Abstract
We have demonstrated a novel sensing strategy employing single-stranded probe DNA, unmodified gold nanoparticles, and a positively charged, water-soluble conjugated polyelectrolyte to detect a broad range of targets including nucleic acid (DNA) sequences, proteins, small molecules, and inorganic ions. This nearly “universal” biosensor approach is based on the observation that, while the conjugated polyelectrolyte specifically inhibits the ability of single-stranded DNA to prevent the aggregation of gold-nanoparticles, no such inhibition is observed with double-stranded or otherwise “folded” DNA structures. Colorimetric assays employing this mechanism for the detection of hybridization are sensitive and convenient—picomolar concentrations of target DNA are readily detected with the naked eye, and the sensor works even when challenged with complex sample matrices such as blood serum. Likewise, by employing the binding-induced folding or association of aptamers we have generalized the approach to the specific and convenient detection of proteins, small molecules, and inorganic ions. Finally, this new biosensor approach is quite straightforward and can be completed in minutes without significant equipment or training overhead.
Keywords: biosensor, aptamer, visual detection, thrombin detection, cocaine detection
Gold nanoparticle colorimetric biosensors have seen significant applications in diagnostics, environmental monitoring, and antibioterrorism supporting unaided, visual readout (1–12). Commonly, the relevant nanoparticles are covalently modified with either a probe DNA or an aptamer such that hybridization (13–16) or aptamer-target interactions (17–27), for example the scanometric method developed by Mirkin (25), which is a very sensitive and specific tool, crosslink them, inducing aggregation. The second broad approach utilizes unmodified nanoparticles. (28–30) These two approaches, however, suffer from time-consuming (20–40 h of assembly) and relatively poor (low nanomolar) detection limits, respectively. Here, a unique, colorimetric sensing strategy employing a simple but selective combination of a single-stranded DNA probe, a positively charged, water-soluble conjugated polyelectrolyte, and unmodified gold nanoparticles is demonstrated. The universality of this method allows detection of a broad range of targets, including nucleic acid (DNA) sequences, proteins, small molecules, and inorganic ions. Our approach is rapid (turnaround time is 5–10 min) and sensitive (picomolar concentrations of target DNA are readily detected with the naked eye, even in complex sample matrices like blood serum). Hence, an operator with minimum scientific overhead can easily employ this technique.
Generally, the gold nanoparticle applications typically rely on a quantitative coupling between target recognition and the aggregation of the nanoparticles, which, in turn, leads to a dramatic change in the photonic properties—and thus the color—of the nanoparticle solution. This colorimetric “readout” avoids the relative complexity inherent in optical imaging/detection methodologies and thus may prove suitable for point-of-care and developing world applications (9–12). Examples for the covalently modified nanoparticles include the work of Mirkin and collaborators, who pioneered the use of gold nanoparticle-DNA conjugates (22, 23) for the sensitive detection of DNA and proteins (24–27). For the unmodified nanoparticles, Li and Rothberg have shown that, at high ionic strength, single-stranded—but not double-stranded—DNA protects unmodified gold nanoparticles from aggregating, thus modifying their color (28–30). Employing this mechanism, these authors have demonstrated a colorimetric assay for specific DNA sequences.
Our approach to the application of gold nanoparticles in optical biosensing relies on the recent observations that (i) both single-stranded DNA and double-stranded DNA prevent aggregation of gold nanoparticles at low salt concentrations (28–30); (ii) conjugated polyelectrolytes lead to the ready aggregation of such nanoparticles (31), and (iii) previous work has demonstrated that, at certain conditions, the cationic conjugated polyelectrolyte poly [(9,9-bis (6′-N,N,N-trimethylammonium) hexyl) fluorene-alt-1,4-phenylene] bromide (PFP-Br) binds single-stranded DNA preferentially to double-stranded or otherwise “folded” DNA, (32–34). This appears to arise due to the greater hydrophobicity of single-stranded DNA. Upon reducing the strength of the hydrophobic interactions, the electrostatic attraction becomes the important interaction that regulates the binding between the water-soluble conjugated polymer and DNA (33). The different affinities between the cationic conjugated polymer and various forms of DNA (single-stranded DNA and double-stranded DNA and single-stranded DNA and complex DNA folds) can be used to design a variety of biosensors (32–34).
Results
Inspired by the above, we have developed a unique colorimetric assay for the detection of nucleic acids, small-molecules, proteins, and inorganic ions that relies on unmodified gold nanoparticles but that employs the conjugated, positively charged, water-soluble polymer poly [(9,9-bis (6′-N,N,N-trimethylammonium) hexyl) fluorene-alt-1,4-phenylene] bromide). We have employed cationic conjugated polyelectrolytes to efficiently sequester single-stranded—but not double-stranded or otherwise “folded”—DNA from nanoparticles, leading to their aggregation and color change (Fig. S1) (32–34). We have then used this effect as the basis for an assay for the sensitive, colorimetric detection of a wide range of molecular analytes.
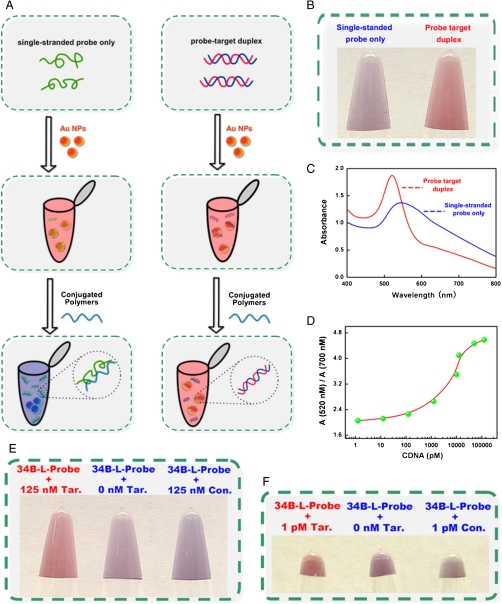
For the detection of DNA (Fig. 1A), we first prepare a control sample containing a single-stranded probe DNA and a test sample containing the probe DNA and its complementary DNA target. A solution of 20 nm gold nanoparticles is added to both, producing a readily apparent red color due to the intense surface plasmon resonance (SPR) absorption of the nanoparticles at 520 nm. The subsequent addition of the conjugated polyelectrolyte sequesters most of single-stranded DNA, leaving it unable to stabilize the nanoparticles against aggregation and thus leading to a characteristic blue color for the control sample (which only contains single-stranded DNA). In the presence of the complementary target sequence a significant concentration of double-stranded DNA is formed, which only weakly binds the conjugated polyelectrolytes and thus remains largely free to stabilize the nanoparticles against aggregation. This, in turn, causes the sample to retain the red color associated with dispersed nanoparticles (Fig. 1B).
Fig. 1.
Here we demonstrate a sensitive colorimetric assay for the detection of DNA. We find that, whereas a mixture of a positively charged, water-soluble, conjugated polyelectrolyte and single-stranded DNA leads to the aggregation of gold nanoparticles (and, consequently, a readily detectable change in their color), admixtures of this polymer with double-stranded DNA does not similarly produce a color change. (A) Here we have used this effect as the basis of a ready colorimetric assay for the detection of specific oligonucleotides. The assay is rapid and sensitive: (< 125 attomoles of target in 10 μL solution differentiates a color change observable with the naked eye in less than 10 min. (B) According to the scheme, the solution only contains a single-stranded probe that makes the gold nanoparticles aggregate (blue), while the probe–target duplex keeps the gold nanoparticles stable (red). (C) The ratio A520/A700 for a perfect match target is much larger than no target indicating the specificity of our assay. (D) By calculating the ratio A520/A700 form the UV-Vis test we could measure the target concentration, which is also crucial for detection. Shown is our ability to specifically detect target DNA molecules at both E, high (125 nM), and F, (1.25 pM) concentrations.
The color change associated with the lack of double-stranded DNA allows us to easily differentiate the single-stranded DNA probe and the double-stranded probe-target duplex via visual inspection at concentrations as low as 1.25 pM (Fig. 1 E and F), a detection limit comparable to those of many previously reported fluorescent DNA assays and enzyme-linked amplified assays (35). A more quantitative analysis can be made using UV-Vis spectroscopy. Specifically, the absorbance of the solution at 520 nm increases and the absorbance at 700 nm decreases when the concentration of double-stranded DNA is increased from 1.25 pM to 125 nM (Fig. 1 C and D). A series of control experiments confirm the specificity of the assay: As expected, duplexes between the 34-base pair probe and targets containing 3-, 5- or 7 base-pair mismatches do not stabilize the nanoparticles against aggregation, leading to the characteristic blue color (Fig. S2).
A key feature of our method is that, unlike approaches that utilize functionalized nanoparticles, hybridization and binding occur under conditions that can be independently regulated and optimized. For example, our approach relies on an ability to detect double-stranded probe–target duplexes. Thus, a potential disturbance is that any double-stranded DNA contaminating the sample will also stabilize the nanoparticles. Because we can optimize test conditions, the pretreatment of the sample with Exonuclease III, which provides a ready and convenient solution degrading any double-stranded DNA initially present in the sample (Fig. S3). Thus our assay can be made quite specific. Likewise the assay is selective and performs well even when challenged directly in complex biological media, such as target-doped blood serum (Fig. S4).
Discussion
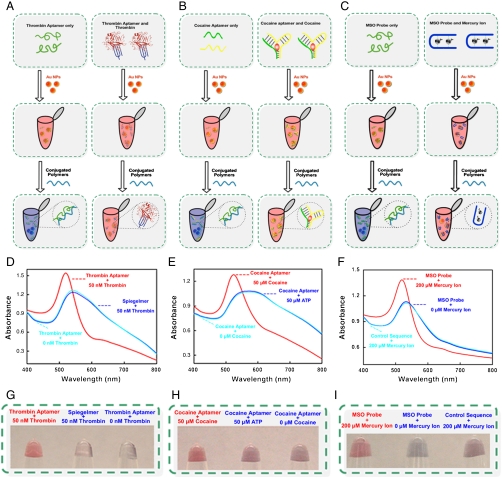
While the detection of DNA is of significant clinical utility in its own right, by employing aptamers, nucleic acid molecules that have been selected in vitro to bind to specific molecular targets (36–39), our colorimetric assay can also be expanded to the detection of nonnucleic acid targets. Specifically, because it is relatively easy to reengineer aptamers such that they undergo a transition from largely single-stranded to a folded, compact structure upon target binding (40–44), and because the conjugated polyelectrolyte only weakly binds with these compact, folded structures (31–34), we can adapt our assay to the detection of the aptamer’s binding partner (Fig. 2).
Fig. 2.
Our assay also could easily be expanded to the detection of proteins, small molecules, and ions. (A) A thrombin aptamer has been used in our protein detection. (B) We use a sandwich approach based on single aptamer sequences in our cocaine detection. (C) We use a mercury-responsive sequence that folds in the presence of Hg (II) to detect the Hg (II). (D) When the sample contains thrombin, it folded the aptamer into G-quadruplex structure together, which stabilized the gold nanoparticles remaining red. The sample containing control probe or no target induced the aggregation, which shifts the absorption from 520 nM to the longer wavelength, resulting in the characteristic red-blue color change. The detect limit for thrombin is 10 nM. (E) When the sample contains cocaine, it associate two part aptamer together, which stabilize the gold nanoparticles remaining red. The sample containing ATP or no target induced the aggregation, which shifts the absorption from 520 nM to the longer wavelength, resulting in the characteristic red-blue color change. The detect limit for cocaine is 10 μM. (F) The sample containing no target induced the aggregation, which shifts the absorption from 520 nM to the longer wavelength, resulting in the characteristic red-blue color change. The detect limit for Hg (II) is 50 μM. (G, H, and I) This color change allows us to easily differentiate between target-containing and target-free samples by visual inspection.
As a first example of the broader applicability of our approach, we developed an assay for the small molecule cocaine using the anticocaine aptamer of Stojanovic (39–42). To do so we modified the aptamer sequence slightly (see Methods) to prevent interstrand binding and then cut the sequence into two short, single-stranded pieces (40, 41). We then prepared two samples: a control sample containing only these fragments and a test sample containing the two aptamer fragments and cocaine. Upon adding a solution of ∼20 nm gold nanoparticles, both samples turn red due to the intense surface plasmon resonance absorption of the nanoparticles. Upon the addition of the conjugated polyelectrolyte, the nanoparticles in the control solution aggregate and the solution turns blue. The test sample, in contrast, retains its red color, presumably because cocaine binding drives the two pieces to associate (40–42) to form a well-folded, largely double-stranded aptamer that only weakly binds with the conjugated polyelectrolytes and thus remains free to inhibit nanoparticle aggregation (Fig. 2). This color change supports the ready detection of cocaine via visual inspection. As expected, this sensor does not respond when challenged with other small molecules, such as ATP (Fig. 2).
Abundant literature suggests that it is relatively easy to modify aptamers such that they only fold upon target binding (40–45), an effect that also supports our assay. As examples we have used Bock’s thrombin-binding aptamer, which folds from a single-stranded state to a largely G-quadruplex structure upon target binding (43, 44) and a mercury-responsive sequence that folds in the presence of Hg (II) (45–49). Once again, we employ control (aptamer-only) and test (aptamer-plus-target) solutions, which, upon mixing with ∼20 nm gold nanoparticles, produce the characteristic absorption at 520 nm. In the absence of target, both aptamers are single-stranded and thus do not stabilize the gold nanoparticles against aggregation in the presence of the polymer. Upon target binding, however, the aptamers fold, reducing their interactions with the conjugated polyelectrolytes and allowing them to prevent nanoparticle aggregation (Fig. 2). The resultant color change supports the easy differentiation between target-containing and target-free samples via visual inspection.
The strategy described in this report offers advantages over many existing sensing methodologies. First, our approach is likely universal; here, for example, we have shown that it is applicable to the detection of nucleic acids, small molecules, proteins, and inorganic ions. Second, our approach is convenient, requiring only the mixing of several solutions at room temperature to achieve rapid, semiquantitative detection via visual inspection or quantitative detection via visible light absorbance spectroscopy. Third, the detection of DNA using this approach is sensitive. We achieve, for example, low picomolar sensitivity for DNA detection, comparing very favorably with previously reported methods (Table 1) (28, 35, 50–52). Fourth, each step in the process (pretreatment with Exonuclease, the binding of the target to the DNA probe, the addition of conjugated polyelectrolytes) is performed separately, allowing each step to be optimized independently. Finally, the detection process is rapid and convenient.
Table 1.
Comparison of sensors for DNA detection
| Detection method | Strategy | Detection limit | Existing problems |
| Colorimetric (this work) | AuNP & conjugated polyelectrolyte | 1 pM (naked eyes) | Not applicable for colored samples |
| Colorimetric (28) | AuNP & salt | 4.3 nM (naked eyes) | Detect limit not applicable for colored samples |
| Colorimetric (50) | Enzyme amplification magnetic separation | 100 pM (naked eyes) | Multistep process, cumbersome preparation of functionalized gold nanoparticles and magnetic bead. Time-consuming (> 1 day), not applicable for colored samples |
| Electrochemistry (51) | Electron transfer | 10 pM | Dual-labeled oligos. Time-consuming (≈5 hours) |
| Electrochemistry (52) | Enzyme Amplification & PNA Probe & AgNP | 10 fM | Multistep process, stability of enzyme, dual label of DNA, reagent intensive. Time-consuming (≈5 hours) |
| Fluorescence (35) | Conjugated Polyelectrolyte FRET | 2.14 pM | Using expensive and complicated instruments for detection. Time-consuming (≈3 hours) |
Methods
In a typical gold nanoparticles/CP assay, the DNA probes were dissolved in a salt solution (112.5 mM NaCl) to form 1.25 μM probe solution at room temperature. Such solution (2.5 μL) was added to gold nanoparticles solution (25 μL), and the obtained solution was incubated for 1 min at room temperature. The concentrations of probes are 125 nM in this solution unless specially indicated. After that CP (5 μL, 1 μM) was added to the above solution, followed by either visual observation or UV/Vis characterization. The total volume of the final solutions in this work may differ, but the volume ratio of the above DNA, gold nanoparticles solution, and CPs is 1∶10∶2. In cocaine assays, the 3′ fragment and 5′ fragment were dissolved in salt solution (112.5 mM NaCl) to form 0.625 uM 3′ fragment and 0.625 uM 5′ fragment probe solution at room temperature. Then cocaine was added to the above solution. This solution (2.5 μL) was added to gold nanoparticles solution (25 μL), and the obtained solution was incubated for 2 min at room temperature. After that CP (5 μL, 1 μM) was added, followed by either visual observation or UV/Vis characterization. For the thrombin assay, the thrombin aptamer (or spiegelmer) was dissolved in a salt solution (112.5 mM NaCl) to form 1.25 μM probe solution at room temperature. Then thrombin was added followed by incubation for 20 min. After that CP (5 μL, 1 μM) was added, followed by either visual observation or UV/Vis characterization.
Supplementary Material
Acknowledgments.
Research supported by the Heeger Presidential Chair Funds, the National Science Foundation under NSF- DMR 0602280, Institute for Collaborative Biotechnologies through Grant DAAD19-03-D-0004 from the U.S. Army Research Office, and by the National Institutes of Health (EB007689-02). The authors thank Aaron Rowe, Minghong Tong, Hongbo Zeng, Hongwei Xia, Wei Lin and Zihang Xia.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005632107/-/DCSupplemental.
References
- 1.Thompson DG, Enright A, Faulds K, Smith WE, Graham D. Ultrasensitive DNA detection using oligonucleotide-silver nanoparticle conjugates. Anal Chem. 2008;80(8):2805–2810. doi: 10.1021/ac702403w. [DOI] [PubMed] [Google Scholar]
- 2.Cho M, Han MS, Ban C. Detection of mismatched DNAs via the binding affinity of MutS using a gold nanoparticle-based competitive colorimetric method. Chem Commun. 2008;(38):4573–4575. doi: 10.1039/b811346g. [DOI] [PubMed] [Google Scholar]
- 3.Béra-Abérem M, Ho H-A, Leclerc M. Functional polythophenes as optical chemo- and biosensors. Tetrahedron. 2004;60(49):11169–11173. [Google Scholar]
- 4.Reynolds RA, Mirkin CA, Letsinger RL. Homogeneous, nanoparticle-based quantitative colorimetric detection of oligonucleotides. J Am Chem Soc. 2000;122(15):3795–3796. [Google Scholar]
- 5.Ho H-A, et al. Colorimetric and fluorometric detection of nucleic acids using cationic polythiophene derivatives. Angew Chem Int Edit. 2002;41(9):1548–1551. doi: 10.1002/1521-3773(20020503)41:9<1548::aid-anie1548>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 6.Storhoff JJ, Mirkin CA. Programmed materials synthesis with DNA. Chem Rev. 1999;99(7):1849–1862. doi: 10.1021/cr970071p. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, et al. Bioinspired colorimetric detection of calcium(II) ions in serum using calsequestrin-functionalized gold nanoparticles. Angew Chem Int Edit. 2009;48(23):4138–4141. doi: 10.1002/anie.200900071. [DOI] [PubMed] [Google Scholar]
- 8.Peter K, Nilsson R, Inganäs O. Chip and solution detection of DNA hybridization using a luminescent zwitterionic polythiophere derivative. Nat Mater. 2003;2(6):419–424. doi: 10.1038/nmat899. [DOI] [PubMed] [Google Scholar]
- 9.Liu JW, Cao ZH, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109(5):1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn DJ, Lee S, Kim JM. Rational design of conjugated polymer supramolecules with tunable colorimetric responses. Adv Funct Mater. 2009;19(10):1483–1496. [Google Scholar]
- 11.Ali MM, Li YF. Colorimetric sensing by using allosteric-DNAzyme-coupled rolling circle amplification and a peptide nucleic acid-organic dye probe. Angew Chem Int Edit. 2009;48(19):3512–3515. doi: 10.1002/anie.200805966. [DOI] [PubMed] [Google Scholar]
- 12.Pu KY, Liu B. Intercalating dye harnessed cationic conjugated polymer for real-time naked-eye recognition of double-stranded DNA in serum. Adv Funct Mater. 2009;19(9):1371–1378. [Google Scholar]
- 13.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277(5329):1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 14.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc. 1998;120(9):1959–1964. [Google Scholar]
- 15.Demers LM, et al. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal Chem. 2000;72(22):5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 16.Storhoff JJ, et al. What controls the optical properties of DNA-linked gold nanoparticle assemblies? J Am Chem Soc. 2000;122(19):4640–4650. [Google Scholar]
- 17.Liu J, Lu Y. Stimuli-responsive disassembly of nanoparticle aggregates for light-up colorimetric sensing. J Am Chem Soc. 2005;127(36):12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 18.Liu JW, Lu Y. Optimization of a Pb2+ -directed gold nanoparticle/DNAzyme assembly and its application as a colorimetric biosensor for Pb2+ Chem Mater. 2004;16(17):3231–3238. [Google Scholar]
- 19.Liu JW, Lu Y. Accelerated color change of gold nanoparticles assembled by DNAzymes for simple and fast colorimetric Pb2+ detection. J Am Chem Soc. 2004;126(39):12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]
- 20.Liu JW, Lu Y. Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Anal Chem. 2004;76(6):1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 21.Liu JW, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew Chem Int Edit. 2006;45(1):90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 22.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 23.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105(4):1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 24.Cao YWC, Jin RC, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297(5586):1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 25.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289(5485):1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Song SP, Wang LH, Pan D, Fan C. A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat Protoc. 2007;2(11):2888–2895. doi: 10.1038/nprot.2007.419. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie F, Faulds K, Graham D. Sequence-specific DNA detection using high-affinity LNA-functionalized gold nanoparticles. Small. 2007;3(11):1866–1868. doi: 10.1002/smll.200700225. [DOI] [PubMed] [Google Scholar]
- 28.Li HX, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci USA. 2004;101(39):14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li HX, Rothberg LJ. Label-free colorimetric detection of specific sequences in genomic DNA amplified by the polymerase chain reaction. J Am Chem Soc. 2004;126(35):10958–10961. doi: 10.1021/ja048749n. [DOI] [PubMed] [Google Scholar]
- 30.Wang LH, Liu XF, Hu XF, Song SP, Fan CF. Unmodified gold nanoparticles as a colorimetric probe for potassium DNA aptamers. Chem Commun. 2006;(36):3780–3782. doi: 10.1039/b607448k. [DOI] [PubMed] [Google Scholar]
- 31.Polavarapu L, Xu QH. Water-soluble conjugated polymer-induced self-assembly of gold nanoparticles and its application to SERS. Langmuir. 2008;24(19):10608–10611. doi: 10.1021/la802319c. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Bazan GC. Homogeneous fluorescence-based DNA detection with water-soluble conjugated polymers. Chem Mater. 2004;16(23):4467–4476. [Google Scholar]
- 33.Xia F, et al. On the binding of cationic, water-soluble conjugated polyelectrolytes to DNA: Electrostatic and hydrophobic interactions. J Am Chem Soc. 2010;132(4):1252–1254. doi: 10.1021/ja908890q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Liu B, Gaylord & Bazan GC. Size-specific interactions between single- and double-stranded oligonucleotides and cationic water-soluble oligofluorenes. Adv Funct Mater. 13(6):463–467. [Google Scholar]
- 35.He F, et al. Selective and homogeneous fluorescent DNA detection by target-induced strand displacement using cationic conjugated polyelectrolytes. Anal Chem. 2008;80(6):2239–2243. doi: 10.1021/ac702415p. [DOI] [PubMed] [Google Scholar]
- 36.Willner I, Zayats M. Electronic aptamer-based sensors. Angew Chem Int Edit. 2007;46(34):6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 37.You KM, Lee SH, Im A, Lee SB. Aptamers as functional nucleic acids: In vitro selection and biotechnological applications (Translated from English) Biotechnol Bioproc E. 2003;8(2):64–75. (in English) [Google Scholar]
- 38.Luzi E, Minunni M, Tombelli S, Mascini M. New trends in affinity sensing: aptamers for ligand binding. Trac-Trend Anal Chem. 2003;22(11):810–818. [Google Scholar]
- 39.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J Am Chem Soc. 2001;123(21):4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 40.Zuo XL, Xiao Y, Plaxco KW. High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices. J Am Chem Soc. 2009;131(20):6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, et al. Visual cocaine detection with gold nanoparticles and rationally engineered aptamer structures. Small. 2008;4(8):1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- 42.Freeman R, Sharon E, Tel-Vered R, Willner I. Supramolecular cocaine-aptamer complexes activate biocatalytic cascades. J Am Chem Soc. 2009;131(14):5028–5029. doi: 10.1021/ja809496n. [DOI] [PubMed] [Google Scholar]
- 43.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992;355(6360):564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 44.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew Chem Int Edit. 2005;44(34):5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 45.Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. A highly selective DNAzyme sensor for mercuric ions. Angew Chem Int Edit. 2008;47(23):4346–4350. doi: 10.1002/anie.200800960. [DOI] [PubMed] [Google Scholar]
- 46.Li D, Wieckowska A, Willner I. Optical analysis of Hg2+ ions by oligonucleotide-gold-nanoparticle hybrids and DNA-based machines. Angew Chem Int Edit. 2008;47(21):3927–3931. doi: 10.1002/anie.200705991. [DOI] [PubMed] [Google Scholar]
- 47.Song FL, Watanabe S, Floreancig PE, Koide K. Oxidation-resistant fluorogenic probe for mercury based on alkyne oxymercuration. J Am Chem Soc. 2008;130(49):16460–16461. doi: 10.1021/ja805678r. [DOI] [PubMed] [Google Scholar]
- 48.Nolan EM, Lippard SJ. Tools and tactics for the optical detection of mercuric ion. Chem Rev. 2008;108(9):3443–3480. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]
- 49.Wang LH, et al. Gold nanoparticle-based optical probes for target-responsive DNA structures. Gold Bull. 2008;41(1):37–41. [Google Scholar]
- 50.Li J, et al. Enzyme-based multi-component optical nanoprobes for sequence-specific detection of DNA hybridization. Adv Mater. 2008;20(3):497–500. [Google Scholar]
- 51.Fan CH, Plaxco KW, Heeger AJ. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc Natl Acad Sci USA. 2003;100(16):9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Ting BP, Jana NR, Gao ZQ, Ying JY. Ultrasensitive electrochemical DNA biosensors based on the detection of a highly characteristic solid-state process. Small. 2009;5(12):1414–1417. doi: 10.1002/smll.200900073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.