Abstract
After each round of protein biosynthesis, the posttermination complex (PoTC) consisting of a ribosome, mRNA, and tRNA must be disassembled into its components for a new round of translation. Here, we show that a Saccharomyces cerevisiae model PoTC was disassembled by ATP and eukaryotic elongation factor 3 (eEF3). GTP or ITP functioned with less efficiency and adenosine 5γ′-(β,γ-imido)triphosphate did not function at all. The kcat of eEF3 was 1.12 min-1, which is comparable to that of the in vitro initiation step. The disassembly reaction was inhibited by aminoglycosides and cycloheximide. The subunits formed from the yeast model PoTC remained separated under ionic conditions close to those existing in vivo, suggesting that they are ready to enter the initiation process. Based on our experimental techniques used in this paper, the release of mRNA and tRNA and ribosome dissociation took place simultaneously. No 40S•mRNA complex was observed, indicating that eEF3 action promotes ribosome recycling, not reinitiation.
Keywords: yeast cytoplasm, ribosome recycling factor, ribosomal subunits, kinetics, antibiotics
During the termination step in protein synthesis, the synthesized polypeptide is released from the ribosome by release factors, forming the posttermination complex (PoTC). The PoTC consisting of the ribosome, mRNA, and tRNA must be disassembled for its components to participate in a new round of protein synthesis. In bacteria, EF-G•GTP and ribosome recycling factor (RRF) were shown to release mRNA and tRNA from PoTC (1) and to split the 70S ribosome into its subunits (2–5). Recent structural studies suggest that RRF binds to the ribosomal A/P site (aminoacyl/peptidyl site) (6, 7), after which this RRF is moved through the 70S intersubunit space, resulting in PoTC disassembly (8). Eukaryotes have a homolog of RRF, only in their organelles (9, 10), and not in their cytoplasm. The mechanism of ribosome recycling during eukaryotic cytoplasmic protein synthesis has been a long-standing unsolved event. Recently, eIF3 (eukaryotic initiation factor 3) was proposed as a major factor in ribosome recycling in the rabbit reticulocyte system (11), but whether the same mechanism operates in yeast cytoplasm, was not known.
In addition to the elongation factors, eEF1A and eEF2, yeast and other fungi have another essential ribosome-associated elongation factor, eEF3 (12), which is a ribosome-dependent ATPase (13). eEF3 stimulates eEF1A-dependent binding of a cognate aminoacyl-tRNA to the A site, and is involved in the release of deacylated tRNA from the ribosomal E (exit) site (14).
In this paper, we show that eEF3 and ATP are used to disassemble a yeast model PoTC. The kinetic parameters of mRNA release from the PoTC by eEF3 suggest that the reaction is sufficiently fast to account for in vitro polypeptide synthesis by yeast extracts. The 80S ribosomes are split into their subunits simultaneously with the release of ribosome-associated mRNA. In addition, deacylated tRNA is released from the PoTC at a time comparable with that of the release of mRNA. These observations suggest that the recycling of PoTC in yeast cytoplasm is catalyzed by eEF3/ATP.
Results
eEF3/ATP Releases mRNA from a Yeast Model PoTC.
The model PoTC examined was prepared using polysomes isolated from growing yeast cells (Figs. S1 and S2). Polysomes were washed with high salt (0.5 M KCl and 25 mM MgCl2), treated with eEF2 and GTP to translocate the ribosome-bound peptidyl-tRNA from the A site to the P site, followed by a second high-salt wash (Fig. S1 A–C). These procedures removed most (90%) of the eEF2 and eEF3 from the ribosomes (Fig. S2 D and E). The washed polysomes, with nascent peptidyl-tRNAs, were then treated with 1 mM puromycin (PUR) to form model PoTC (Fig. S1 C and D). Similar PoTC model complexes isolated from Escherichia coli were used to discover RRF (1). Throughout this paper, this substrate is designated “model PoTC” or “the PoTC.”
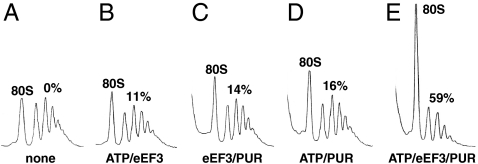
In Fig. 1, we show that mRNA was released from the PoTC by the addition of eEF3 and ATP. Each polysome consists of multiple 80S ribosomes associated with an mRNA. Hence, the dissociation (release) of mRNA should result in the increase of 80S ribosomes or their subunits. In this experiment, the added ATP/eEF3/PUR caused 59% of 80S ribosomes to be released from the polysomes. As shown later, the 80S ribosomes were formed from the subunits due to 25 mM MgCl2 addition to stop the reaction. Fig. 1 C and D show that both ATP and eEF3 were necessary to release mRNA from the PoTC. Maximum eEF3-dependent mRNA release occurred with 3 mM MgCl2 and 150 mM KCl (Fig. S3), which are close to the ionic concentrations (2.5 ∼ 3 mM Mg2+ and 150 ∼ 170 mM K+) required for in vitro yeast cell-free translation (15, 16).
Fig. 1.
eEF3/ATP releases mRNA from the PoTC. The purified polysomes (Fig. S1C) (15 pmol) were incubated for 3 min at 30 °C in buffer 3/150 with additions, as shown below each sedimentation profile. MgCl2 was added to 25 mM to stop the reaction. The percentage of polysome bound mRNA that was released is indicated above the polysome region. The complete reaction mixture (E) contained 50 μM ATP, 0.5 μM eEF3, and 1 mM PUR in buffer 3/150.
The nucleotide specificity (Table 1) is consistent with the concept that the reaction is dependent on energy because adenosine 5γ′-(β,γ-imido)triphosphate (ADPNP) alone did not work. In contrast to the known eEF3 function, stimulation of the aminoacyl tRNA binding to the A site (17), UTP functioned reasonably well for the release of mRNA.
Table 1.
Nucleotide specificity and energy-dependence of mRNA release by eEF3
| Nucleotide | mRNA released, % of the ATP reaction |
| ATP | 100 |
| GTP | 85 |
| CTP | 7 |
| UTP | 47 |
| ITP | 58 |
| ADP | 8 |
| ADPNP | 7 |
| GDP | 6 |
| GDPCP | 5 |
Disassembly reactions were carried out for 30 s with 50 μM of each nucleotide, as in Fig. 1. The release of mRNA is expressed as the percentage of the release with ATP (30% of polysomes were disassembled).
Kinetic Studies of eEF3/ATP-Dependent mRNA Release from the PoTC.
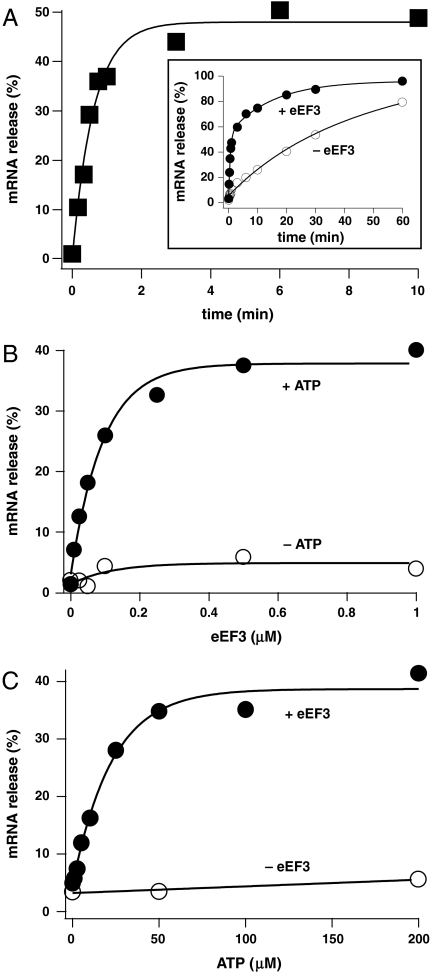
The data in Fig. 2A show the time course of mRNA release from the PoTC. Approximately 50% of the mRNA (corresponding to almost 100% of the PoTC, see below) was released within 2 min by added eEF3 (0.5 μM) and ATP (50 μM). It should be noted that the PUR reaction is not the rate-determining step of the overall reaction (Fig. S4). In the inset figure, the time course of the disassembly was followed until disassembly was almost complete in the presence or absence of eEF3. We also measured the extent of mRNA release at 30 s, with various amounts of eEF3 in the presence or absence of ATP (Fig. 2B). Taking into consideration the amount of endogenous eEF3 associated with the polysomes (Fig. S2E), we estimate that the KM value of eEF3 for the PoTC was 50.9 nM and the kcat was 1.12 molecules of mRNA released per min. This rate is similar to the rate of 43S complex formation in the yeast initiation pathway [1.17 per minute (18)]. The kcat/KM of eEF3 for the PoTC estimated from the mRNA release reaction (22.0 μM-1 min-1) is comparable to that for the ribosome-dependent ATPase activity of eEF3 [6.9 μM-1 min-1 (19)]. In the experiment shown in Fig. 2C, the KM value for ATP was estimated to be 15 μM, which is similar to that of other ATP-dependent reactions, such as yeast valyl-tRNA synthetase [40 μM, (20)].
Fig. 2.
Kinetic analysis of eEF3/ATP-dependent release of mRNA from the PoTC. (A) Time course of mRNA release by added eEF3/ATP, as in Fig. 1. The percentage of mRNA released from the PoTC without eEF3 was subtracted to show the time course of enzymatic release. (Inset) Actual values for mRNA release with or without eEF3 are shown. (B) eEF3 dose-response curve as in A except that incubation was for 30 s. (C) ATP dose-response curve as in B.
As shown in Fig. 2A, only about 50% of the total polysomes were disassembled by ATP/eEF3/PUR in 2 min, and the enzymatic reaction leveled off at that point. This is due to the fact that about 50% of the polysomes prepared were the pretranslocation complex with the peptidyl-tRNA at the A site as indicated in Fig. S1D, making them unreactive with PUR (Fig. S2A). The data presented in Fig. S2A are consistent with the known fact that yeast ribosomes retain significant portions of the bound peptidyl-tRNA at the A site even after the translocation process promoted by eEF2/GTP (21). In further support of this fact, the pretreatment of polysomes with eEF2 (Fig. S1 B and C) moved all the translocatable peptidyl-tRNA from the A site to the P site (Fig. S2 B and C).
Inhibitors of eEF3/ATP-Dependent mRNA Release from the PoTC.
Various reagents were examined for their effects on the mRNA release reaction (Table 2). Paromomycin, an inhibitor of bacterial ribosome recycling (22), effectively prevented mRNA release. Other aminoglycosides, Neomycin (23) and Hygromycin (24), also inhibited the disassembly reaction. Two translocation inhibitors, sordarin and fusidic acid, which specifically bind to eEF2 (25, 26), were less effective. This is consistent with our finding that eEF2/ATP did not disassemble the PoTC (see Discussion). On the other hand, cycloheximide, which also inhibits translocation, inhibited the reaction significantly. Because translocation may cause simultaneous release of E-site-bound tRNA (27), which requires the open form of L1 at the E site, involvement of L1 in both translocation and disassembly may be the reason for inhibition of these reactions by cycloheximide. This view is further supported by the finding that cycloheximide binds to the E site of the 60S subunit (28, 29).
Table 2.
Effect of various compounds on mRNA release promoted by eEF3 + ATP
| Compound | mM | Inhibition of release, % |
| None | 0 | |
| Paromomycin | 0.2 | 88 |
| Neomycin | 0.1 | 63 |
| Hygromycin | 0.38 | 54 |
| Sordarin | 0.1 | 25 |
| Fusidic acid | 1 | 8 |
| Cycloheximide | 0.36 | 86 |
| ADP | 0.05 | 2 |
| ADPNP | 0.05 | 4 |
| GDPCP | 0.05 | 1 |
| Vanadate | 0.8 | 0 |
| Spermine | 0.8 | 90 |
| Spermidine | 0.8 | 0 |
| Putrescine | 2 | −4 |
The release of mRNA from the PoTC was determined as in Fig. 1 except for the incubation time, which was 1 min. The percentage inhibition was calculated from the extent of the release of mRNA by each compound, in comparison with the control with no inhibitor added (42% of polysomes were disassembled).
Spermine, at a concentration much higher than found in vivo (30), effectively inhibited the reaction. This is perhaps due to the fact that spermine action is similar to that of high Mg2+ concentration (30). Other polyamines, such as spermidine and putrescine at concentrations stimulatory for in vitro protein synthesis (31, 32), did not inhibit mRNA release. Vanadate (33) and ADPNP also did not inhibit the release of mRNA. This does not mean that hydrolysis of ATP is not required because ADPNP alone did not function as shown earlier in Table 1. This may be related to the observation that, in the crystal structure of the eEF3/ADPNP complex, the bound nucleotide is 14 Å away from the putative ATP binding site (34).
eEF3/ATP Splits the PoTC into Subunits.
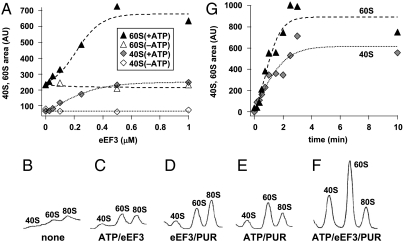
If the mRNA release reaction described above is a part of the recycling step, it should accompany the dissociation of ribosomes. In the mRNA release experiment described above, the reaction was stopped by the addition of 25 mM MgCl2 which reassociates split subunits into 80S ribosomes. To observe subunits, this reassociation has to be stopped by adding eIF6 (antiassociation factor). As shown in Fig. 3A, the amount of both subunits observed upon the addition of eIF6 increased in an eEF3 dose-dependent manner. The dissociation reaction was clearly ATP-dependent. eIF6 alone had very little effect on the ribosomes in the presence of 25 mM MgCl2 (Fig. S5).
Fig. 3.
eEF3/ATP splits the PoTC into subunits. (A) eEF3 dose-response curve. Experiments were performed as in Fig. 1 except that the reaction mixtures contained 5 μM eIF6 and the incubation was for 1 min. The amounts of 60S and 40S subunits are expressed as arbitrary units. (B–F) As in A, except that the reaction mixture did not contain eIF6 but the reaction was stopped by formaldehyde addition at 3 min. Sedimentation profiles of the split subunits with the reaction components described below each profile are shown. (G) Time course of the splitting of the PoTC. The reaction was stopped at various time points by formaldehyde addition. The background value without eEF3 was subtracted.
Formaldehyde is known to freeze subunits as they are in solution (35). In confirmation of Fig. 3A, ribosome splitting was observed when the reaction was stopped by formaldehyde, and this was also eEF3/ATP dependent (Fig. 3 B–F). Furthermore, the enzymatic splitting reaction proceeded in a time-dependent manner (Fig. 3G). The increase in appearance of both subunits was completed at about 2 min, which is comparable to the mRNA release time course, suggesting that both reactions may occur simultaneously. The eEF3-dependent splitting was further confirmed by the experiment shown in Fig. S6 where no stabilizer was added, but the reaction mixture was sedimented with low centrifugal force to preserve the integrity of residual polysomes (36). These data show that the eEF3-dependent disassembly of the PoTC produces subunits, not 80S ribosomes, as final products in the reaction mixture.
eEF3/ATP Releases tRNA from the PoTC.
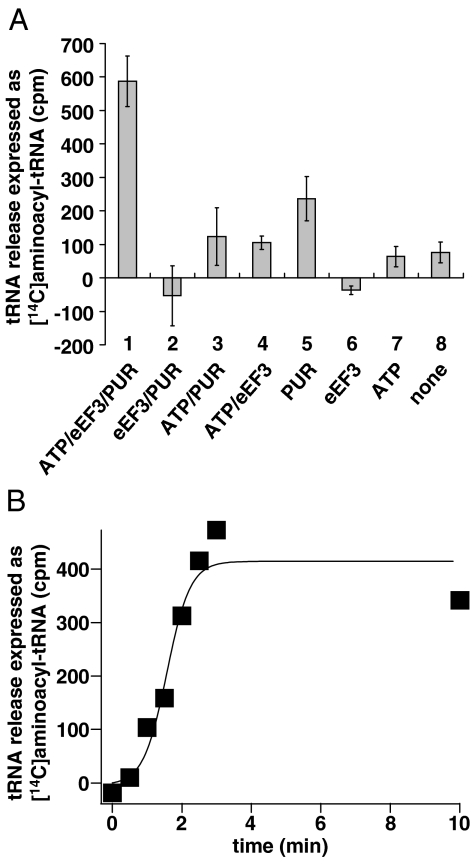
PoTC harbors the tRNA corresponding to the C-terminal amino acid of the protein that the ribosome just synthesized. This tRNA should also be released from the PoTC by eEF3 if the eEF3 reaction is a part of the disassembly process. To examine this possibility, the reaction mixture described in Fig. 1 was filtered through a Millipore filter, and tRNA which had been released from the ribosomes was measured in the filtrate by charging with a mixture of [14C]amino acids and aminoacyl-tRNA synthetases. The data in Fig. 4 show that the release of tRNA indeed takes place from the PoTC by the action of eEF3. It is clear from Fig. 4A that the release of tRNA was dependent on both ATP (compare 1 and 2) and eEF3 (compare 1 and 3). In the absence of PUR, nominal tRNA release was observed (compare 1 and 4), implying again that eEF3/ATP disassembles the PoTC. The relatively high background release (column 5) is due to the unstable nature of the PoTC. To measure the total bound tRNA, the polysomes were treated with PUR in high-salt buffer, where all tRNA, including peptidyl-tRNA, is considered to be released from ribosomes as deacylated tRNA due to nonenzymatic translocation (37) followed by the peptidyl-PUR reaction (38). The amount of released tRNA in the complete system (column 1, 588 ± 75 cpm) was 32% of the total tRNA bound in the polysomes (1829 ± 298 cpm). Assuming that ≈50–60% of the polysomes consist of the PoTC containing one tRNA and the rest are pretranslocation complexes containing two tRNAs (see Fig. 2A and Fig. S2A), we estimate that 32% of the total bound tRNA represents the major portion of deacylated tRNA in the PoTC. Thus, eEF3/ATP releases most of the bound tRNA of the PoTC. Fig. 4B indicates that the enzymatic tRNA release reaction was completed at about 2 min, which is comparable to the mRNA release (Fig. 2A) and the ribosome splitting reaction (Fig. 3G), suggesting that these three reactions may occur simultaneously.
Fig. 4.
eEF3/ATP releases tRNA from the PoTC. (A) [14C]aminoacyl-tRNA formed from the tRNA released under various conditions are shown. The background value (tRNA released without factors and incubation, 272 ± 68 cpm) was subtracted. The error bars represent SD. (B) Time course of tRNA release by eEF3/ATP. The value corresponding to tRNA released without eEF3 was subtracted.
Discussion
There are two possible pathways for yeast ribosomes to follow, after the termination step (39, 40). The first pathway is reinitiation, i.e., the 40S subunit would remain on the same mRNA and engage in the translation of the next ORF on the same mRNA. It occurs efficiently after translation of short ORFs, under certain conditions such as amino acid starvation (41). Recently, in rabbit reticulocyte lysates, ABCE1 protein and nucleotide triphosphate have been shown to split PoTC into a 60S subunit and an mRNA/tRNA/40S subunit complex (42). Hence, ABCE1 must catalyze reinitiation where the 40S subunit remains on the mRNA. The second pathway is ribosome recycling, in which the ribosome is released from the mRNA. The released ribosome can then be channeled to the initiation site of the same mRNA (39).
In this paper, we show that eEF3 catalyzes the ribosome recycling step in yeast cytoplasmic extracts. This is based on the observation that mRNA and tRNA are released from the PoTC which is split into subunits. The kinetic studies on the disassembly reaction suggest that these three reactions occur simultaneously within the 2 min limit of our analytical method. Furthermore, the ribosomal subunits remain apart under buffer conditions that are close to those occurring under physiological conditions. These findings are in contrast to the disassembly of the bacterial PoTC, where the PoTC is disassembled into tRNA, mRNA, and ribosomal subunits in a stepwise manner (4, 5, 8, 43). The fact that we did not observe intermediate polysomes termed “halfmers” that have extra 40S subunits in addition to multiple 80S ribosomes (44) suggests that the eEF3 reaction studied above represents recycling, not reinitiation.
We recently presented evidence that eEF2/ATP dissociates 80S ribosomes into subunits (45). However, the yeast model PoTC was not disassembled by eEF2/ATP (Fig. S7) under conditions where eEF3 induces dissociation. Therefore, splitting of 80S ribosomes and disassembly of PoTC may be performed differently. On the other hand, Pisarev et al. reported that eIF3 is mostly responsible for the recycling (11). However, under their ionic conditions, eEF3/ATP-dependent disassembly was not observed (Fig. S3C). Hence, we tested yeast eIF3 under the conditions where eEF3/ATP dissociated the PoTC, but it did not release mRNA from the PoTC (Fig. S8A). The eIF3 we used was active in protein synthesis initiation (Fig. S8 B and C). Because the yeast homolog of eEF3 does not exist in higher eukaryotes (46), ribosome recycling in yeast must be different from that in higher eukaryotes.
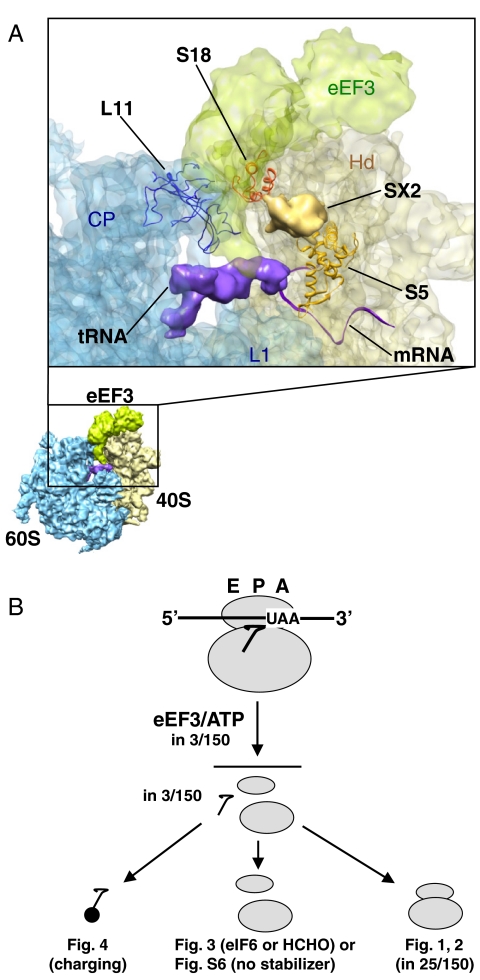
According to cryo-EM data, eEF3 binds to the ribosome at a position suitable for disassembly, covering both subunits near the E site (Fig. 5A). This position is close to SX2 (probably corresponding to the N-terminal domain of S5), L11, and S18 (34). We propose that these components are influenced by the eEF3 conformational change, resulting in the disassembly. S5 (S7 in bacteria) is probably involved in the release of mRNA by eEF3 for the following reasons: (i) It is a part of the mRNA exit channel (47). This channel is closed in 80S ribosomes containing mRNA (48) but open in ribosomes without mRNA (49). (ii) The residues of the bacterial S7 interact with the upstream bases of the mRNA in elongating 70S ribosomes (50). (iii) S5 interacts with cricket paralysis virus internal ribosome entry site mRNA (51). (iv) Yeast ribosomes with human S5 have lower affinity to eEF3 (52). Regarding the ribosome splitting by eEF3, involvement of L11 and S18 is likely, because they constitute a conserved bridge, B1b/c (48). On the other hand, L1 must be involved in the release of tRNA. Thus, the L1 stalk is an E site mobile domain of the 60S subunit. eEF3 stabilizes L1 in an open conformation, resulting in the release of tRNA from the ribosome during translocation by eEF2 (34). tRNA release during disassembly may occur similarly. PoTC assumes the ratcheted form (7) that normally keeps the position of L1 closed (53, 54). We postulate that, similarly, eEF3 functions to open L1, resulting in the release of tRNA from PoTC. In further support of this model, the elbow of the tRNA in the hybrid P/E state has continuous contact with the head of L1 (53, 54). In addition, L11 interacts with the elbow of the P site bound tRNA (48).
Fig. 5.
Model of eEF3 action. (A) eEF3 binds to the ribosomal site suitable for its action—the cryo-EM map of single particle reconstruction (34). Atomic models of S5 (orange), S18 (red), and L11 (blue) shown as ribbon representations are docked to the cryo-EM density of 40S subunit (shown in light yellow), 60S subunit (light blue), eEF3 (light green), P site bound tRNA (purple), and unassigned SX2 (gold). The atomic structure of mRNA (purple ribbon) was superimposed onto a cryo-EM map by using UCSF Chimera 1.4 (http://www.cgl.ucsf.edu/chimera/). Landmark of 40S subunit, Hd (head); Landmarks of 60S subunit, CP (central protuberance), L1 (L1 stalk). Cryo-EM map and coordinate of mRNA were obtained from EMD-1233 and PDB-2HGP, respectively. (B) eEF3/ATP disassembles PoTC into its components simultaneously and the components remain apart until the initiation process begins.
These considerations and the data presented in this paper suggest that recycling of ribosomes in the yeast cytoplasm must result in simultaneous release of mRNA, tRNA, and ribosomal subunits by eEF3/ATP, as shown in Fig. 5B. Conversely, one must ask why eEF3 does not split ribosomes and release mRNA during the elongation cycle? We suggest two possible reasons: First, an elongating ribosome is much more stable because of bound peptidyl-tRNA, which is known to stabilize subunit association (55) and the ribosome-mRNA interaction (56). Second, PoTC has an empty A site, whereas that of the elongating ribosome is occupied with either peptidyl-tRNA or eEF1A-aminoacyl-tRNA. In support of this hypothesis, the eEF3–eEF1A interaction appears to be important for eEF3 elongation activity (24).
Materials and Methods
Buffers.
Buffer X/Y indicates that the buffer contains 20 mM Hepes-KOH (pH 7.6), X mM MgCl2, Y mM KCl, and 2 mM DTT.
Preparation of the PoTC, eEF3, eEF2, and eIF6.
The PoTC was prepared as described in Fig. S1. Details are provided in the SI Text. The histidine-tagged factors were prepared as described previously (34, 45).
Release of mRNA from the PoTC.
The reaction mixture (150 μL) containing 0.75 A260 units of polysomes (15 pmol of ribosomes) were preincubated in buffer 3/150 for 10 min at 30 °C. The disassembly reaction was then started by the addition of 1 mM PUR, 50 μM ATP (potassium salt), and 0.5 μM eEF3, and incubated as indicated in legends. The reaction was stopped by the addition of MgCl2 to 25 mM. The mixture was loaded onto 4.5 mL of 15–45% sucrose gradient prepared in buffer 25/150 and sedimented for 75 min at 4 °C (Beckman SW50.1 rotor, 150,000 × g). The sedimentation behavior of polysomes and 80S ribosomes was monitored using an ISCO UA-6 spectrophotometer at 254 nm. The polysome area was measured using ImageJ 1.38x software and the background (without polysomes) was subtracted. Percentages of mRNA release (z) were calculated: z = 100 × [1 - (y/w)], where (y) is the polysome area remaining after the reaction and (w) is that without any factors.
Splitting of the PoTC into Ribosomal Subunits.
In Fig. 3A, the PoTC was disassembled as described above, except that the reaction mixture contained 5 μM eIF6. After the reaction, the mixture was loaded onto 4.5 mL of 5–30% sucrose gradient in buffer 25/150 and sedimented for 128 min at 4 °C (SW50.1, 150,000 × g). In Fig. 3 B–G, the conditions were identical to Fig. 3A except that the reaction was stopped by formaldehyde (4%, vol/vol), placed on ice for 5 min, and sedimented in buffer 3/150. The third method (Fig. S6) was not to use the subunit stabilization agents and the reaction was stopped by cooling to 0 °C. Sedimentation of the subunits was with less gravity force as described in the SI Text.
Release of tRNA from the PoTC.
The PoTC (1.5 A260 units) was disassembled in the reaction mixture (300 μL) as described above. The reaction was stopped by filtering through a Millipore filter (pore size 0.45 μm) prewashed with 300 μL of buffer 3/150. The filter was washed with 400 μL of buffer 3/150 and tRNA in the filtrate was concentrated by ethanol precipitation with 20 μg/mL glycogen. The recovered tRNA was incubated for 30 min at 30 °C in 20 μL of buffer 3/150 containing 1 mM ATP, 0.55 mg/mL aminoacyl-tRNA synthetases prepared as described in the SI Text, and 50 nCi of [14C]amino acids (Moravek Biochemicals, > 500 mCi/mol). The cold (4 °C) trichloroacetic acid-insoluble radioactivity thus formed was regarded as a mixture of [14C]aminoacyl-tRNA derived from the released tRNA. To obtain the value of total bound tRNA, the polysomes were incubated with 1 mM PUR in buffer 5/500 at 37 °C for 10 min (38) and the released tRNA was measured as above.
Supplementary Material
Acknowledgments.
The authors thank Drs. Roland Beckmann and Thomas Becker for supplying the Protein Data Bank file for the eEF3/ribosome complex and the strain WCGα harboring the plasmid used for overexpression of His-tagged eEF3, Dr. Terri G. Kinzy for supplying the strain TKY675 for eEF2 production and antibody against yeast eEF2, and Dr. Takeshi Yokoyama for constructing Fig. 5A. We also thank Drs. Charles Yanofsky and Eric Wickstrom for critical reading of the manuscript. This work was supported by the Alfred Benzon Foundation (K.H.N.), National Institutes of Health Grant GM60429 (A.K.), the Nippon Paint Research Fund (H.K.), and the Creative Biomedical Research Institute (H.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006247107/-/DCSupplemental.
References
- 1.Hirashima A, Kaji A. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J Biol Chem. 1973;248:7580–7587. [PubMed] [Google Scholar]
- 2.Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 3.Hirokawa G, et al. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal RK, et al. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pai RD, et al. Structural insights into ribosome recycling factor interactions with the 70S ribosome. J Mol Biol. 2008;376:1334–1347. doi: 10.1016/j.jmb.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barat C, et al. Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Rolland N, et al. Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc Natl Acad Sci USA. 1999;96:5464–5469. doi: 10.1073/pnas.96.10.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teyssier E, et al. Temperature-sensitive mutation in yeast mitochondrial ribosome recycling factor (RRF) Nucleic Acids Res. 2003;31:4218–4226. doi: 10.1093/nar/gkg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skogerson L, Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci USA. 1976;73:73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasmahapatra B, Chakraburtty K. Protein Synthesis in Yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J Biol Chem. 1981;256:9999–10004. [PubMed] [Google Scholar]
- 14.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 15.Beckmann R, et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 16.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath A, Chakraburtty K. Role of yeast elongation factor 3 in the elongation cycle. J Biol Chem. 1989;264:15423–15428. [PubMed] [Google Scholar]
- 18.Algire MA, et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarthy AV, et al. Identification and kinetic analysis of a functional homolog of elongation factor 3, YEF3 in Saccharomyces cerevisiae. Yeast. 1998;14:239–253. doi: 10.1002/(SICI)1097-0061(199802)14:3<239::AID-YEA219>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 20.Lagerkvist U, Waldenstrom J. Purification and some properties of valyl ribonucleic acid synthetase from yeast. J Biol Chem. 1967;242:3021–3025. [PubMed] [Google Scholar]
- 21.Nilsson L, Nygard O. Reduced puromycin sensitivity of translocated polysomes after the addition of elongation factor 2 and non-hydrolysable GTP analogues. FEBS Lett. 1992;309:89–91. doi: 10.1016/0014-5793(92)80746-4. [DOI] [PubMed] [Google Scholar]
- 22.Hirokawa G, et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalchuke O, Kambampati R, Pladies E, Chakraburtty K. Competition and cooperation amongst yeast elongation factors. Eur J Biochem. 1998;258:986–993. doi: 10.1046/j.1432-1327.1998.2580986.x. [DOI] [PubMed] [Google Scholar]
- 24.Anand M, Chakraburtty K, Marton MJ, Hinnebusch AG, Kinzy TG. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J Biol Chem. 2003;278:6985–6991. doi: 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen R, et al. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 26.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stocklein W, Piepersberg W. Binding of cycloheximide to ribosomes from wild-type and mutant strains of Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1980;18:863–867. doi: 10.1128/aac.18.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter AR, Farrell PJ, Jackson RJ, Hunt T. The role of polyamines in cell-free protein synthesis in the wheat-germ system. Eur J Biochem. 1977;75:149–157. doi: 10.1111/j.1432-1033.1977.tb11512.x. [DOI] [PubMed] [Google Scholar]
- 32.Tuite MF, Plesset J, Moldave K, McLaughlin CS. Faithful and efficient translation of homologous and heterologous mRNAs in an mRNA-dependent cell-free system from Saccharomyces cerevisiae. J Biol Chem. 1980;255:8761–8766. [PubMed] [Google Scholar]
- 33.Pezza RJ, Villarreal MA, Montich GG, Argarana CE. Vanadate inhibits the ATPase activity and DNA binding capability of bacterial MutS. A structural model for the vanadate-MutS interaction at the Walker A motif. Nucleic Acids Res. 2002;30:4700–4708. doi: 10.1093/nar/gkf606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen CB, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 35.Zitomer RS, Flaks JG. Magnesium dependence and equilibrium of the Escherichia coli ribosomal subunit association. J Mol Biol. 1972;71:263–279. doi: 10.1016/0022-2836(72)90350-6. [DOI] [PubMed] [Google Scholar]
- 36.Infante A, Baierlein R. Pressure-induced dissociation of sedimenting ribosomes: Effect on sedimentation patterns. Proc Natl Acad Sci USA. 1971;68:1780–1785. doi: 10.1073/pnas.68.8.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. 3. The formation of peptide bonds by ribosomes in the absence of supernatant enzymes. J Biol Chem. 1968;243:2810–2820. [PubMed] [Google Scholar]
- 38.Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci USA. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JEG. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J Mol Biol. 2004;335:71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- 40.Szamecz B, et al. eIF3a cooperates with sequences 5’ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 42.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A. The ribosome-recycling step: Consensus or controversy? Trends Biochem Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Helser TL, Baan RA, Dahlberg AE. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol. 1981;1:51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demeshkina N, Hirokawa G, Kaji A, Kaji H. Novel activity of eukaryotic translocase, eEF2: Dissociation of the 80S ribosome into subunits with ATP but not with GTP. Nucleic Acids Res. 2007;35:4597–4607. doi: 10.1093/nar/gkm468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraburtty K. Translational regulation by ABC systems. Res Microbiol. 2001;152:391–399. doi: 10.1016/s0923-2508(01)01210-4. [DOI] [PubMed] [Google Scholar]
- 47.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 48.Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 49.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Jenner L, Rees B, Yusupov M, Yusupova G. Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep. 2007;8:846–850. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 52.Galkin O, et al. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–2128. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlessinger D, Mangiarotti G, Apirion D. The formation and stabilization of 30S and 50S ribosome couples in Escherichia coli. Proc Natl Acad Sci USA. 1967;58:1782–1789. doi: 10.1073/pnas.58.4.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uemura S, et al. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.