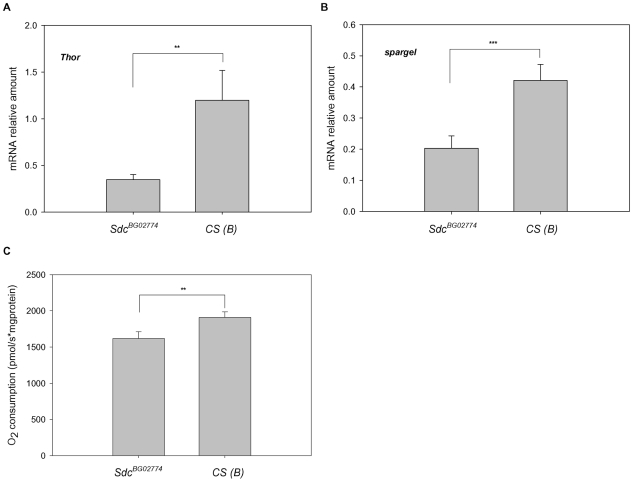
Reduced expression of dilps should lead to systemic reduction of the insulin signaling pathway. To assess whether the insulin signaling is indeed reduced in the dSdc mutants, we first investigated the insulin-dependent transcriptional response induced by the Drosophila forkhead transcription factor (dFoxO). Reduced insulin receptor/PI3K/Akt signaling in Drosophila induces nuclear accumulation of the dFoxO, which in turn activates the transcription of target genes, such as the translational regulator Thor, the Drosophila homolog of mammalian EIF4EBP1 [36]. We measured Thor mRNA levels in dSdc mutants and controls and found no differences between male and female flies; therefore we pooled the data for the analysis. Interestingly, in contrast to our prediction, we observed that Thor expression was significantly reduced in dSdc mutants (71%) compared to control flies (Fig. 4A), despite the fact that mutants exhibited reduced levels of DILPs (Fig. 3A,B).
Figure 4. Homozygous SdcBG02774 flies have lower expression levels of spargel and reduced mitochondrial function.
(A–B) Despite reduced levels of DILPs, homozygous SdcBG02774 flies do not show increased levels of Thor, but display lower levels of spargel mRNA than controls. Gene expression levels were measured by RT-qPCR using mRNA extracted from whole body of SdcBG02774 and CS (B) flies. Transcript levels of Thor and spargel were normalized to rp49. Values represent average of 17 (panel A) and 12 (panel B) independent replicates. (C) O2 consumption by whole-fly isolated mitochondria was measured in the presence of saturating amounts of NAD+-linked respiratory substrates (pyruvate plus proline) and ADP (state 3 respiration rate). Values represent average of 20 independent replicates. In all panels, pooled male and female data was used for the analysis. Error bars represent SEM. ** P≤0.01, *** P≤0.001 compared to control.
Peroxisome-proliferator-activated receptor-gamma co-activator-1 (PGC-1) family members play a pivotal role in the control of energy homeostasis in mammals [37]–[39]. Recently, Tiefenböck et al. [40] have shown that the Drosophila homologue of PGC-1, Spargel, is a critical downstream component for insulin signaling. The authors reported that Spargel acts in parallel to dFOXO and mediates mitochondrial respiration in response to insulin signaling [40]. To determine whether reduced levels of dilps in SdcBG02774 flies might lead to a decrease in the expression of spargel gene, we compared mRNA levels of spargel in SdcBG02774 and CS (B) flies. Once again, we did not observe differences in transcript levels between male and females and therefore we pooled the data for the statistical analysis. We found that mRNA levels of spargel were reduced 52% in the mutant flies relative to controls (Fig. 4B).
Finally, we analyzed respiration rates in mitochondria isolated from mutant and control flies using NAD+-linked respiratory substrates (pyruvate plus proline) that deliver electrons into complex I of the mitochondrial electron transport chain. In the analysis averaged across sexes, we observed that the ADP-dependent state 3 respiration rate (a measure of oxidative phosphorylation capacity) was reduced by approximately 15% in SdcBG02774 compared with the controls (Fig. 4C).
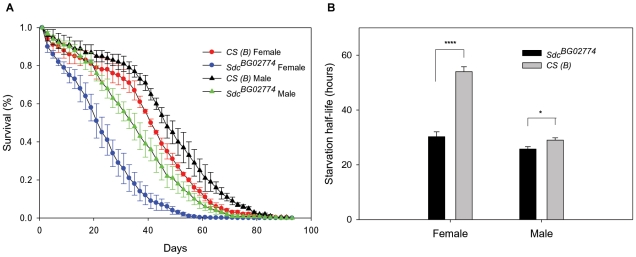
Homozygous SdcBG02774 flies have reduced survival and are more sensitive to starvation than controls
Ablation of the IPCs in the pars intercerebralis of the final instar larval brain results in adult flies with reduced fecundity, increased resistance to starvation, and extended life span [29]. An increase in survival has also been observed in dilp2 mutant flies [34] and in flies with reduced expression levels of dilp2 as a result of genetic manipulation of dFoxO [41], c-Jun N-Terminal Kinase [42], or p53 [43]. Thus, we asked whether the reduced dilps transcript levels observed in the dSdc mutant flies would affect survival under normal feeding and water-only starvation conditions. We found significant differences in survival between SdcBG02774 mutants and controls. However, contrary to our prediction, median life span of female and male SdcBG02774 flies was decreased by 46% and 28%, respectively, under normal feeding conditions (Fig. 5A). Female and male mutant flies were also less resistant to starvation than controls by 45% and 11%, respectively (Fig. 5B).
Figure 5. Homozygous SdcBG02774 flies have reduced lifespan and are more sensitive to starvation.
(A) Life span assays were carried out using population cages. We used three replicates of each sex and genotype combination with initial population sizes ranging from 183 to 295 individuals. Survival was monitored every other day until all flies in a population cage had died. Mutants of both sexes had significantly reduced survival compared with controls (Females and Males: P<0.0001). (B) Values represent the average starvation half-life of n = 5 independent replicates. In both panels, error bars represent SEM. *P≤0.05, **** P≤0.0001 compared to control.
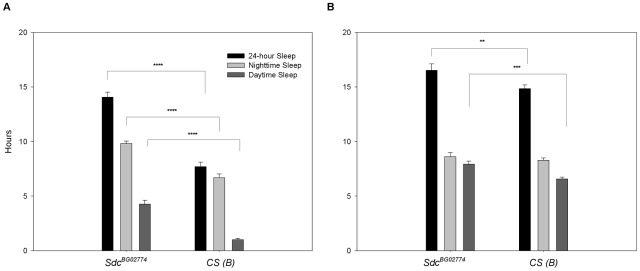
Homozygous SdcBG0277 4 flies sleep longer than controls
Previously, Harbison and collaborators [44] reported a significant correlation between two genetic variants in the dSdc gene and daytime sleep in 40 wild-derived Drosophila lines, suggesting that dSdc may also be involved in sleep duration. To test this hypothesis, we assessed the sleep-wake cycle in dSdcBG 02774 and CS (B) flies over the course of a 24 h day. As expected, sleep was increased by 32% during nighttime and by 76% during daytime in dSdcBG 02774 female flies compared to controls (Fig. 6A). A 17% increase in daytime sleep was also observed in mutant males (Fig. 6B).
Figure 6. Homozygous SdcBG02774 flies sleep longer.
(A–B) Sleep parameters were measured counting the number of times a given fly crosses an infrared beam during a one-minute interval. Sleep was defined as any period 5 minutes or longer without an activity count. Values represent average hours of sleep of two independent replicates of 16 female (panel A) and male (panel B) flies. In both panels, error bars represent SEM. ** P≤0.01, *** P≤0.001, **** P≤0.0001 compared to control.
Variation in human SDC4 is associated with energy metabolism, body composition, and sleep
We conducted a population-based association study on 252 European American (EA), African American (AA) and Hispanic American (HA) children. Anthropometric and metabolic characteristics of the study subjects are shown in Table 1. We selected three haplotype-tagging SNPs (htSNPs) that map within the SDC4 gene from the International Haplotype Map (HapMap) Project (http://www.hapmap.org): rs1981429 (T/G) and rs2267871 (T/A), which map in the intron 2, and rs4599 (C/T) in the 3′UTR region. All genotype groups were in Hardy-Weinberg equilibrium (Table 2). The Minor Allele Frequencies ranged from 0.11 to 0.49 (Table 2). There was no difference in genotype frequencies between ethnic groups for rs2267871. However, there was a significant difference in genotype frequencies between ethnic groups for rs1981429 (P<0.0001) and rs4599 (P = 0.0002), which is consistent with genotypic data reported by the HapMap Project. Low to moderate pair-wise linkage disequilibrium estimates were observed among the SNPs in the three ethnic groups (Fig. 7).
Table 1. Characteristics of human subjects.
| All (n = 252) | Hispanic Americans (n = 58) | European Americans (n = 108) | African Americans (n = 86) | |
| Sex (%male) | 51.6 | 46.6 | 52.8 | 53.5 |
| Age (yrs) | 9.7±1.6 | 9.4±1.5 | 9.7±1.7 | 9.7±1.5 |
| BMI * | 18.5±2.9 | 19.4±2.7 | 18.0±2.6 | 18.6±3.3 |
| Weight (kg) | 36.9±9.1 | 37.1±8.5 | 35.9±8.8 | 37.9±10.0 |
| Total Lean Mass (kg) ** | 26.0±5.2 | 24.7±4.7 | 8.4±5.1 | 27.5±5.5 |
| Total Fat Mass (kg) * | 8.8±5.5 | 10.4±5.1 | 8.4±5.1 | 8.17±6.05 |
| IAAT (cm2)** | 33.6±22.7 | 42.1±24.7 | 34.9 ±22.8 | 26.7±18.7 |
| SI [×10−4 min−1/(µU/mL)]*** | 5.8±3.6 | 5.4±3.8 | 7.0 ±3.9 | 4.55±2.6 |
| Fasting Glucose (mg/dL)*** | 97.0±6.4 | 99.6±6.1 | 97.5±6.2 | 94.5±5.9 |
| REE (kcal/day) | 1195±243 | 1191±292 | 1194 ±242 | 1199±204 |
| Hours of sleep/night *** | 8.9±1.1 | 8.85±1.22 | 9.3±0.8 | 8.53±1.15 |
Values are means ± SD. BMI: body mass index. IAAT: Intra-abdominal adipose tissue. SI: Insulin sensitivity index. REE: Resting energy expenditure. P values for difference between ethnic groups are obtained using ANOVA.
p<0.05,
p<0.01 and
p<0.001.
Table 2. SNP marker information at the human SDC4 locus in the study cohort.
| dbSNP rs # | Alleles | Genomic Position* | Heterozygosity | HWE P-value | MAF | |||||||
| HA | EA | AA | HA | EA | AA | HA | EA | AA | ||||
| rs1981429 | T/G | 43409107 | 0.43 | 0.50 | 0.47 | 0.74 | 1.00 | 0.66 | 0.27 | 0.49 | 0.36 | |
| rs2267871 | T/A | 43405784 | 0.31 | 0.28 | 0.22 | 1.00 | 0.34 | 0.59 | 0.19 | 0.19 | 0.11 | |
| rs4599 | C/T | 43387821 | 0.29 | 0.31 | 0.53 | 0.26 | 1.00 | 0.17 | 0.23 | 0.18 | 0.36 | |
*Position in [91]. HWE: Hardy-Weinberg Equilibrium. MAF: Minor Allele Frequency.
Figure 7. Pair-wise linkage disequilibrium (D') pattern between three SDC4 SNPs in the human cohort.
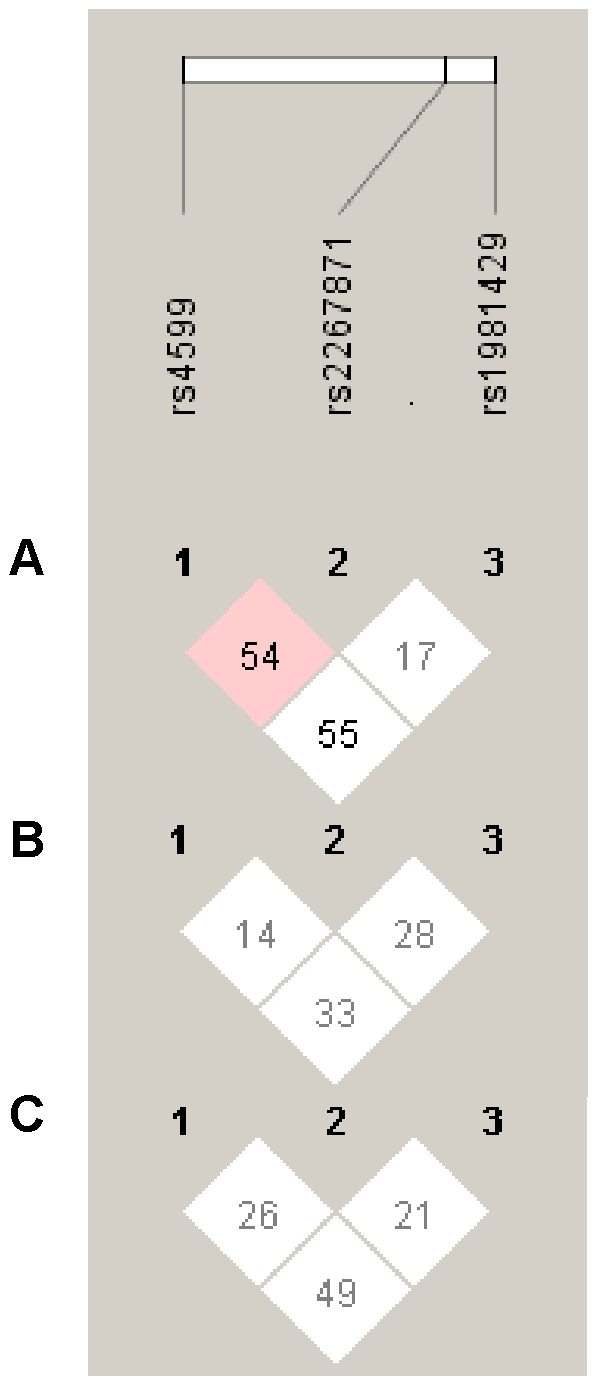
Plots generated using Haploview v3.2 [89]. The color code on the plots follows the Haploview Standard Color D'/LOD Scheme: shades of pink/red (D'<1; LOD ≥2); white (D'<1; LOD<2). The numbers represent D' values expressed as a percentage. (A) European Americans. (B) African Americans. (C) Hispanic Americans.
We next examined whether the SNPs were independently associated with each trait. To account for the confounding effects of population stratification, we used estimates of genetic admixture as a covariate in statistical models (see Materials and Methods below). Age, gender, Tanner status, and appropriate potential confounding variables were also included in the analysis as covariates (see Table 3 for details). We found that SNP rs4599 was significantly associated with REE (the conservative Bonferroni corrected significance threshold is P = 0.0038) and nominally associated with fasting glucose levels and sleep duration (Table 3). On average, children homozygous for the C allele had 4% lower levels of glucose, 8% higher REE, and slept 7% less than children homozygous for the T allele. The association with sleep duration was also observed with SNP rs2267871 (Table 3). In this case, the P value was smaller which suggests that SNP rs2267871 is the site with the largest association with the true causal polymorphism.
Table 3. Least square means (95% lower and upper confidence intervals) for body composition, metabolic, and sleep traits of study subjects stratified according to SDC4 polymorphisms.
| P* | ||||
| rs4599 | C/C | T/C | T/T | |
| n | 16 | 96 | 140 | |
| BMI | 18.94 (17.66–20.30) | 18.57 (18.03–19.13) | 18.16 (17.72–18.61) | 0.126a, 0.156d, 0.347r |
| Weight (kg) | 37.24 (34.8–39.84) | 36.26 (35.25–37.3) | 35.59 (34.47–36.43) | 0.164, 0.228, 0.305 |
| TFM (kg) | 7.74 (6.09–9.83) | 7.63 (6.92–8.42) | 7.32 (6.75–7.94) | 0.534, 0.526, 0.788 |
| LTM (kg) | 26.22 (25.14–27.34) | 25.74 (25.3–26.18) | 25.38 (25.02–25.75) | 0.087, 0.131, 0.232 |
| IAAT (cm2) | 30.78 (24.97–37.94) | 27.85 (25.49–30.42) | 28.85 (26.74–31.14) | 0.919, 0.778, 0.441 |
| SI [×10−4 min−1/(µU/mL)] | 4.65 (3.65–5.91) | 5.28 (4.80–5.80) | 4.70 (4.35–5.09) | 0.274, 0.112, 0.619 |
| Fasting Glucose (mg/dL) | 93.21 (90.02–96.40) | 97.53 (96.27–98.79) | 97.57 (96.53–98.60) | 0.104, 0.463, 0.01 |
| REE (kcal/day) | 1240 (1132–1358) | 1217 (1176–1261) | 1135 (1103–1167) | 0.002, 0.001, 0.242 |
| Hours of sleep/night | 8.35 (7.85–8.87) | 8.83 (8.61–9.06) | 8.95 (8.77–9.13) | 0.056, 0.187, 0.044 |
BMI: body mass index; TFM: total fat mass; LTM: lean tissue mass; IAAT: Intra-abdominal adipose tissue; SI: Insulin sensitivity index; REE: resting energy metabolism. Sex, age, Tanner stage, and African and European genetic admixture were used as covariates in all the analyses. Weight, TFM, and LTM were also adjusted for height. IATT, SI, fasting glucose, and REE were also adjusted for TFM. *P values represent the significance of the comparison among genotypes.
, d, and
indicate P values calculated assuming additive, dominant, and recessive models, respectively. P values <0.05 are highlighted in bold case.
Nominal associations were also observed between alternative alleles at SNP rs1981429 and variation in lean tissue mass and intra-abdominal adipose tissue (Table 3), with individuals homozygous for the G allele having ∼3% less lean mass and 6% more intra-abdominal fat than those homozygous for the T allele.
Finally, we found a nominal association between alternative alleles at rs2267871 and variation in insulin sensitivity (Table 3). On average, children homozygous for the A allele were 15% more insulin sensitive than heterozygous children.
Discussion
The syndecans are a family of cell-surface heparan sulphate proteoglycans that are expressed on the surface of all adherent cells [12]. In this study, we used quantitative complementation tests with two mutations of dSdc (Sdc10608 and SdcBG01305) to show that variation in this gene affects inter-individual variability in TAG storage between two strains of D. melanogaster. We then confirmed the effect of dSdc on TAG storage using a hypomorphic dSdc mutant (SdcBG02774) and showed that flies homozygous for the mutation had significantly lower TAG storage than control flies under ad libitum feeding conditions. They also displayed significantly reduced metabolic rate and lower mitochondrial ADP-stimulated (state 3) respiration rate. Furthermore, female mutants had lower body weight, were leaner, had reduced glycogen levels, and were less fecund than controls. DILPs produced by IPCs in the brain of Drosophila have been shown to regulate body size, energy metabolism, and fecundity in several studies [29]–[31], [34], [45]. For example, Grönke et al. [34] reported that dilp2-3,5 mutant flies had significantly reduced body weight, higher levels of lipids and glycogen, and were less fecund than controls. Another study found that flies homozygous for a deletion of dilps1-5 not only were smaller and less fertile but also displayed reduced metabolic activity and TAG storage than controls [30]. Recent work has also suggested that activation of the insulin signaling in the fat body of adult flies leads to increased TAG storage via activation of shaggy, the fly ortholog of glycogen synthase kinase 3 [46]. In our study we observed that female SdcBG02774 flies had significantly lower expression of dilp2-3,5 genes compared to controls. A significant reduction in dilp2 expression was also observed in males. Thus, these data suggest that some of the effects associated with reduced dSdc function may be mediated by the reduced levels of DILPs. Further studies however will be required to determine not only if reduced expression of DILPs is causal in these effects, but also to understand how dSdc regulates the expression levels of dilps in Drosophila. We also observed that female mutants had significantly higher expression levels of Akh, which encodes a hormone involved in mobilization of glycogen reserve from the fat body [33], [47]. This finding could offer a plausible explanation for why female SdcBG02774 flies have lower levels of glycogen than controls.
Reduced insulin receptor/PI3-kinase/Akt signaling pathway results in nuclear accumulation of the transcription factor dFoxO and transcriptional activation of its targets, such as Thor [36]. However, despite reduced DILPs we observed a significant reduction in Thor expression in SdcBG02774 flies. This result is quite surprising considering that previous work reported that Thor transcript levels are up-regulated in dilp2-3,5 mutants [34]. A possible explanation for our findings is that dSdc mutant flies have a defect in the mechanism(s) involved in dFoxO translocation and transcriptional regulation. This hypothesis is supported by the observation that, despite reduced dilp2 transcript levels, male dSdc mutant flies did not show a compensatory increase in dilp3 mRNA as a result of the reduced insulin signaling in the IPCs, an autocrine regulatory mechanism proposed by previous studies [34], [48]. These same studies have also shown that dilp3 transcript in the IPCs was significantly reduced in dFoxO null flies [48] and suggested that dilp3 acts as a positive regulator of dilp2 and dilp5 expression [34]. Hence, the hypothesis of a defect in dFoxo function may help explain the reduced levels of dilp2-3,5 observed in female SdcBG02774 flies. In this context, another unanticipated finding in our study is that reduced expression of dilps did not lead to extended life span and increased starvation resistance as observed in several other studies [29], [34], [41]–[43]. On the contrary, homozygous SdcBG02774 flies showed significantly reduced survival. The mechanism behind this finding is unknown at present. However, it has been previously reported that Thor null flies had an impaired response to nutrient deprivation and shorter lifespan than controls [49], [50]. Based on these observations, it is tempting to speculate that the reduced levels of Thor transcript observed in the SdcBG02774 flies could be responsible for these phenotypes. Experiments are currently underway in our laboratory to test this hypothesis.
In mammals, PGC-1α and PGC-1β have been shown to play a pivotal role in the control of energy homeostasis. They regulate glucose and fat oxidation in muscle and fat tissue [37] as well as gluconeogenesis in liver [38] and glucose-regulated insulin secretion in pancreatic β cells [38]. In addition, they play an essential role in mitochondria biogenesis and function [39]. In Drosophila, a recent study suggested that Spargel, the fly homologue of PGC-1, is required for the stimulation of mitochondrial respiration, but not biogenesis [40]. spargel gene expression is induced by insulin signaling via a pathway parallel to dFOXO [40]. Consistent with these observations and the reduced levels of DILPs, we observed a significant decrease in the expression of spargel in SdcBG0277 4 mutant flies compared to controls. SdcBG0277 4 flies also showed a significant reduction in mitochondrial state-3 respiration rate, suggesting reducing mitochondrial oxidative phosphorylation capacity. In mammals, skeletal muscle metabolism influences whole-body metabolic rate [51], we thus speculate that dSdc might control Drosophila whole-body metabolic rate via the insulin signaling in skeletal muscle, which in turn regulates spargel expression and mitochondrial respiration.
In the present study we also found that dSdc mutant flies slept longer than controls. This increased sleep length is due largely to increases in the average duration of sleep bouts rather than their number (data not shown). Previous microarray analyses of fly heads revealed three genes with predicted functions in lipid metabolism that increased expression during sleep [52]. Furthermore, P-element insertions in metabolic pathway genes impacted sleep duration and bout number [53]. Like these recent studies, our results demonstrate a molecular link between energy stores and sleep. Though the nature of that link has yet to be elucidated, we hypothesize that the increased sleep in SdcBG02774 mutants in combination with reduced TAGs may be indicative of a strategy to conserve energy, an idea long postulated as a possible function of sleep [54]. This hypothesis is also supported by the significant reduction in metabolic rate of dSdc mutant flies.
One of the interesting findings of this component of the study was the sex-specific nature of the effects of SdcBG02774 on several phenotypes. Sex-specific effects on TAG storage were also observed in our initial complementation tests with different mutant alleles of dSdc. Such sex-specific allelic effects are commonly observed in Drosophila [55]–[57] and highlight the fact that genetic influences on phenotypes are sensitive to environmental conditions (in this case, differences in the internal physiological environment between sexes). As dSdc can act to modulate cell signaling, alternative alleles could be highly sensitive to different levels of signaling molecules that naturally occur between the sexes. Along these same lines, in the comparison of the control CS (B) flies, the baseline expression of dSdc was much higher in control females compared with the control males. Thus, we would expect to observe greater phenotypic effects of the mutation in females compared with males and this is what we observed in most traits. Understanding the mechanism underlying these sex specific allelic effects is an important goal for future study.
The results of the human study parallel the results of the experiments in flies. Overall, we observed that genetic variation in the human SDC4 gene is associated with REE, sleep duration, and insulin dynamics in children. These results may shed light on physiological pathways underlying disease-related phenotypes in humans, perhaps by mechanisms that involve REE and/or sleep duration. Low REE has previously been associated with obesity- and diabetes-related traits in adults [58]–[60] and children [61]. Population differences in REE have been found, but the underlying reasons for these differences have not been elucidated. The role of genetics has been suggested in some studies [62]. Absolute levels of energy expenditure in humans are positively correlated with the amount of lean mass. However, individuals with higher levels of African genetic admixture tend to have lower levels of REE after adjustment for lean mass, suggesting a mechanism beyond simple energy balance [63]. This observation of lower REE is supported by research demonstrating a lower muscle oxidative capacity in AA women, suggesting that although they have higher levels of lean mass, the efficiency of fuel utilization that impacts energy expenditure is reduced [63]. In the current study, the SDC4 genetic associations with REE and lean mass highlight the inter-relationship between these two traits. Previous studies reported that syndecan-4 is essential in skeletal muscle development and regeneration [64]. Moreover, syndecan-4 has been shown to play a direct role in the regulation of focal adhesion kinase phosphorylation [65], which in turn mediates the insulin sensitivity of skeletal muscle cells [66]. Thus, it is plausible that syndecan-4 is involved in energy regulation via an insulin-signaling mechanism that impacts overall body metabolism. This idea is corroborated by our Drosophila data.
Another point of potential interest is the relationship between the SDC4 polymorphisms and sleep duration in early pubertal children. Previous reports have demonstrated that “sleep need” for this age group is approximately 9 hours per night [67], [68], and that a reduction in sleeping hours is a risk factor for obesity, due to its postulated effect on energy balance. Interestingly, the extent to which short sleep impacts obesity risk factors appears to be greater in children than adults [69]. Although a number of mechanisms have been proposed [67], some of which have a genetic basis [70], [71], no mechanism has been clearly identified to be in the causal pathway linking sleep and obesity. Given the strong similarity in results in Drosophila and humans, further exploration of the relationship between genes of the syndecan family, energy metabolism, and sleep is warranted.
Materials and Methods
Drosophila study
Fly stocks
The unrelated isogenic laboratory lines 2b and Ore were used to establish the recombinant inbred lines in which QTL affecting TAG were previously mapped [10]. Mutant stocks were obtained from the Bloomington Drosophila Stock Center and from Trudy Mackay at North Carolina State University. The SdcBG01305 and SdcBG02774 lines were established by the Berkeley Drosophila Gene Disruption (BDGD) Project via P-element insertion into the second intron of the dSdc gene in the w1118;Canton S (CS) strain [26]. Sdc10608 line is a P-element mutation that was established by the BDGD Project and maintained over a balancer (Bal) chromosome [72].
Experimental Design
To control for larval density, we allowed the parents of the experimental flies to mate for 3 hours to generate egg collections on apple juice/agar medium in laying plates. After 24 hours, we picked groups of 100 first-instar larvae from the surface of the medium and put into replicate vials. To minimize the influence of genetic variation in reproduction on energy metabolism, we performed all the metabolic assays on 3–5 day old virgin flies that were randomly collected from the replicate vials. We reared flies in vials containing 10 ml of standard cornmeal, agar, sugar, and yeast medium at a constant temperature of 25°C, 60–75% relative humidity, and a 12-hr light-dark cycle.
Quantitative complementation tests with dSdc mutations
We performed quantitative complementation tests by crossing virgin Ore and 2b female flies to males of dSdc mutation stocks. These crosses produced four F1 genotypes: M/2b, Bal or CS/2b, M/Ore, and Bal or CS/Ore, where M denotes the Sdc mutation. We measured TAG storage in each genotypic class for each sex using the same experimental design described in [7]. We analyzed quantitative complementation test data for each sex separately, using the two-way mixed model factorial analysis of covariance (ANCOVA): y = µ + L + G + W + L×G + E, where L and G are the fixed cross-classified main effects of line (Ore, 2b) and genotype (M, Bal or CS), W is the covariate body weight, and E is the within-vial variance. We inferred significant failure of the mutation to complement quantitative TAG phenotypes of Ore and 2b alleles if the main effect of the L×G interaction term was significant, the contrast [M/Ore – M/2b]) was significant, and the contrast [Bal or CS/Ore – Bal or CS/2b] was not significant [73].
Body weight, TAG, protein, and glycogen measurements
We measured body weight, TAG and total protein levels using the protocol described in [7]. Briefly, groups of 10 single-sexed individuals were weighed to 0.01 mg accuracy with an analytical balance and homogenized in ice-cold KH2PO4 buffer. TAG content was measured for each homogenate spectrophotometrically using a commercially available kit (Sigma-Triglyceride Assay Kit) following the manufacturer's suggested protocol. Total protein levels were measured using a standard Lowry protein assay. Glycogen content was measured from the same homogenates using the protocol described in [74]. Briefly, aliquots of 1.67 µl of homogenate were added to 250 µl of a reagent containing 0.1 U/ml of amyloglucosidase. After 30-minute incubation period at 37°C, OD450 was measured. Concentration of glycogen was determined from glucose and glycogen standards run with each replicate. Each sample was assayed twice and the mean was used in the analysis. Analysis of variance (ANOVA) was used to determine statistical significance between mutant and control flies in body weight. Statistical differences in TAG, proteins, and glycogen were assessed by ANCOVA, with body weight used as covariate.
Female fecundity
We estimated female fecundity by standard procedures [75] using 20 females per genotype. Females were placed in egg laying chambers containing standard fly food and fecundity measured by counting the total number of eggs laid over a five day period. Statistical significance was determined by one-way ANOVA.
Metabolic rate
We measured metabolic rate as CO2 production using a flow-through respirometry system (Qubit System Research, Kingston, Ontario, Canada) and a modification of the method described in [76]. CO2 was measured for 10 minutes/chamber with a 30 second flush period between measurements. The amount of CO2 produced by each group of flies was calculated using C950 Data Acquisition software (Qubit System Research, Kingston, Ontario, Canada). Each group of flies was sampled at the same time of the day. The data analysis was performed using ANCOVA, with body weight used as covariate.
Sleep and waking activity
We maintained adult virgins at 30 flies to a single-sex vial to ensure that each line was exposed to identical levels of social [77] and had equal access to food. Sleep parameters for each fly were measured with the Drosophila Activity Monitoring System (Trikinetics, Waltham, MA), which counts the number of times a given fly crosses an infrared beam during a specified time interval. Here, we used one-minute intervals to record activity counts. Seven continuous days of sleep and activity were recorded for each experimental block. Sleep was defined as any period 5 minutes or longer without an activity count [78]. An in-house C++ program was used to calculate duration of sleep in minutes, numbers of sleep bouts, average sleep bout duration in minutes, and the number of activity counts per waking minute-waking activity. We used Wilcoxon T test to assess statistical significance between mutant and control in sleep and waking activity.
Survival assay
To examine the effects of the dSdc mutation on the lifespan of virgin flies, we measured survival using population cages. To obtain virgin offspring for each population cage, we standardized the parental density of each genotype (10 pairs) and allowed them to lay eggs for five days. Virgin offspring were collected over four successive days and added to a population cage at the end of this period. We set up three population cages of each sex and genotype (initial population sizes ranged from 183–295) and removed and counted dead flies every other day. We used Cox regression [79] as implemented by SAS (V9.1.3) to compare survival of mutants and controls. We analyzed survival for each sex separately, with replicate cage used as a covariate.
Starvation assay
We measured survival under starvation conditions by placing 10 flies per genotype/sex on 1.5% agarose medium and recording the number of flies alive at 8-h intervals until all were dead. Statistical significance was determined by one-way ANOVA.
Mitochondrial respiration rate
We placed live flies into 200 µl of ice-cold isolation buffer [250 mM sucrose, 5 mM Tris-HCl, 2 mM EGTA, 1% (w/v) bovine serum albumin (BSA), pH 7.4 at 4°C] supplemented with protease inhibitors (leupeptin 1 mg/ml, aprotinin 1 mg/ml and pepstatin 1 mg.ml) in a 1.5 ml Eppendorf tube. The samples were pounded gently 126 times over a 2 minute period, using a motorized micromortar, and filtered through a 5 µm nylon mesh. We then raised the volume to 400 µl by washing the nylon membrane with additional isolation buffer. After a cycle of low-speed centrifugation followed by a centrifugation of the filtered solution for 10 min at 3000 g, at 4°C, the pellet was re-suspended in 100 µl of isolation buffer. Protein concentrations in the mitochondrial fractions were determined using a Lowry assay. We performed mitochondrial respiration assays using freshly isolated mitochondria by measuring oxygen consumption in a two-chamber polarographic oxygen sensor (Oroboros oxygraph, OROBOROS® INSTRUMENTS, Innsbruck, Austria). We measured state 2, state 3 and state 4 respiration rates using NAD+-linked substrates (5 mM pyruvate + 5 mM proline) as implemented in [80]. Data were analyzed using the software DatLab Version 4.1.0.8. Statistical significance was determined by the two-tailed Student's t test.
Quantitative RT-PCR
We isolated total RNA using the TriPure RNA isolation kit (Roche). Isolated RNA was then used to make cDNA, using the First Strand Synthesis kit (Invitrogen). We performed RT-qPCR using a Syber Green Master mix and 50 ng total of cDNA per reaction and run in a Stratagene Mx3000P® qPCR machine. Statistical significance was determined by the two-tailed Student's t test.
Human study
Subjects
A total of 252 children between the ages of 7 and 12 (52% male) were evaluated for the human association study. Subjects were participants from an ongoing study conducted at the University of Alabama at Birmingham. Race was determined by self-reported African-American, Caucasian or Hispanic ancestry in both parents and grandparents.
The study protocol was approved by the Institutional Review Board for human studies at the University of Alabama at Birmingham. A written informed consent was obtained from all study participants before enrolling in the study.
Phenotype measurements
Height and body weight in human subjects were measured in light indoor clothes and without shoes. Body composition (total fat mass and lean tissue mass) was measured by DXA using either a Lunar DPX-L densitometer (LUNAR Radiation Corp., Madison, WI) or a LUNAR Prodigy densitometer in the Department of Nutrition Sciences at UAB. Subjects were scanned in light clothing while lying flat on their backs with arms at their sides. Intra-abdominal adipose tissue was analyzed in the Department of Radiology by computed tomography scanning with a HiLight/Advantage Scanner (General Electric, Milwaukee).
Following an overnight fast, blood samples were obtained to establish the basal levels of glucose and insulin, and a frequently sampled intravenous glucose tolerance test (FSIGTT) was performed as described elsewhere [81]–[83]. Insulin sensitivity index (the increase in fractional glucose disappearance per unit of insulin increase) was estimated from the FSIGTT using minimal modeling [84].
REE was measured in the morning immediately after awakening during the overnight visit. A computerized, open-circuit, indirect calorimetry system with a ventilated canopy (Delta Trac II; Sensor Medics, Yorba Linda, CA) was used. While lying supine on a bed, the head of the subject was enclosed in a plexiglass canopy. Subjects were instructed not to sleep and remain quiet and still, breathing normally. One-minute average intervals of oxygen (VO2) uptake and VCO2 production were measured continuously for thirty minutes.
Sleep patterns were assessed by a questionnaire that was administered to the parents of the subjects. Parents were inquired about the amount of hours their child spent sleeping at night. We used hours sleeping at night as continuous phenotype.
Genotyping
We determined the genotypes of each SDC4 polymorphism by Pyrosequencing technology [85] at the NORC Genetics Core at UAB. The genetic admixture estimates were obtained from the genotyping of ancestry informative markers across the human genome. Genotyping for the measures of genetic admixture was performed at Prevention Genetics (www.preventiongenetics.org). Approximately 100 ancestry informative markers (AIMs) were utilized for the study. Information regarding marker sequences, experimental details, and parental population allele frequencies has been submitted to dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) under the handle PSU-ANTH [86], [87]. Individual West African, Amerindian, and European genetic admixture estimates were obtained by maximum likelihood approaches as reported in [88]. Briefly, the individual's genotypes at each AIM and the estimated allele frequencies of the AIMs in the three ancestral parental populations were used to estimate the admixture proportion that corresponds to the maximum combined probability across all loci for West African, Amerindian and European proportions. These estimates were used as covariates in all statistical models to control for population stratification.
Statistical analyses
We assessed Hardy-Weinberg equilibrium, allelic frequencies, and D' linkage disequilibrium coefficients using Haploview v3.2 [89]. We performed genotype frequency comparisons between EA, AA, and HA samples by ANOVA. To test the effect of each genotyped SNP on trait variation, we performed genotypic associations for dominant, additive, and recessive models using linear regression analysis. Dummy variables were assigned to code the three genotypes in each model. In the additive model, we used 0, 1 and 2 to code for individuals homozygous for the major allele, heterozygous, and homozygous for the minor allele, respectively. In the dominant and recessive models, we used 0 to code for individuals homozygous for the major and minor alleles, respectively, and 1 to code for individuals carrying at least one copy of the other allele. For all regression models, studentized residuals were evaluated for normality and logarithmic transformations of the dependent variable was performed to improve normality. When normality of the residuals was not obtained after transformations, the observations that were above and below three standard deviations were removed from the analyses. Analyses were performed using PLINK [90] and SAS 9.1 software (SAS Institute, Cary, NC).
Acknowledgments
We thank Kerry H. Lok for his technical support in SNP genotyping. We thank Tashauna Felix and Lucas Horn for help with the survival experiments and Hir Dalwadi, Anne Logie and Cynthia O'Rourke for help with the fecundity assay. We are grateful to Carlos Krumdieck for the designing and making of the motorized micro-mortar used for the mitochondrial assays and Douglas Moellering for technical assistance with the mitochondrial assays.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by U.S. National Institutes of Health Grants P30-DK56336 (Nutrition Obesity Research Center), P60-DK0797626 (Diabetes Research and Training Center's Bioanalytical Redox Biology Core), R01HL80812 (to MD and JL), R01DK084219 (to MD and JL), and R01DK067426 (to JF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Korner J, Woods SC, Woodworth KA. Regulation of energy homeostasis and health consequences in obesity. Am J Med. 2009;122:S12–S18. doi: 10.1016/j.amjmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303:235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 3.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, et al. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 4.Serdula MK, Ivery D, Coates RJ, Freedman DS, Williamson DF, et al. Do obese children become obese adults? A review of the literature. Prev Med. 1993;22:167–177. doi: 10.1006/pmed.1993.1014. [DOI] [PubMed] [Google Scholar]
- 5.Silventoinen K, Rokholm B, Kaprio J, Sorensen TI. The genetic and environmental influences on childhood obesity: a systematic review of twin and adoption studies. Int J Obes. 2010;34:29–40. doi: 10.1038/ijo.2009.177. [DOI] [PubMed] [Google Scholar]
- 6.Comuzzie AG, Allison DB. The search for human obesity genes. Science. 1998;280:1374–1377. doi: 10.1126/science.280.5368.1374. [DOI] [PubMed] [Google Scholar]
- 7.De Luca M, Chambers MM, Casazza K, Lok KH, Hunter GR, et al. Genetic variation in a member of the laminin gene family affects variation in body composition in Drosophila and humans. BMC Genet. 2008;9:52. doi: 10.1186/1471-2156-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pospisilik JA, Schramek D, Schnidar H, Cronin SJ, Nehme NT, et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140:148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Seo J, Fortuno ES, 3rd, Suh JM, Stenesen D, Tang W, et al. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Luca M, Yi N, Allison DB, Leips J, Ruden DM. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes Res. 2005;13:1596–1605. doi: 10.1038/oby.2005.196. [DOI] [PubMed] [Google Scholar]
- 11.Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tkachenko E, Rhodes JM, Simons M. Syndecans: new kids on the signaling block. Circ Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 13.Woods A. Syndecans: transmembrane modulators of adhesion and matrix assembly. J Clin Invest. 2001;107:935–941. doi: 10.1172/JCI12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Klass C, Woods A. Syndecan-2 regulates transforming growth factor-beta signaling. J Biol Chem. 2004;279:15715–15718. doi: 10.1074/jbc.C300430200. [DOI] [PubMed] [Google Scholar]
- 15.Couchman JR, Chen L, Woods A. Syndecans and cell adhesion. Int Rev Cytol. 2001;207:113–150. doi: 10.1016/s0074-7696(01)07004-8. [DOI] [PubMed] [Google Scholar]
- 16.Kolset SO, Salmivirta M. Cell surface heparan sulfate proteoglycans and lipoprotein metabolism. Cell Mol Life Sci. 1999;56:857–870. doi: 10.1007/s000180050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotte M. Syndecans in inflammation. FASEB. 2003;J.17:575–591. doi: 10.1096/fj.02-0739rev. [DOI] [PubMed] [Google Scholar]
- 18.Rostand KS, Esko JD. Microbial adherence to and invasion through proteoglycans. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spring J, Paine-Saunders SE, Hynes RO, Bernfield M. Drosophila syndecan: conservation of a cell-surface heparan sulfate proteoglycan. Proc Natl Acad Sci U S A. 1994;91:3334–3338. doi: 10.1073/pnas.91.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steigemann P, Molitor A, Fellert S, Jackle H, Vorbruggen G. Heparan sulfate proteoglycan syndecan promotes axonal and myotube guidance by slit/robo signaling. Curr Biol. 2004;14:225–230. doi: 10.1016/j.cub.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Kojima T, Inazawa J, Takamatsu J, Rosenberg RD, Saito H. Human ryudocan core protein: molecular cloning and characterization of the cDNA, and chromosomal localization of the gene. Biochem Biophys Res Commun. 1993;190:814–822. doi: 10.1006/bbrc.1993.1122. [DOI] [PubMed] [Google Scholar]
- 22.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, et al. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 23.Ji L, Malecki M, Warram JH, Yang Y, Rich SS, et al. New susceptibility locus for NIDDM is localized to human chromosome 20q. Diabetes. 1997;46:876–881. doi: 10.2337/diab.46.5.876. [DOI] [PubMed] [Google Scholar]
- 24.Zouali H, Hani EH, Philippi A, Vionnet N, Beckmann JS, et al. A susceptibility locus for early-onset non-insulin dependent (type 2) diabetes mellitus maps to chromosome 20q, proximal to the phosphoenolpyruvate carboxykinase gene. Hum Mol Genet. 1997;6:1401–1408. doi: 10.1093/hmg/6.9.1401. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S, Watanabe RM, Hauser ER, Valle T, Magnuson VL, et al. Type 2 diabetes: evidence for linkage on chromosome 20 in 716 Finnish affected sib pairs. Proc Natl Acad Sci U S A. 1999;96:2198–2203. doi: 10.1073/pnas.96.5.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Divers J, Vaughan LK, Padilla MA, Fernandez JR, Allison DB, et al. Correcting for measurement error in individual ancestry estimates in structured association tests. Genetics. 2007;176:1823–1833. doi: 10.1534/genetics.107.075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 29.Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Liu J, Li CR, Momen B, Kohanski RA, et al. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci U S A. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, et al. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 32.Leopold P, Perrimon N. Drosophila and the genetics of the internal milieu. Nature. 2007;450:186–188. doi: 10.1038/nature06286. [DOI] [PubMed] [Google Scholar]
- 33.Buch S, Melcher C, Bauer M, Katzenberger J, Pankratz MJ. Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab. 2008;7:321–332. doi: 10.1016/j.cmet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 36.Miron M, Verdú J, Lachance PE, Birnbaum MJ, Lasko PF, et al. The translational inhibitor 4E-BP is an effector of PI3K/Akt signalling and cell growth in Drosophila. Nat Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- 37.Goto M, Terada S, Kato M, Katoh M, Yokozeki T, et al. cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 38.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 39.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, et al. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiefenbock SK, Baltzer C, Egli NA, Frei C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 2010;29:171–183. doi: 10.1038/emboj.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 42.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 43.Bauer JH, Chang C, Morris SN, Hozier S, Andersen S, et al. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci U S A. 2007;104:13355–13360. doi: 10.1073/pnas.0706121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, et al. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet. 2009;41:371–375. doi: 10.1038/ng.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 46.DiAngelo JR, Birnbaum MJ. Regulation of Fat Cell Mass by Insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broughton S, Alic N, Slack C, Bass T, Ikeya T, et al. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS ONE. 2008;3:e3721. doi: 10.1371/journal.pone.0003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev. 2005;19:1840–1843. doi: 10.1101/gad.1311805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zurlo F, Larson K, Bogardus C, Ravussin E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest. 1990;86:1423–1427. doi: 10.1172/JCI114857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 53.Harbison ST, Sehgal A. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics. 2008;178:2341–2360. doi: 10.1534/genetics.107.081232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zepelin H. Mammalian sleep. In: Kryger MH, Roth T, Dement WC, editors. Principles and practice of sleep medicine. Philadelphia: Saunders; 1994. pp. 69–80. [Google Scholar]
- 55.Nuzhdin SV, Pasyukova EG, Dilda CL, Zeng ZB, Mackay TF. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1997;94:9734–9739. doi: 10.1073/pnas.94.18.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Luca M, Roshina NV, Geiger-Thornsberry GL, Lyman RF, Pasyukova EG, et al. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat Genet. 2003;34:429–433. doi: 10.1038/ng1218. [DOI] [PubMed] [Google Scholar]
- 57.Cho I, Horn L, Felix TM, Foster L, Gregory G, Starz-Gaiano M, et al. Age- and Diet-Specific Effects of Variation at S6 Kinase on Life History, Metabolic, and Immune Response Traits in Drosophila melanogaster. DNA Cell Biol. 2010 doi: 10.1089/dna.2009.0997. 2010 May 22. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinsier RL, Hunter GR, Desmond RA, Byrne NM, Zuckerman PA, et al. Free-living activity energy expenditure in women successful and unsuccessful at maintaining a normal body weight. Am J Clin Nutr. 2002;75:499–504. doi: 10.1093/ajcn/75.3.499. [DOI] [PubMed] [Google Scholar]
- 59.Weinsier RL, Hunter GR, Zuckerman PA, Redden DT, Darnell BE, et al. Energy expenditure and free-living physical activity in black and white women: comparison before and after weight loss. Am J Clin Nutr. 2000;71:1138–1146. doi: 10.1093/ajcn/71.5.1138. [DOI] [PubMed] [Google Scholar]
- 60.Hunter GR, Weinsier RL, Darnell BE, Zuckerman PA, Goran MI. Racial differences in energy expenditure and aerobic fitness in premenopausal women. Am J Clin Nutr. 2000;71:500–506. doi: 10.1093/ajcn/71.2.500. [DOI] [PubMed] [Google Scholar]
- 61.Ten S, Bhangoo A, Ramchandani N, Mueller C, Vogiatzi M, et al. Resting energy expenditure in insulin resistance falls with decompensation of insulin secretion in obese children. J Pediatr Endocrinol Metab. 2008;21:359–367. doi: 10.1515/JPEM.2008.21.4.359. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, et al. Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res. 2003;11:904–911. doi: 10.1038/oby.2003.124. [DOI] [PubMed] [Google Scholar]
- 63.Roy JL, Hunter GR, Fernandez JR, McCarthy JP, Larson-Meyer DE, et al. Cardiovascular factors explain genetic background differences in VO2max. Am J Hum Biol. 2006;18:454–460. doi: 10.1002/ajhb.20509. [DOI] [PubMed] [Google Scholar]
- 64.Cornelison DD, Wilcox-Adelman SA, Goetinck PF, Rauvala H, Rapraeger AC, et al. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004;18:2231–2236. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilcox-Adelman SA, Denhez F, Goetinck PF. Syndecan-4 modulates focal adhesion kinase phosphorylation. J Biol Chem. 2002;277:32970–32977. doi: 10.1074/jbc.M201283200. [DOI] [PubMed] [Google Scholar]
- 66.Bisht B, Goel HL, Dey CS. Focal adhesion kinase regulates insulin resistance in skeletal muscle. Diabetologia. 2007;50:1058–1069. doi: 10.1007/s00125-007-0591-6. [DOI] [PubMed] [Google Scholar]
- 67.Carskadon MA, Acebo C. Regulation of sleepiness in adolescents: update, insights, and speculation. Sleep. 2002;25:606–614. doi: 10.1093/sleep/25.6.606. [DOI] [PubMed] [Google Scholar]
- 68.Knutson KL, Van CE. Associations between sleep loss and increased risk of obesity and diabetes. Ann N Y Acad Sci. 2008;1129:287–304. doi: 10.1196/annals.1417.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel SR, Hu FB. Short sleep duration and weight gain: a systematic review. Obesity (Silver Spring) 2008;16:643–653. doi: 10.1038/oby.2007.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bray MS, Young ME. Circadian rhythms in the development of obesity: potential role for the circadian clock within the adipocyte. Obes Rev. 2007;8:169–181. doi: 10.1111/j.1467-789X.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- 71.Bray MS, Young ME. The role of cell-specific circadian clocks in metabolism and disease. Obes Rev. 2009;2:6–13. doi: 10.1111/j.1467-789X.2009.00684.x. [DOI] [PubMed] [Google Scholar]
- 72.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, et al. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasyukova EG, Vieira C, Mackay TF. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics. 2000;156:1129–1146. doi: 10.1093/genetics/156.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clark AG, Keith LE. Variation among extracted lines of Drosophila melanogaster in triacylglycerol and carbohydrate storage. Genetics. 1988;119:595–607. doi: 10.1093/genetics/119.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leips J, Gilligan P, Mackay TF. Quantitative trait loci with age-specific effects on fecundity in Drosophila melanogaster. Genetics. 2006;172:1595–1605. doi: 10.1534/genetics.105.048520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Voorhies WA, Khazaeli AA, Curtsinger JW. Testing the “rate of living” model: further evidence that longevity and metabolic rate are not inversely correlated in Drosophila melanogaster. J Appl Physiol. 2004;97:1915–1922. doi: 10.1152/japplphysiol.00505.2004. [DOI] [PubMed] [Google Scholar]
- 77.Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Waking experience affects sleep need in Drosophila. Science. 2006;313:1775–1781. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 78.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, et al. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 79.Cox DR, Oakes D. London: Chapman and Hall; 1984. Analysis of Survival Data. [Google Scholar]
- 80.Ferguson M, Mockett RJ, Shen Y, Orr WC, Sohal RS. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem J. 2005;390:501–511. doi: 10.1042/BJ20042130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matthews DR, Edge JA, Dunger DB. An unbiased glucose clamp method using a variable insulin infusion: its application in diabetic adolescents. Diabet Med. 1990;7:246–251. doi: 10.1111/j.1464-5491.1990.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 82.Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986;23:113–122. doi: 10.1016/0169-2607(86)90106-9. [DOI] [PubMed] [Google Scholar]
- 83.Casazza K, Phadke RP, Fernandez JR, Watanabe RM, Goran MI, et al. Obesity Attenuates the Contribution of African Admixture to the Insulin Secretory Profile in Peripubertal Children: A Longitudinal Analysis. Obesity (Silver Spring) 2009;17:1313–1325. doi: 10.1038/oby.2008.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergman RN, Ider YZ, Bowden CR, Cobelli C. Quantitative estimation of insulin sensitivity. Am J Physiol. 1979;236:E667–E677. doi: 10.1152/ajpendo.1979.236.6.E667. [DOI] [PubMed] [Google Scholar]
- 85.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 86.Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 87.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 88.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 89.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 90.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]