Abstract
Aromatase converts androgens to estrogens. Although third-generation aromatase inhibitors (AIs) are important drugs in hormonal therapy for breast cancer in postmenopausal women, there are concerns about the side effects associated with the estrogen deprivation achieved with AIs. Expression of aromatase in breast cancer tissue is driven by different promoters than those in noncancer tissues; thus, suppression of aromatase expression in cancer tissues through the down-regulation of breast tumor–specific promoters would reduce the side effects associated with whole-body suppression of estrogen biosynthesis by AIs. We report that histone deacetylase inhibitor LBH589 (panobinostat) is a potent inhibitor of aromatase expression (with an IC50 value < 25 nM). LBH589 selectively suppresses human aromatase gene promoters I.3/II, which are preferentially used in breast cancer tissue. Furthermore, using the H295R cell culture model, we found that achieving the same degree of inhibition of aromatase activity required only one-fifth as much letrozole (an AI) in the presence of 25 nM LBH589 as in the absence of LBH589. We also used an H295R/MCF7 coculture model to demonstrate the synergistic interaction of LBH589 + letrozole in suppressing the proliferation of hormone-responsive breast cancer cells. Finally, our results also indicate that LBH589 down-regulates the activity of promoters I.3/II in an epigenetic fashion. LBH589 reduces the levels of C/EBPδ, decreases the binding of C/EBPδ, and increases the levels and binding of acetyl-histones to the promoters I.3/II. These findings provide an important basis for future clinical evaluations of LBH589 in hormone-dependent breast cancer.
A major strategy for treatment of hormone-dependent breast cancers is the suppression of estrogen receptor (ER) action that can be achieved by antiestrogens or aromatase inhibitors (AIs). Aromatase is the enzyme that catalyzes the final step of estrogen synthesis. This enzyme is expressed at higher levels in breast cancer tissue than in normal breast tissues (1–3). The estrogen produced in situ due to the overexpression of aromatase in breast cancer cells is thought to play a more crucial role than circulating estrogen in stimulating cancer cell growth (4). The increased efficacy of AIs compared with antiestrogen (tamoxifen) therapy has recently been demonstrated by clinical trials showing a significant increase in disease-free survival using three third-generation AIs (5–7). These three FDA-approved third-generation AIs—two nonsteroidal derivatives [anastrozole (Arimidex) and letrozole (Femara)] and one steroidal derivative [exemestane (Aromasin)] are now widely used as first-line drugs in the endocrine treatment of estrogen-dependent breast cancer in postmenopausal patients. Anastrozole and letrozole act as competitive inhibitors with respect to the androgen substrates. Exemestane is a mechanism-based inhibitor that is catalytically converted into a chemically reactive species, leading to irreversible inactivation of aromatase, as well as degradation of aromatase protein (8).
AIs are thought to be of value in treating estrogen-dependent breast cancer, especially in postmenopausal women. In these women, estrogens are produced mainly in peripheral adipose tissues and in cancer cells, and peripheral aromatase is not under gonadotropin regulation (9). In premenopausal women, luteinizing hormone and follicle-stimulating hormone stimulate the synthesis of aromatase in ovaries and may counteract the effects of AIs.
Although AI therapy for hormone-dependent breast cancer in postmenopausal women has been shown to be effective in the clinic, some patients demonstrate resistance to these endocrine therapies. In addition, AI treatment is a “whole-body” treatment, and significant side effects associated with estrogen depletion have been reported (e.g., refs. 10 and 11). In response to the recognition of the side effects and resistance associated with AI treatment, several laboratories, including ours, have been searching for methods to selectively suppress aromatase level/expression in breast tumors. We were one of the three research groups that cloned human aromatase cDNA (12). The human aromatase gene contains nine translated exons (II–X) and at least 10 tissue-specific untranslated exon Is (I.1, I.2, 2a, I.3, I.4, I.5, I.6, I.7, I.f, and PII). The various exon Is are present at different levels in the different aromatase-expressing tissues and cells (13–15). The specific promoter is located immediately upstream of the corresponding exon I, and each promoter is regulated by different mechanisms. Studies conducted in our laboratory and other laboratories have revealed that exons I.3 and PII are the major exon Is in aromatase mRNA isolated from breast cancer tissue, indicating that aromatase expression in breast cancer is driven mainly by promoters I.3 and II (which are ∼200 bp apart from each other) (1, 14, 16, 17). In normal breast stromal cells and bone tissue, promoter I.4 is the major promoter driving aromatase expression (14, 17). Thus, finding a way to selectively suppress promoters I.3/II, but not promoter I.4, would be valuable. Such a treatment would have fewer side effects than the AI treatment. In a recent breakthrough, we found that the histone deacetylase (HDAC) inhibitor LBH589 (panobinostat) can selectively suppress promoters I.3/II at nM ranges. We believe that these exciting preclinical results will help design new treatment strategies for hormone-dependent breast cancer.
There are three major classes of HDAC (18). Class I and class II HDACs have structural homology to yeast RPD3 and HDA1, respectively. Both of these HDAC classes require zinc for catalytic activity and are inhibited by compounds such as trichostatin A (TSA) and suberoyl anilide hydroxamic acid (SAHA, or vorinostat). Class III HDACs include sirtuins, which have homology to yeast Sir2 and are not inhibited by such compounds as TSA or SAHA. HDAC6 belongs to class II but it is unique in that it has two catalytic sites, and thus is classified as class IIa. Although HDAC inhibitors are recognized as relatively nonspecific agents, they have been shown to be useful in treating several types of cancer. They are thought to be more effective in inhibiting the proliferation of cancer cells compared with normal cells. Cancer cells have been shown to have more multiple defects than normal cells, and to be less tolerant to the inhibition of one or more prosurvival factors or activation of a prodeath pathway (19).
HDAC6 has been shown to enhance oncogenic transformation (20) and to modulate epithelial-mesenchymal transition in cancer cells (21). Thus, selective inhibitors of HDAC6 are thought to be useful for cancer therapy (22). The HDAC6 inhibitor LBH589 induces expression of DNA damage response genes and apoptosis in Ph− acute lymphoblastic leukemia cells (23). This inhibitor is well tolerated and induces clinical responses in cutaneous T cell lymphoma patients (24). HDAC6 also has been identified as a major deacetylase of a-tubulin as well as heat shock protein (Hsp) 90 (25). In addition, HDAC6 can interact via its zinc finger with ubiquitin to modulate aggresome function and autophagy (26). Microarray analyses of tumor samples indicate that LBH589 induces rapid changes in gene expression, and, surprisingly, more genes are repressed than are activated (27, 28). A unique set of genes that can mediate biological responses, such as apoptosis, immune regulation, and angiogenesis, are commonly regulated in response to LBH589. Ellis et al. (29) reported that damage to the mitochondria is an important event required for LBH589 to mediate tumor cell death and a robust therapeutic response.
For breast cancer, Elsheikh et al. (30) reported a highly significant correlation among histone modifications status, tumor biomarker phenotype, and clinical outcome, with high relative levels of global histone acetylation and methylation associated with favorable prognosis and detected almost exclusively (93%) in luminal-like breast tumors (i.e., ER-positive tumors). As an HDAC6 inhibitor, LBH589 also acetylates Hsp90, leading to the degradation of Hsp90 client proteins, including ER (31), ErbB2 (32), and other key survival signaling proteins (33). The effect of LBH589 on ER expression has not been defined precisely, however. LBH589 also has been reported to reactivate ER expression in ER-negative cells (34). Furthermore, inhibition of HDACs is thought to promote ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells (35).
LBH589 was developed by Atadja (36). Recently, LBH589 was further demonstrated to deplete members of the polycomb repressive complex 2 (EZH2, SUZ12, and EED proteins) and DNMT1 in acute myeloid leukemia cells (37).
Results
LBH589 Suppresses Aromatase Expression Through Promoters I.3/II.
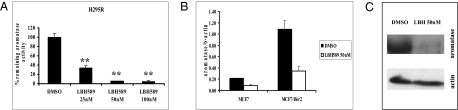
We performed two sets of experiments to demonstrate the selective inhibitory effect of LBH589 against promoters I.3/II of the human aromatase gene. From studies in our laboratory and other laboratories, we have learned that aromatase expression in the following cell lines is driven by different promoters: JAR (using promoter I.1), SKBR3 (using promoter I.1), H295R (using promoters I.3/II), HepG2 (using promoter I.4), and MCF-7aro (generated by transfection using a β-actin promoter–containing plasmid). When treated with LBH589, only aromatase activity in H295R cells was suppressed in a dose-dependent manner with an IC50 value < 25 nM (Fig. 1A). H295R is an aromatase-positive human adrenocortical carcinoma cell line (38). Aromatase activity in other cell lines was not suppressed at these concentrations (Fig. S1). As confirmed by Western blot analysis (Fig. 1C), LBH589-induced suppression of aromatase activity in H295R cells was due to a decrease in aromatase protein expression. Western blot analysis also revealed that LBH589 treatment of H295R cells led to the degradation of both ERα and ERβ proteins, similar to the findings of Fiskus et al. (31).
Fig. 1.
Suppression of aromatase activity/expression in H295R cells by LBH589. (A) H295R cells were treated with LBH589 for 24 h. After treatment, the cells were washed with PBS, and aromatase activity was measured by the [3H] H2O release assay. Student's t test was used for statistical analysis compared with the vehicle control. **P < 0.001. (B) MCF7 and MCF7/Her2 cells were treated with LBH589 (50 nM) or with DMSO as a control for 24 h. After treatment, total RNA was isolated; 5 μg of total RNA was used for real-time qPCR for quantifying aromatase gene expression. β-actin mRNA was amplified as an internal control. All samples were run in triplicate, and SDs were calculated. (C) After treatment with LBH589 (50 nM) for 24 h, H295R cells were lysed and applied for Western blot analysis using aromatase antiserum.
Previous experiments from our laboratory and other laboratories have demonstrated that aromatase expression in MCF7 and MCF7Her2 cells is low, but is driven through promoters I.3/II (17, 39). Real time RT-PCR demonstrated that LBH589 treatment reduced aromatase expression at the transcriptional level in these two cell lines (Fig. 1B). Exon I–specific RT-PCR revealed that LBH589 treatment decreased the levels of exons I.3/II containing mRNA in three cell lines (Fig. S2), confirming that this drug suppresses the activity of promoters I.3/II.
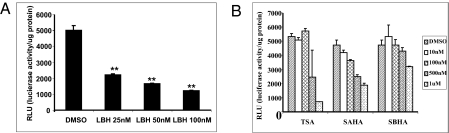
Our laboratory also generated a HeLa cell line stably transfected with a luciferase reporter containing promoters I.3/II. LBH589 was found to suppress luciferase reporter activity (Fig. 2A) with potency similar to its ability to suppress aromatase activity in H295R cells. These results reinforce the finding that LBH589 is very effective in suppressing the activity of human aromatase promoters I.3/II. Interestingly, LBH589 was not able to suppress the luciferase reporter activity when the cells were transiently transfected with the same reporter plasmid, suggesting that the reporter plasmid is integrated into the chromosome in the stably transfected HeLa cell line, and that LBH589 down-regulates the activity of promoters I.3/II involving an epigenomic mechanism. Along with LBH589, we also examined the effects of three other HDAC inhibitors: TSA, SAHA, and suberoyl bishydroxamic acid (SBHA). In agreement with the results of the aromatase activity assay in H295R cells, these three inhibitors effectively inhibited the promoters I.3/II–mediated luciferase reporter assay in stably transfected HeLa cells only at μM ranges (Fig. 2B). These results demonstrate that LBH589 is a potent inhibitor that suppresses aromatase expression through promoters I.3/II.
Fig. 2.
Suppression of promoters I.3/II luciferase reporter activity by HDAC inhibitors. (A) Promoters I.3/II stably transfected HeLa cells were treated with LBH589 at indicated concentrations for 24 h. Cells were then lysed and harvested, and luciferase activity was determined and normalized with protein concentration in triplicate for each treatment condition. Student's t test was used for statistical analysis compared with the vehicle control. **P < 0.001. (B) Promoters I.3/II stably transfected HeLa cells were treated with TSA, SAHA, or SBHA at the indicated concentrations for 24 h. Cells were lysed and harvested. The luciferase activity was determined and normalized with protein concentration.
LBH589 Suppresses C/EBPδ-Mediated Aromatase Expression.
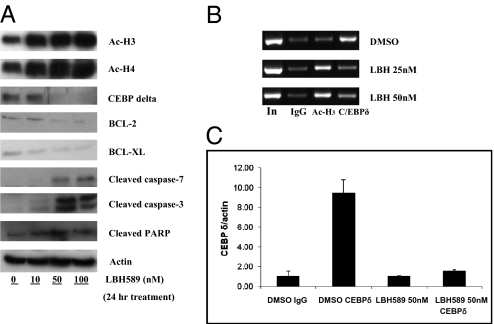
To better design treatment strategies incorporating LBH589, we investigated the mechanism of LBH589-mediated suppression of promoters I.3/II activity. Our laboratory has extensively studied the regulatory mechanisms of aromatase expression in breast cancer cells (40–43). Recently, through in vivo footprinting analysis, we identified an important regulatory site at which C/EBPδ binds and up-regulates breast cancer-specific aromatase promoters I.3/II in breast cancer epithelial cells (44). Our review of the literature on HDAC inhibitors revealed that adiponectin gene expression can be suppressed by the HDAC inhibitor valproic acid, which decreases C/EBPα levels and lessens binding of C/EBPα to the adiponectin promoter (45). Our analysis found that LBH589 decreased C/EBPδ protein levels (Fig. 3A) and reduced the binding of C/EBPδ to promoters I.3/II of the human aromatase gene. This reduced binding of C/EBPδ to the aromatase promoter was demonstrated by ChIP analysis and quantitative ChIP analysis (Fig. 3B and C). Our experiments revealed that LBH589-mediated degradation of C/EBPδ is an important mechanism for inhibiting aromatase expression through the suppression of promoters I.3/II. Treatment with an Hsp90 inhibitor, 17-DMAG, was not able to decrease C/EBPδ levels, suggesting that the LBH589-induced degradation of C/EBPδ is not mediated through acetylation of Hsp90. As expected, LBH589 treatment also increased the levels of acetyl-histone H3 and acetyl-histone H4 and enhanced their binding to the aromatase promoter (Fig. 3A and B), a mechanism that could be critical for the suppression of binding of C/EBPδ to its responsive element. LBH589 treatment had no affect on the level of C/EBPδ mRNA, indicating that the decreased C/EBPδ level was not caused by inhibition of its transcription. This treatment induced apoptosis, as demonstrated by the decreased BCL-2 and BCL-XL levels and increased cleaved caspase-3, cleaved caspase-7, and cleaved PARP levels (Fig. 3A).
Fig. 3.
Effects of LBH589 on the expression of key proteins. (A) Western blot analyses were performed with H295R cells that had treated with LBH589 (50 nM) for 24 h. After treatment, cells were lysed and harvested, and cell lysates were analyzed by immunoblotting with indicated antibodies. (B) ChIP analysis was performed to determine binding of C/EBPδ to the aromatase promoter. H295R cells were treated with LBH589 (25 nM or 50 nM) for 24 h. Cells were harvested after cross-linking with 1% formaldehyde. Immunoprecipitation was conducted with normal rabbit IgG, anti–acetyl-histone H3, and anti-C/EBPδ antibody. PCR was performed using primers to amplify the C/EBPδ-binding region of promoters I.3/II. (C) Quantitive ChIP analysis was performed to quantify the binding of C/EBPδ to the aromatase promoter. H295R cells were treated with LBH589 (50 nM) for 24 h. The cells were then harvested after cross-linking with 1% formaldehyde. Real-time qPCR was performed using primers to amplify the C/EBPδ-binding region of promoters I.3/II.
LBH589 and Letrozole Synergistically Inhibit Aromatase-Positive Breast Cancer Cell Proliferation.
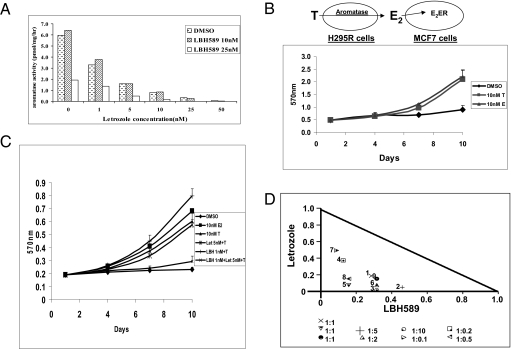
Our experiments have demonstrated that LBH589 suppresses aromatase expression through promoters I.3/II, an inhibitory mechanism different from that of letrozole, which inhibits aromatase activity. Consequently, we examined the combined effect of LBH589 and letrozole. In the presence of 25 nM LBH589, we could achieve the same degree of inhibition of aromatase activity in H295R cells with letrozole at concentrations only one-fifth of those used in the absence of LBH589 (Fig. 4A). As a proof-of-principle study, these results suggest that it is possible to reduce the side effects of letrozole or LBH589 by using lower levels of letrozole plus LBH589.
Fig. 4.
(A) Synergistic inhibitory effect of LBH589 and letrozole on aromatase activity in H295R cells. The cells were treated with LBH589 and letrozole at indicated concentrations for 24 h. After treatment, the cells were washed with PBS, and aromatase activity was measured using the [3H] H2O release assay. (B) Proliferation assay of H295R/MCF7 coculture model. MCF-7 and H295R cells were cocultured in 96-well plates with hormone-deprived media at a concentration of 1 × 103 and 5 × 102 per well, respectively. The next day, hormone-deprived media containing 10 nM T or E2 was added to the cells to induce proliferation, with DMSO as a vehicle control. Cells were cultured for 9 d, and media were replaced every 72 h. Cell viability was assessed by the MTT assay and measured at 570 nm on a SpectraMax M5 microplate reader. Three replicates were performed for each measurement, and the mean and SD were calculated. (C) Combined effect of LBH589 and letrozole on the H295R/MCF7 coculture model. With the conditions described above, cells were cultured for 9 d with hormones with or without the two drugs. Three replicates were performed for each measurement, and the mean and SD were calculated. (D) Isobologram analysis of the combined effect of LBH589 and letrozole on the H295R/MCF7 coculture model. The cells were cultured with the two drugs at different concentrations at conditions described above. Normalized isobolograms were produced by CalcuSyn software. Values below the threshold line represent synergistic combination.
A cell culture model is essential to further confirm the beneficial effect of LBH589. We recently developed an H295R/MCF7 coculture model in which the aromatase in H295R cells can convert androgen to estrogen, which drives the proliferation of ER-positive MCF7 cells. As shown in Fig. 4B, both testosterone (T) and 17β-estradiol (E2) (at 10 nM) were able to stimulate the proliferation of these cells. As in the controls, the proliferation of MCF7 could be stimulated only by estrogen, and the proliferation of H295R cells was not affected by either androgen or estrogen. We also found that letrozole and LBH589 could suppress the proliferation of H295R/MCF7 in a dose-dependent manner. More importantly, as shown in Fig. 4C, neither 5 nM letrozole nor 1 nM LBH589 alone was able to reduce the androgen-mediated cell proliferation; rather, a combination of these drugs was required to effectively suppress this proliferation. The synergistic reduction of cell proliferation when the two inhibitors were used in different combinations was further demonstrated by isobologram analysis (Fig. 4D). These results confirm that this H295R/MCF7 coculture model will be very useful for evaluating the combined effect of LBH589 and letrozole. Furthermore, our results indicate that LBH589 and letrozole are able to suppress the proliferation of ER-positive and aromatase-positive breast cancer cells in a synergistic manner.
Discussion
AIs have been demonstrated to be superior to tamoxifen in terms of disease progression, incidences of locoregional and distant relapses, and contralateral breast cancers. Although these findings are exciting, there are concerns about the side effects associated with the use of AIs and the possibility of developing resistance to AI treatment. A selective suppression of aromatase expression/estrogen biosynthesis in breast cancer tissue through the down-regulation of breast tumor–specific promoters would be an approach to reduce the side effects associated with whole-body reduction of estrogen (through the use of AIs) and to delay the time to progression (i.e., resistance). After many attempts at searching for such drugs, we recently found that LBH589 is a potent inhibitor of aromatase expression (with an IC50 value < 25 nM; LBH589 is 40 times more potent than SAHA in its ability to suppress aromatase expression) through the suppression of human aromatase gene promoters I.3/II, the promoters selectively used in breast cancer tissue.
In numerous in vitro and preclinical studies, HDAC inhibitors have demonstrated their vast potential as single-agent anticancer therapies; unfortunately, equivalent responses have not always been observed in patients, however (46). Given the nonselective action of HDAC inhibitors on malignant cells, these agents’ true therapeutic potential most likely lies in combination with other anticancer drugs. For instance, the synergy of LBH589 plus doxorubicin in acute myeloid leukemia has been demonstrated (47). Furthermore, as in any new antitumor therapy, development of resistance to HDAC inhibitors can occur (48). In preclinical studies, resistance to HDAC inhibitor–induced transformed cell death has been observed in human bladder carcinoma cells (T24) and prostate cancer cells (PC3) (49–51). HDAC inhibitors induce a very broad, pleiotropic anticancer drug resistance phenotype in acute myeloid leukemia cells through modulation of multiple ABC transporter genes (52). Given these findings, and based on our results, we hypothesize that the combined use of letrozole and LBH589 can increase the specificity and delay the development of resistance to either drug. It is our belief that this combined approach will produce critical results for the development of new treatment strategies with fewer side effects, delayed time to progression (i.e., resistance), and improved effectiveness of AIs.
An H295R/MCF7 coculture model has been developed in our laboratory as a direct way to study the effect of the drugs on aromatase expression that is driven by breast cancer–selective promoters. At the present time, we do not have a breast cancer cell line that is ER-positive, aromatase-positive and aromatase exons I.3/II-mRNA positive. This newly developed coculture model will be very important to evaluate antiaromatase expression drugs on breast cancer cell proliferation.
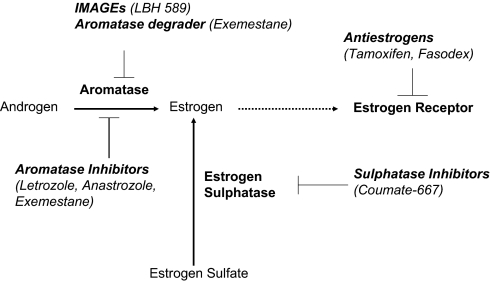
Currently, two major types of drugs (antiestrogens and AIs) are used to treat hormone-dependent breast cancer (Fig. 5). The proliferation of hormone-dependent breast cancer also can be suppressed by sulphatase inhibitors (53) and aromatase degraders (8). We now report that LBH589 can suppress aromatase promoters I.3/II in a selective and potent manner. Our findings indicate that LBH589 has little effect on aromatase expression in non–breast cancer cells and bone. It represents a class of drugs termed “selective aromatase modulators” (SAMs) or, more appropriately, “inhibitory modulators of aromatase gene expression” (IMAGE). LBH589 is currently in use in clinical trials; thus, our findings can be translated as soon as preclinical studies are completed. Combination therapy with LBH589 and letrozole is thought to be a way to increase selectivity at the target tissue (ER-positive/aromatase-positive breast tumor), reduce the side effects of these drugs, and delay endocrine resistance/recurrence.
Fig. 5.
Different treatment strategies for hormone-dependent breast cancer.
Materials and Methods
Reagents.
LBH589 and letrozole were provided by Novartis Pharmaceuticals. SAHA, SBHA, and TSA were purchased from Sigma-Aldrich.
Cell Culture.
MCF7, HeLa, HepG2, SK-BR-3, and H295R cells were purchased from American Type Culture Collection. MCF-7, HeLa, and HepG2 cells were maintained in Eagle's MEM containing 2 mM L-glutamine, 1× penicillin-streptomycin, 1× nonessential amino acid, 1 mM sodium pyruvate, and 10% FBS. SK-BR-3 cells were maintained in McCoy's 5A medium containing 2 mM L-glutamine, 1× penicillin-streptomycin, 1× nonessential amino acid, 1 mM sodium pyruvate, and 15% FBS. H295R cells were maintained in DMEM/F-12 medium supplemented with 2.5 mM L-glutamine, 1 mM sodium pyruvate, 1× penicillin-streptomycin, 2.5% Nu-Serum, and ITS + Premix (BD Biosciences). MCF-7aro cells, aromatase-overexpressing MCF7 cells (54, 55), were maintained in the same medium used for MCF7 cells plus 100 μg/mL of G418. All of the cell lines were cultured at 37 °C with 5% CO2.
To perform the coculture experiments, the ERα-positive breast cancer cell line MCF7 and the aromatase-positive adrenal gland carcinoma cell line H295R cell line were used in proliferation experiments. MCF7 and H295R cells were cultured in MEM with Earle's salts supplemented with 10% charcoal dextran-treated FBS.
‘‘In-Cell’’ Aromatase Assay.
H295R cells were treated with LBH589 for 24 h at varying concentrations. Aromatase activity was measured in a [3H] H2O release assay as described previously (55). Tritiated [1β-3H (N)] androstenedione (final concentration, 100 nM) and 0.5 μL of 1 mM progesterone per mL of medium were mixed with FBS-free culture medium. The substrate, androst-4-ene-3,17-dione [1h-3H (N)] (specific activity, 24.7 Ci/mmol), was purchased from New England Nuclear.
Aromatase Promoter Analysis.
Plasmid preparation.
The genomic DNA fragment of human aromatase promoters I.3/II (−329/+284 bp) was subcloned into KpnI/XhoI sites of pGL4.14 containing firefly luciferase reporter gene. This construct is referred as to pGL4.14-firefly-pI.3/II. The pGL4 plasmid was purchased from Promega.
Stable transfection.
For stable transfection, 1 mg/mL of neomycin (Omega Scientific) was introduced to a pGL4.14-firefly-pI.3/II overexpressing HeLa cell line after 24 h of posttransfection incubation. After 4 wk of culture with the selection agent, individual cells formed colonies. Each clone was checked for luciferase activity using the Luciferase Reporter Assay System (Promega).
Reporter gene assay.
Cells were lysed in Passive Lysis Buffer (Promega) with a 15-min incubation on a shaker. For samples in 96-well plates, 400 μL of the lysis buffer was used, and 20 μL of each sample was transferred into new Eppendorf tubes. After 50 μL of Luciferase Assay Reagent (Promega) was added to the tubes, and firefly luciferase activity was immediately read manually in a luminometer (TD-20/20; Turner Designs). Protein concentration was measured using Bradford's method (56) (Bio-Rad). The relative luciferase activity was calculated by dividing the light unit of luciferase activity by the protein concentration of each sample.
RNA Isolation and Semiquantitative Exon I-Specific RT-PCR Analysis.
MCF7, MCF7/Her2, and H295R cells were treated with 50 nM LBH589 or DMSO as a control. After a 24-h incubation, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The reverse-transcription and aromatase exon I–specific PCR procedures have been described previously (57). As an internal control to normalize aromatase mRNA expression in each sample, a set of human β-actin–specific primers was used. Each PCR product was electrophoresed on 1.8% agarose gel and stained with ethidium bromide.
Real-Time Quantitative PCR.
Quantitative analyses of CYP19A1 and C/EBPδ mRNA expression levels were assessed using a real-time quantitative PCR (qPCR) technique. For this, 1 μL of each cDNA sample prepared from LBH589-treated MCF7, MCF7/Her2, and H295R cells was mixed in a 25-μL reaction volume with 1× iQ SYBR Green Supermix (Bio-Rad) and 0.2 μmol/L of each primer (CYP19A1: forward, 5′-AAATCCAGACTGTTATTGGTGAGAG-3′; reverse, 5′-GTAGCCATCGATTACATCATCTTCT-3′; C/EBPδ: forward, 5′-TCAACGATGCCCAAGAAATG-3′; reverse, 5′-CATTCCCAATTGAAAGCCAAA-3′). All real-time qPCR reactions were performed on an iCycler IQ5PCR machine (Bio-Rad). Each sample was analyzed in triplicate. The relative expression level of each gene was normalized by the β-actin expression level for each sample.
Chromatin Immunoprecipitation Assay.
ChIP assays were performed according to the manufacturer's protocol using a kit purchased from Upstate Biotechnology. H295R cells were treated with 50 nM LBH589 for 24 h. DNA samples were sonicated using Digital Sonifier (Branson). Immunoprecipitation was performed with normal rabbit IgG (Santa Cruz Biotechnology) or anti-C/EBPδ antibody (Santa Cruz Biotechnology) anti–Ac-H3 or anti–Ac-H4 antibodies or normal rabbit serum (control). DNA (1 μL), dissolved in 50 μL of TE, was subjected to PCR amplification in a 25-μL reaction mixture containing 0.5 units of HotstarTaq DNA polymerase (Qiagen), 1× PCR reaction buffer, 0.2 μM of forward/reverse primers, and 0.2 mM of deoxynucleotide triphosphate mix. Primers were designed to flank the C/EBPδ-binding site in aromatase promoters I.3/II (forward, 5′-TCAACGATGCCCAAGAAATG-3′; reverse, 5′-CATTCCCAATTGAAAGCCAAA-3′). The immunoprecipitated promoter DNA fragments were quantitated by real-time qPCR. The fold difference value in each assay compares the antibody-treated specific sample to the corresponding control sample (normal IgG).
Western Blot Analysis.
After the treatment with 50 nM LBH589 for 24 h, H295R cells were lysed and applied for Western blotting using anti-C/EBPδ (Santa Cruz Biotechnology), anti–Bcl-XL, anti–Bcl-2, anti–cleaved caspase 3, anti–cleaved caspase 7, and anti–poly[ADP ribose] polymerase antibodies (Cell Signaling), antiacetylated histone H3 and H4 antibodies (Upstate Biotechnology), as described previously (57). The blot was reprobed with antiactin antibody (Santa Cruz Biotechnology) as a loading control. The expression of aromatase, ERα, and ERβ in H295R cells was also analyzed using aromatase antiserum (generated in the Chen laboratory with functionally active recombinant aromatase protein as the antigen), anti-ERα (HC-20; Santa Cruz Biotechnology), and anti-ERβ antibodies (H-150; Santa Cruz Biotechnology), respectively.
Cell Proliferation Assay.
MCF7 and H295R cells were cocultured in 96-well plates with hormone-deprived media at a concentration of 1 × 103 and 5 × 102 per well. The next day, hormone-deprived media containing 10 nM T or E2 was added to the cells to induce proliferation. Vehicle control or compound was added simultaneously and left for 9 d. Media, E2, T, and inhibitors were renewed every 72 h. Cell viability was assessed by the MTT assay and measured at 570 nm on a SpectraMax M5 microplate reader (Molecular Devices). Three replicates were used for each measurement, and the mean and SD were calculated.
Statistical Analysis.
To assess statistical significance, values were compared with controls with either Student's t test or one-way ANOVA, followed by Dunnett's multiple range test (α = 0.05) using Prism GraphPad 4 software (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Dr. Dujin Zhou for her help in the Western blot analysis of ERα and ERβ expression in H295R cells before and after LBH589 treatment. This research was supported by Susan G. Komen for the Cure Grant KG080161 and National Institutes of Health Grant CA44735.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000917107/-/DCSupplemental.
References
- 1.Harada N. Aberrant expression of aromatase in breast cancer tissues. J Steroid Biochem Mol Biol. 1997;61:175–184. [PubMed] [Google Scholar]
- 2.Utsumi T, Harada N, Maruta M, Takagi Y. Presence of alternatively spliced transcripts of aromatase gene in human breast cancer. J Clin Endocrinol Metab. 1996;81:2344–2349. doi: 10.1210/jcem.81.6.8964875. [DOI] [PubMed] [Google Scholar]
- 3.Bulun SE, Price TM, Aitken J, Mahendroo MS, Simpson ER. A link between breast cancer and local estrogen biosynthesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J Clin Endocrinol Metab. 1993;77:1622–1628. doi: 10.1210/jcem.77.6.8117355. [DOI] [PubMed] [Google Scholar]
- 4.James VH, et al. Aromatase activity in normal breast and breast tumor tissues: In vivo and in vitro studies. Steroids. 1987;50:269–279. doi: 10.1016/0039-128x(83)90077-6. [DOI] [PubMed] [Google Scholar]
- 5.Coombes RC, et al. for the Intergroup Exemestane Study A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350:1081–1092. doi: 10.1056/NEJMoa040331. and erratum (2004) 355: 1746. [DOI] [PubMed] [Google Scholar]
- 6.Goss PE, et al. Randomized trial of letrozole following tamoxifen as extended adjuvant therapy in receptor-positive breast cancer: Updated findings from NCIC CTG MA.17. J Natl Cancer Inst. 2005;97:1262–1271. doi: 10.1093/jnci/dji250. [DOI] [PubMed] [Google Scholar]
- 7.Howell A, et al. ATAC Trialists’ Group Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365:60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Chen S. Aromatase destabilizer: Novel action of exemestane, a Food and Drug Administration–approved aromatase inhibitor. Cancer Res. 2006;66:10281–10286. doi: 10.1158/0008-5472.CAN-06-2134. [DOI] [PubMed] [Google Scholar]
- 9.Ackerman GE, Smith ME, Mendelson CR, MacDonald PC, Simpson ER. Aromatization of androstenedione by human adipose tissue stromal cells in monolayer culture. J Clin Endocrinol Metab. 1981;53:412–417. doi: 10.1210/jcem-53-2-412. [DOI] [PubMed] [Google Scholar]
- 10.Lester J, Coleman R. Bone loss and the aromatase inhibitors. Br J Cancer. 2005;93(Suppl 1):S16–S22. doi: 10.1038/sj.bjc.6602691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breast Cancer Action Side effects revisited: Women's experiences with aromatase inhibitors. [Accessed May 17, 2010];2008 Available at: http://bcaction.org/uploads/PDF/AIReport.pdf. [Google Scholar]
- 12.Chen SA, et al. Human aromatase: cDNA cloning, Southern blot analysis, and assignment of the gene to chromosome 15. DNA. 1988;7:27–38. doi: 10.1089/dna.1988.7.27. [DOI] [PubMed] [Google Scholar]
- 13.Bulun SE, et al. The human CYP19 (aromatase P450) gene: Update on physiologic roles and genomic organization of promoters. J Steroid Biochem Mol Biol. 2003;86:219–224. doi: 10.1016/s0960-0760(03)00359-5. [DOI] [PubMed] [Google Scholar]
- 14.Harada N, Utsumi T, Takagi Y. Tissue-specific expression of the human aromatase cytochrome P-450 gene by alternative use of multiple exons 1 and promoters, and switching of tissue-specific exons 1 in carcinogenesis. Proc Natl Acad Sci USA. 1993;90:11312–11316. doi: 10.1073/pnas.90.23.11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada N. A unique aromatase (P-450AROM) mRNA formed by alternative use of tissue-specific exons 1 in human skin fibroblasts. Biochem Biophys Res Commun. 1992;189:1001–1007. doi: 10.1016/0006-291x(92)92303-f. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal VR, Bulun SE, Leitch M, Rohrich R, Simpson ER. Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J Clin Endocrinol Metab. 1996;81:3843–3849. doi: 10.1210/jcem.81.11.8923826. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, et al. Aromatase gene expression and its exon I usage in human breast tumors: Detection of aromatase messenger RNA by reverse transcription-polymerase chain reaction. J Steroid Biochem Mol Biol. 1996;59:163–171. doi: 10.1016/s0960-0760(96)00100-8. [DOI] [PubMed] [Google Scholar]
- 18.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: Overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 19.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan B, et al. Requirement of HDAC6 for transforming growth factor-beta1–induced epithelial-mesenchymal transition. J Biol Chem. 2008;283:21065–21073. doi: 10.1074/jbc.M802786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrump DS. Cytotoxicity mediated by histone deacetylase inhibitors in cancer cells: Mechanisms and potential clinical implication. Clin Cancer Res. 2009;15:3948–3957. doi: 10.1158/1078-0432.CCR-08-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scuto A, et al. The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph− acute lymphoblastic leukemia cells. Blood. 2008;111:5093–5100. doi: 10.1182/blood-2007-10-117762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis L, et al. Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin Cancer Res. 2008;14:4500–4510. doi: 10.1158/1078-0432.CCR-07-4262. [DOI] [PubMed] [Google Scholar]
- 25.Banerji U. Heat shock protein 90 as a drug target: Some like it hot. Clin Cancer Res. 2009;15:9–14. doi: 10.1158/1078-0432.CCR-08-0132. [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 27.LaBonte MJ, et al. DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines. BMC Med Genomics. 2009;2:67. doi: 10.1186/1755-8794-2-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiso P, et al. The histone deacetylase inhibitor LBH589 is a potent antimyeloma agent that overcomes drug resistance. Cancer Res. 2006;66:5781–5789. doi: 10.1158/0008-5472.CAN-05-4186. [DOI] [PubMed] [Google Scholar]
- 29.Ellis L, et al. The histone deacetylase inhibitors LAQ824 and LBH589 do not require death receptor signaling or a functional apoptosome to mediate tumor cell death or therapeutic efficacy. Blood. 2009;114:380–393. doi: 10.1182/blood-2008-10-182758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elsheikh SE, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 31.Fiskus W, et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-α levels and transcriptional activity: A result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin Cancer Res. 2007;13:4882–4890. doi: 10.1158/1078-0432.CCR-06-3093. [DOI] [PubMed] [Google Scholar]
- 32.Chinnaiyan P, et al. Enhancing the antitumor activity of ErbB blockade with histone deacetylase (HDAC) inhibition. Int J Cancer. 2006;118:1041–1050. doi: 10.1002/ijc.21465. [DOI] [PubMed] [Google Scholar]
- 33.Edwards A, Li J, Atadja P, Bhalla K, Haura EB. Effect of the histone deacetylase inhibitor LBH589 against epidermal growth factor receptor–dependent human lung cancer cells. Mol Cancer Ther. 2007;6:2515–2524. doi: 10.1158/1535-7163.MCT-06-0761. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer Biol Ther. 2007;6:64–69. doi: 10.4161/cbt.6.1.3549. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res. 2008;6:873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atadja P. Development of the pan-DAC inhibitor panobinostat (LBH589): Successes and challenges. Cancer Lett. 2009;280:233–241. doi: 10.1016/j.canlet.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Fiskus W, et al. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006;5:3096–3104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- 38.Nicol MR, et al. Estrogen biosynthesis in human H295 adrenocortical carcinoma cells. Mol Cell Endocrinol. 2009;300:115–120. doi: 10.1016/j.mce.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subbaramaiah K, Norton L, Gerald W, Dannenberg AJ. Cyclooxygenase-2 is overexpressed in HER-2/neu–positive breast cancer: Evidence for involvement of AP-1 and PEA3. J Biol Chem. 2002;277:18649–18657. doi: 10.1074/jbc.M111415200. [DOI] [PubMed] [Google Scholar]
- 40.Yang C, Yu B, Zhou D, Chen S. Regulation of aromatase promoter activity in human breast tissue by nuclear receptors. Oncogene. 2002;21:2854–2863. doi: 10.1038/sj.onc.1205386. [DOI] [PubMed] [Google Scholar]
- 41.Yang C, Zhou D, Chen S. Modulation of aromatase expression in the breast tissue by ERR alpha-1 orphan receptor. Cancer Res. 1998;58:5695–5700. [PubMed] [Google Scholar]
- 42.Zhou D, Chen S. Identification and characterization of a cAMP-responsive element in the region upstream from promoter 1.3 of the human aromatase gene. Arch Biochem Biophys. 1999;371:179–190. doi: 10.1006/abbi.1999.1454. [DOI] [PubMed] [Google Scholar]
- 43.Okubo T, et al. Down-regulation of promoter 1.3 activity of the human aromatase gene in breast tissue by zinc-finger protein, snail (SnaH) Cancer Res. 2001;61:1338–1346. [PubMed] [Google Scholar]
- 44.Kijima I, Ye J, Glackin C, Chen S. CCAAT/Enhancer binding protein δ up-regulates aromatase promoters I.3/II activity in breast cancer epithelial cells. Cancer Res. 2008;68:4455–4464. doi: 10.1158/0008-5472.CAN-07-3249. [DOI] [PubMed] [Google Scholar]
- 45.Qiao L, Schaack J, Shao J. Suppression of adiponectin gene expression by histone deacetylase inhibitor valproic acid. Endocrinology. 2006;147:865–874. doi: 10.1210/en.2005-1030. [DOI] [PubMed] [Google Scholar]
- 46.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15:3970–3977. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 47.Maiso P, et al. The synergy of panobinostat plus doxorubicin in acute myeloid leukemia suggests a role for HDAC inhibitors in the control of DNA repair. Leukemia. 2009;23:2265–2274. doi: 10.1038/leu.2009.182. [DOI] [PubMed] [Google Scholar]
- 48.Fantin VR, Richon VM. Mechanisms of resistance to histone deacetylase inhibitors and their therapeutic implications. Clin Cancer Res. 2007;13:7237–7242. doi: 10.1158/1078-0432.CCR-07-2114. [DOI] [PubMed] [Google Scholar]
- 49.Butler LM, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci USA. 2002;99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu W, Ngo L, Perez G, Dokmanovic M, Marks PA. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proc Natl Acad Sci USA. 2006;103:15540–15545. doi: 10.1073/pnas.0607518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 52.Hauswald S, et al. Histone deacetylase inhibitors induce a very broad, pleiotropic anticancer drug resistance phenotype in acute myeloid leukemia cells by modulation of multiple ABC transporter genes. Clin Cancer Res. 2009;15:3706–3715. doi: 10.1158/1078-0432.CCR-08-2048. [DOI] [PubMed] [Google Scholar]
- 53.Foster PA, et al. A new therapeutic strategy against hormone-dependent breast cancer: the preclinical development of a dual aromatase and sulfatase inhibitor. Clin Cancer Res. 2008;14:6469–6477. doi: 10.1158/1078-0432.CCR-08-1027. [DOI] [PubMed] [Google Scholar]
- 54.Sun XZ, Zhou D, Chen S. Autocrine and paracrine actions of breast tumor aromatase: A three-dimensional cell culture study involving aromatase-transfected MCF-7 and T-47D cells. J Steroid Biochem Mol Biol. 1997;63:29–36. doi: 10.1016/s0960-0760(97)00068-x. [DOI] [PubMed] [Google Scholar]
- 55.Zhou DJ, Pompon D, Chen SA. Stable expression of human aromatase complementary DNA in mammalian cells: A useful system for aromatase inhibitor screening. Cancer Res. 1990;50:6949–6954. [PubMed] [Google Scholar]
- 56.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 57.Kijima I, Phung S, Hur G, Kwok SL, Chen S. Grape seed extract is an aromatase inhibitor and a suppressor of aromatase expression. Cancer Res. 2006;66:5960–5967. doi: 10.1158/0008-5472.CAN-06-0053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.