Abstract
Light is the source of energy for photosynthetic organisms; when in excess, however, it also drives the formation of reactive oxygen species and, consequently, photoinhibition. Plants and algae have evolved mechanisms to regulate light harvesting efficiency in response to variable light intensity so as to avoid oxidative damage. Nonphotochemical quenching (NPQ) consists of the rapid dissipation of excess excitation energy as heat. Although widespread among oxygenic photosynthetic organisms, NPQ shows important differences in its machinery. In land plants, such as Arabidopsis thaliana, NPQ depends on the presence of PSBS, whereas in the green alga Chlamydomonas reinhardtii it requires a different protein called LHCSR. In this work, we show that both proteins are present in the moss Physcomitrella patens. By generating KO mutants lacking PSBS and/or LHCSR, we also demonstrate that both gene products are active in NPQ. Plants lacking both proteins are more susceptible to high light stress than WT, implying that they are active in photoprotection. These results suggest that NPQ is a fundamental mechanism for survival in excess light and that upon land colonization, photosynthetic organisms evolved a unique mechanism for excess energy dissipation before losing the ancestral one found in algae.
Keywords: LHCSR, nonphotochemical quenching, photosystem, plants evolution, PSBS
Sunlight provides energy supporting the life of photosynthetic organisms but also leads to the formation of reactive oxygen species when in excess (1, 2). During early Devonian, when plants first colonized terrestrial habitats, they underwent no competition by other organisms. However, they had to adapt to harsher physicochemical conditions than in the original water ecosystem (3) because in the atmosphere concentration of oxygen, an inhibitor of photosynthesis, is higher (4) and concentration of carbon dioxide, the final acceptor of electrons extracted from water by photosystems, is lower. Moreover, the sessile form of life acquired on land prevented escape from rapid changes in light intensity by swimming deeper, a behavior typical of algae (5). The combination of these conditions makes it more likely that light is harvested in excess with respect to the maximal rate of photochemical reactions, and a fast and efficient photoprotection response is essential for survival.
The fastest response to high light stress is provided by nonphotochemical quenching (NPQ), which consists of the thermal dissipation of the chlorophyll excited singlet states (1Chl*) (6–8). NPQ has two major components: energy quenching (qE), which is activated within seconds on an increase in light intensity, and inhibitory quenching, which is slower and relaxes within 1–2 h in the dark (7, 9). In vascular plants, qE activation requires PSBS, a protein homologous to light harvesting antenna subunits of photosystems (Lhc) (10), which is activated by the accumulation of protons in the chloroplast lumen and the protonation of two glutamate residues (11). Activated PSBS induces a decrease in excited state lifetime in the pigment-binding subunits of the antenna system consequent to a reorganization of photosystem subunits in the thylakoid membrane (12–14). NPQ was proven to be particularly important in field conditions, wherein plants lacking PSBS are rapidly counterselected (8).
The green alga Chlamydomonas reinhardtii shows different requirements for NPQ activation, and the process is induced by acclimation to high light, whereas it is constitutive in plants. Also, this green alga does not need PSBS (15–17) but a distinct Lhc-like protein, called LHCSR (or Li818), implying important differences in the mechanism for activation of heat dissipation (17). LHCSR has been found in many taxa, such as brown and green algae (18), but not in vascular plants. Conversely, although genes encoding PSBS have been identified in the genome (19), the protein itself has not been detected in algae to date (16), consistent with their NPQ being dependent on LHCSR. Further evidence that NPQ activation is different in algae and vascular plants (20) is provided by the lack in the former of Lhcb6, an antenna system component involved in PSBS-dependent NPQ activation of plants (12, 21).
The organization of photosystems is very similar in vascular plants and green algae but for the absence in the latter of Lhcb6 and Lhcb3 (18, 20). Grana stacks only developed in less ancient taxa of the Streptophyta line (Coleochaetales and Charales) (22, 23), whereas other algae, at most, show stacking between two thylakoid membranes.
Studying organisms that diverged from the green lineage leading to vascular plants early after land colonization helps us to understand how photosynthetic organisms adapted to the challenges of different environmental conditions. The moss Physcomitrella patens is a valuable choice as a model organism because its genome has been completely sequenced and tools are available for its genetic manipulation (3, 24). Early studies on the organization of photosynthetic genes showed that P. patens is the only example among plants wherein both LHCSR and PSBS genes are present (3, 18, 20). In this work, we show that both of these genes are expressed and the corresponding polypeptides are accumulated in the thylakoid membranes. By generating specific KO mutants for each of the two LHCSR genes and PSBS in the Physcomitrella genome, we show that both proteins are active in promoting NPQ and contribute to photoprotection under excess light conditions. These results suggest that the PSBS-dependent NPQ of plants evolved before the LHCSR-based mechanism typical of algae was lost. The latter disappeared at later stages of plant evolution, when the newly evolved PSBS-dependent mechanism ensured a sufficient level of photoprotection.
Results
P. patens Expresses Both LHCSR and PSBS and Shows High NPQ.
NPQ in the green alga C. reinhardtii relies on the presence of LHCSR. This group of Lhc-like proteins has been found in several algae but not in land plants, with the remarkable exception of P. patens, a bryophyte that diverged from vascular plants around 400 million years ago, early after land colonization (3, 25). A search of The Institute for Genomic Research (TIGR) sequence databases allowed identification of several ESTs encoding LHCSR and PSBS proteins in another bryophyte, Tortula ruralis, as well (TA794_38588 and CN203629, respectively), suggesting that this is not a unique property of P. patens.
Finding genome sequences does not imply that the corresponding polypeptides are expressed or that they play the expected function, as shown in the case of PSBS in many green algae (16). We therefore searched for LHCSR and PSBS polypeptides in P. patens thylakoids by using specific antibody probes. We detected specific signals at the expected molecular mass for P. patens LHCSR (around 23 kDa) (20) and PSBS (22 kDa), showing that both proteins are actively expressed and accumulate in the membrane of thylakoids (Fig. 1A). A search in Arabidopsis and Chlamydomonas extracts yielded a signal for PSBS and LHCSR only, respectively. In Chlamydomonas, LHCSR is detected only in extracts from high light-acclimated cells, consistent with the recent report that high light acclimation is needed for its accumulation and induction of NPQ activity (17) and at variance with P. patens, in which it is constitutively accumulated.
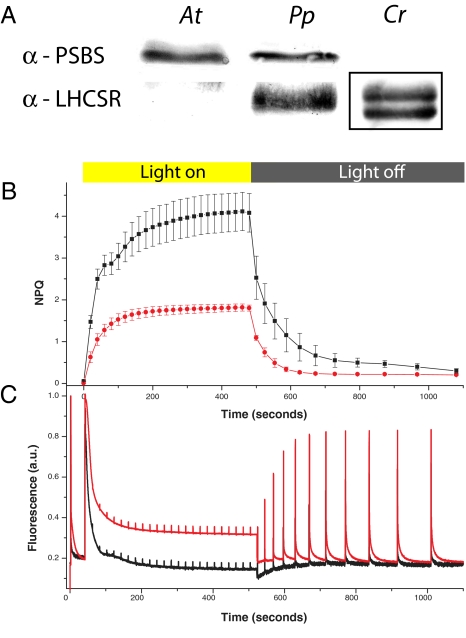
Fig. 1.
Comparison of NPQ A. thaliana, P. patens, and C. reinhardtii. (A) Western blotting against NPQ involved polypeptides, PSBS, and LHCSR in Arabidopsis (At), Physcomitrella (Pp), and Chlamydomonas (Cr). Thylakoids (1 μg of Chl) were loaded in all cases. In the case of Cr, thylakoids were isolated from high light-grown cells; here, two LHCSR bands are recognized as in the study by Peers et al. (17). Bands are squared because LHCSR has a larger molecular mass in Cr with respect to Pp (29 vs. 23 kDa). Anti-LHCSR antibody recognizes other proteins with lower specificity (likely Lhc proteins) at different molecular masses in both At and Pp. NPQ (B) and fluorescence (C) kinetics were measured for Pp (black) compared with At (red). Actinic light intensities were shown to saturate photosynthetic capacity in both organisms, as shown by the fact that pulses induce a very small peak in fluorescence (Fm′), and thus activate maximal NPQ.
Fig. 1B shows that NPQ activation in response to illumination with strong actinic light of dark-adapted plants is higher in P. patens with respect to Arabidopsis. Consistently, the decline of maximal fluorescence yield (Fm′), a parameter closely reflecting the lifetime of chlorophyll (Chl) a singlet excited states in the antenna compartment, is faster and the final intensity is lower in the moss with respect to the vascular plant (Fig. 1C). It should be noted that actinic light intensity during the measurement was chosen to be just sufficient to saturate photosynthesis to ensure maximal NPQ amplitude in both organisms. Despite using lower actinic light intensity (800 vs. 1,200 μE·m−2·s−1), capacity of heat dissipation was higher in P. patens than in Arabidopsis thaliana. Consistently, the stronger NPQ in mosses was observed for a large range of actinic lights (Fig. S1), confirming that the capacity of dissipating excitation energy as heat was higher in the moss with respect to the plant.
Targeted KO of PSBS, LHCSR1, and LHCSR2.
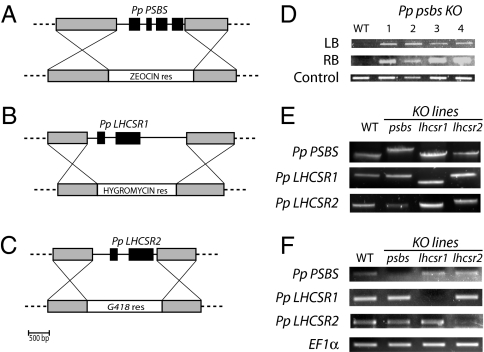
It is tempting to hypothesize that the faster and stronger photoprotection found in mosses relies on the action of both PSBS-dependent and LHCSR-dependent mechanisms of energy dissipation. To assess if this was indeed the case, we produced specific KO mosses depleted in the three genes encoding putative NPQ triggering polypeptides. P. patens is a very valuable organism for gene KO because it performs homologous recombination at high efficiency (24). Disruption constructs for the three genes were created to substitute the entire coding sequences with selection cassettes, as schematized in Fig. 2 A–C. PEG-mediated protoplast transformation led to the isolation of stably resistant colonies that were characterized by PCR to verify the presence of the insertion at the expected target site. A first set of PCR assays on genomic DNA was performed to amplify both left and right borders of the inserted cassette (an example of amplificates for selected lines of psbs mutants is reported in Fig. 2D). At least four independent clones with the insertion in the correct position were isolated and retained for further characterization.
Fig. 2.
KO mutant generation and characterization. Scheme of constructs used for KO generation. Genomic region of PSBS (A) and LHCSR (B and C) genes is schematized, with exons shown in black. Gray boxes represent the genomic regions exploited for homologous recombination. Below are shown the constructs for homologous recombination: genes for antibiotic resistance are located between regions homologous to the genome (all primers used for mutagenesis are reported in Table S1). (D) Example of verification of DNA insertion in the genome by amplification of the right and left borders. An example of four independent lines of Psbs KO is shown. (E) Amplification of genomic DNA using primers external to the target recombination region. Mutants carrying a single insertion are identified by the different size of the amplified band with respect to WT. Amplificates were also sequenced to verify the insertion in the correct position. (F) Evaluation of PSBS and LHCSR gene expression assessed by RT-PCR in WT and different mutant lines. Elongation Factor-1 alpha (EF1α) is also reported as a control.
During transformation in Physcomitrella, multiple insertions at the same site might occur (24, 26, 27). To select single copy insertions, we used a PCR assay with primers annealing to the genomic sequences at either side of the insertion region. PCR amplifications thus are possible only in the case of a single insertion, whereas for multiple insertions, fragments are too large to be efficiently amplified (Fig. 2E). All PCR fragments were sequenced for further control, and we finally retained at least two independent single insertion lines for each transformation. The suppression of gene expression was confirmed by RT-PCR with specific primers: cDNAs were all present in WT and lost in the corresponding mutants (Fig. 2F).
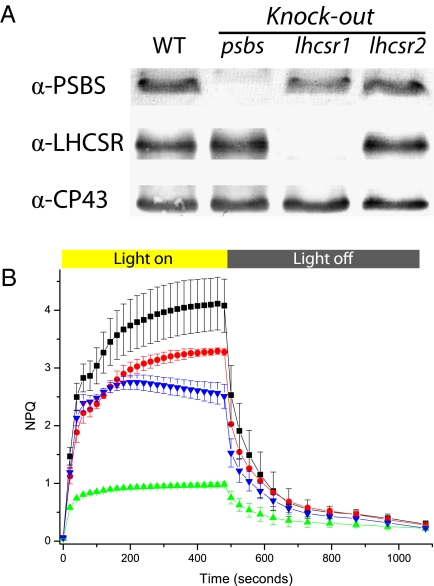
Single-insertion mutants were grown, and 10-day-old plants were harvested and used for thylakoid purification. When analyzed by Western blotting, no signal for PSBS was detected in psbs KO (Fig. 3A). KO for LHCSR showed that most of the immunoblotting signal was lost in lhcsr1 KO, whereas protein level was substantially unaffected in lhcsr2 KO. Red Ponceau staining and anti-CP43 antibody were routinely used to verify equal loading for all samples (Fig. 3A). In all mutants, we observed no significant alterations in Chl a/b and Chl/carotenoid content (Table 1), which indicates that mutations do not affect the composition of the photosynthetic apparatus, which is thus comparable in all mutants. The maintenance of photosystem organization was also confirmed by measuring the functional antenna size, as determined in vivo by measuring fluorescence kinetics in plants treated with dichloromethylurea (DCMU) (Fig. S2). Also, the quantum yield of photosystem II (PSII) in dark-adapted samples was unaffected: All plants showed a variable over maximal fluorescence (Fv/Fm) value of 0.80 ± 0.03 after 10 days of growth in control light conditions.
Fig. 3.
P. patens psbs and lhcsr KO phenotypes. (A) Western blotting using antibodies against LHCSR and PSBS. Western blotting against CP43 is also shown as a loading control. Thylakoids were loaded in all cases, 0.5 μg of Chl for anti-PSBS and 0.3 μg of Chl for all the others. (B) NPQ kinetics of selected lines. Curves for WT and psbs KO, lhcsr1 KO, and lhcsr2 KO are shown in black, blue, green, and red, respectively. Averages and SD are calculated from at least five independent measures.
Table 1.
Pigment-binding properties of isolated mutants
| WT | psbs KO | lhcsr1 KO | lhcsr2 KO | |
| Chl a/Chl b | 2.56 | 2.58 | 2.52 | 2.60 |
| Chl/Car | 4.33 | 4.25 | 4.60 | 4.43 |
| WT | psbs lhcsr1 KO | psbs lhcsr1 lhcsr2 KO | |
| Chl a/Chl b | 2.56 | 2.42 | 2.57 |
| Chl/Car | 4.33 | 4.52 | 4.29 |
Pigment-binding properties of single and multiple mutants depleted in PSBS, LHCSR1, and LHCSR2, respectively. Chl a/b and Chl/carotenoids (Car) ratios are reported. SD is below 0.1 in the case of Chl a/b and 0.3 for Chl/Car.
When NPQ kinetics were recorded instead, KO plants showed large differences with respect to WT (Fig. 3B). The stronger phenotype was observed in the case of lhcsr1 KO, followed by psbs KO and lhcsr2 KO. This pattern is consistent with the Western blotting analysis shown in Fig. 3A: most LHCSR protein immunodetected in WT is synthesized by the PpLHCSR1 gene, and the other isoform is expressed at lower levels, explaining the small phenotype of lhcsr2 KO. It should be pointed out that by overloading lhcsr1 KO thylakoids, we were able to detect LHCSR2 by Western blotting, whose content is below the detection limit in Fig. 3, as shown in Fig. S3. Although the antibody might bind to the two isoforms with different affinity, these results strongly suggest that there is no major compensatory increase of LHCSR2 in the absence of LHCSR1.
It is interesting to observe that all KO mutants are still competent in NPQ, with their activity being comparable to NPQ levels measured in WT Arabidopsis (Fig. S4). Furthermore, the largest fraction of NPQ can be attributed to qE, the component dependent on low luminal pH, from its fast kinetics of relaxation and sensitivity to nigericin. This is different from the case of Arabidopsis, in which residual NPQ in the PSBS-less mutant npq4 is very small and slow in both rise and relaxation (10).
We report data from only one line for each genotype, but results have been replicated for all other selected lines. Also, we did not observe any significant difference in NPQ amplitude or PSBS/LHCSR protein accumulation between mutants having single or multiple insertions, suggesting that the effect of inserting multiple copies of DNA is not relevant for the observed phenotype (an example is shown in Fig. S5). In all cases, there was a strict correlation between the recombination event shown by PCR assay, the absence of the protein on Western blotting, and the amplitude of the NPQ phenotype, implying that the low NPQ phenotype was attributable to the KO mutations (Fig. S5). Moreover, transformations using two different plasmids carrying either zeocin or neomycin resistance were tested in the case of PSBS. In both cases, the phenotype was the same, confirming that effect of the presence/absence of PSBS on NPQ is much stronger than other possible secondary effects (Fig. S6).
Double and Triple KO Mutants.
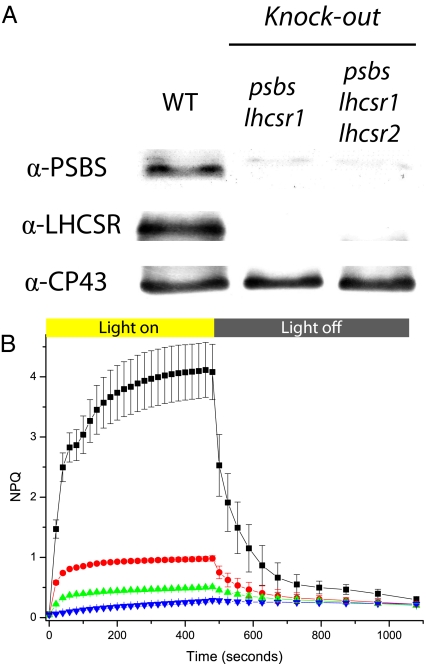
Once the role of each gene in NPQ was assessed, we performed further transformation on mutant backgrounds to obtain double and triple KO mutants. The presence of the mutation was always verified by PCR and Western blotting, as described above (Fig. 4A). The depletion of LHCSR1 in psbs KO background causes a further reduction in NPQ. However, this double mutant still retains a residual NPQ having qE properties, which can be attributed to the residual presence of LHCSR2. This hypothesis was verified by isolating triple mutants, where LHCSR2 was also missing. NPQ in the triple mutant was very low and completely depleted in its fast component, qE (Fig. 4B). In fact, the small residual NPQ was stable during at least 10 min of dark recovery following illumination, as observed in the PSBS-less npq4 mutant of Arabidopsis (10).
Fig. 4.
Phenotypes of P. patens psbs and lhcsr KO double and triple mutants. (A) Western blotting using antibody against LHCSR and PSBS. (B) NPQ kinetics of selected lines. Curves for WT, lhcsr1 KO, psbs lhcsr1 KO, and psbs lhcsr1 lhcsr2 KO are shown in black, red, green, and blue, respectively. Averages and SD are calculated from at least five independent measures.
Role of Individual LHCSR and PSBS Gene Products in Protection from High Light Stress.
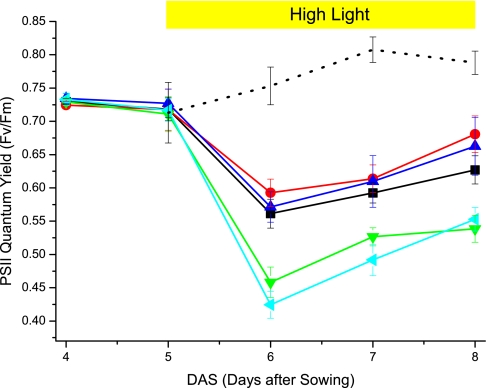
NPQ is a mechanism known to contribute to resistance to light stress in both higher plants and algae (8, 17, 28). To prove this is also the case in P. patens, we treated WT and mutant 5-day-old plants with high light intensities. Fv/Fm was measured every day to monitor PSII photodamage. As shown in Fig. 5, WT plants, on the shift to high light conditions, showed a drastic decrease in PSII efficiency, a clear sign of photoinhibition. PSII efficiency recovers over the following days, suggesting that plants start acclimating to the light conditions. Single KO mutants were similarly affected by the light treatment, whereas the double psbs lhcsr1 and the triple psbs lhcsr1 lhcsr2 KO mutants showed a stronger degree of PSII photoinhibition. This is a clear indication that the shift to high light conditions affects plants depleted in both PSBS and LHCSR more drastically. Despite experiencing a stronger photoinhibition, however, even these plants show partial recovery over the following days, demonstrating that other acclimating mechanisms acting with slower time courses are active in mosses independent of NPQ, as previously reported for plants and algae (29–31). In single KO mutants, phenotype on photoprotection is small, if any, suggesting that PSBS and LHCSR are able to provide a photoprotection capacity similar to WT, even in the absence of the other subunit, in these conditions.
Fig. 5.
Photoprotection capacity of NPQ-affected mutants. The photoprotection capacity of the previously described mutants has been compared with WT by treating 5-day-old plants with an additional 3 days of high light. PSII quantum yield, expressed as Fv/Fm, was monitored daily. WT, psbs KO, and lhcsr1 KO are shown in black, red, and blue, respectively, whereas psbs lhcsr1 and psbs lhcsr1 lhcsr2 KO double and triple mutants are shown in green and cyan. Values for a WT culture that was kept to control light conditions are also reported in black dots. Data from mutants grown in control conditions were omitted for clarity because they were indistinguishable from data on WT culture.
Discussion
PSBS and LHCSR Are Active Independently in P. patens NPQ.
Arabidopsis NPQ is well known to depend on the presence of the PSBS subunit of photosystem II (10, 11). Although the PSBS gene is conserved in the genome of some green algae (19), it was never detected as polypeptide (16). On the contrary, in the green alga C. reinhardtii, it has been shown recently that NPQ requires LHCSR, a different Lhc-like polypeptide previously called Li818. Genes encoding the LHCSR polypeptide are found in many algae groups, including diatoms (32).
PSBS gene and polypeptide have been identified in several plant species that also show comparable NPQ kinetics, and all data available suggest that NPQ depends on PSBS in vascular plants. Algae are a highly diversified and less characterized group of organisms; thus, we should be cautious before generalizing. Nevertheless, so far, PSBS polypeptide has never been detected in any green alga belonging to Viridiplantae, despite the presence of a corresponding gene in the genome (16). Thus, present knowledge is consistent with the hypothesis that NPQ activation relies on different mechanisms. This was proven to be true for Chlamydomonas and Arabidopsis, the two model systems for green algae and plants, respectively, which are the subject of the present study. Although the molecular mechanisms of LHCSR action are still unclear, available data in Arabidopsis suggest that PSBS acts as a modulator for antenna proteins (12, 21, 33, 34). Among different Lhc proteins, Lhcb3 and Lhcb6 were shown to be specifically involved in PSBS-dependent NPQ activation (12, 21) and are present only in plants. So far, no evidence has been obtained for their presence in algae (20), suggesting that the function of PSBS in algae, if any, is substantially different from that in plants.
P. patens is the only plant in which both PSBS and LHCSR gene sequences have been found (20). The report of PSBS and LHCSR EST sequences in T. ruralis suggests that they are present in other mosses as well. There is no indication at present that any vascular plants carry LHCSR-encoding sequences in their genomes, which suggests a change in the mechanisms of photoprotection in Viridiplantae during evolution. In this work, we have analyzed the role of LHCSR and PSBS polypeptides in P. patens by generating specific KO plants: all mutants showed a decreased NPQ phenotype, implying that both LHCSR and PSBS are active in triggering this photoprotection mechanism.
LHCSR and PSBS NPQ activities in P. patens are largely independent; in this study, we have shown that mutants lacking PSBS or most of the LHCSR protein (psbs and lhcsr1 KO, respectively) are still capable of significant NPQ. Although mutual interactions cannot be excluded, their effect appears to be additive, consistent with the observation that in conditions of maximal NPQ, fluorescence quenching in P. patens is faster and stronger than in Arabidopsis (Fig. 1). Consistently, lhcsr1 KO plants, missing most of the LHCSR protein, show an NPQ amplitude comparable to that of Arabidopsis plants (Fig. S4).
The mutant phenotype in high light stress experiments also supports this hypothesis; we observed strong light sensitivity only when PSBS and LHCSR were both absent (or largely depleted as in the case of psbs lhcsr1 double KO mutant). When only one protein was present, it was sufficient to activate enough NPQ to provide protection levels equivalent to WT in the experimental conditions tested.
Evolution of Heat Dissipation Mechanisms.
Presented results allow the proposal that P. patens retains the LHCSR-dependent NPQ mechanism found in Chlamydomonas, although additionally showing the newly evolved PSBS-dependent mechanism typical of land plants. Together, these mechanisms provide optimal photoprotection of Physcomitrella in high light conditions. Our results suggest that the PSBS-dependent mechanism of excess energy dissipation appeared during plant evolution before the ancestral LHCSR-dependent function was lost. This finding highlights the importance of photoprotection in the newly colonized land environment. In fact, because the PSBS-dependent NPQ mechanism evolved later, it could be established only as being superimposed on the ancestral mechanism so as to ensure a sufficient level of continual protection. In the absence of NPQ, in fact, a strong decrease in fitness and fast counterselection would be expected, as shown in the case of psbs-less Arabidopsis mutants (8) and consistent with the effect of the high light treatment in the triple mutant (Fig. 5).
Materials and Methods
Plant Growth.
Protonemal tissue of P. patens, Gransden WT strain, was grown on minimum PpNO3 medium (35) solidified with 0.8% Plant Agar (Duchefa Biochemie). Plants were propagated under sterile conditions on 9-cm Petri dishes overlaid with a cellophane disk (A.A. Packaging Limited), as previously described (20). Plates were placed in a growth chamber under controlled conditions: 24 °C, 16-h light/8-h dark photoperiod, and a light intensity of 40 μE·m−2·s−1 (control conditions). For high light tests, 5-day-old plants were moved from control to 350 μE·m−2·s−1, maintaining temperature and photoperiod.
NPQ Measurements.
In vivo chlorophyll fluorescence in P. patens was measured at room temperature with a Dual PAM-100 fluorometer (Heinz Walz GmbH, Germany), with saturating light of 6,000 μE·m−2·s−1 and actinic light of 830 μE·m−2·s−1. In the case of Arabidopsis, plants were grown at 100 μE·m−2·s−1 in an 8-h light/16-h dark photoperiod and were measured with 1,200 μE·m−2·s−1 of actinic light. Before measurements, plates were dark-adapted for 40 min at room temperature. The parameters Fv/Fm and NPQ were calculated as (Fm−Fo)/Fm and (Fm−Fm′)/Fm′ (36). Antenna size was determined by the rising time of PSII fluorescence in DCMU-treated plants.
Protoplast Transformation and KO Generation.
Genomic P. patens protonemal DNA was extracted (37) and used as starting template for all molecular cloning. All regions up and downstream of target coding sequences were amplified by PCR and subcloned into pGEM-T Vector (catalog no. A3600; Promega). Regions from PSBS (locus XM_001778511), LHCSR1 (locus XM_001776900), and LHCSR2 (locus XM_001768019) genes were successively cloned into BZRf/BNRr, BHRf, and BNRf plasmids (kindly provided by F. Nogue, Institut National de la Recherche Agronomique, Versailles, France), respectively. P. patens transformation was performed as in the study by Schaefer and Zrÿd (24) with minor modifications. Briefly, 5- to 6-day-old protonemal tissue was collected for protoplast generation and PEG-mediated transformation. Resistant colony selection started 6 days after transformation by transferring regenerated plants on culture media supplemented with the appropriate antibiotic [50 μg·L−1 G418 (Sigma-Aldrich); 30 μg·L−1 Hygromicyn B (Sigma-Aldrich), or 50 μg·L−1 zeocin (Duchefa Biochemie)]. Resistant colonies were transferred for 10 days on nonselective media and then again in antibiotic media to isolate stable transformants. Confirmation of DNA insertion was performed by PCR assay, as detailed in Table S1.
Thylakoid Extraction, SDS/PAGE, and Western Blotting Analyses.
Thylakoids from protonemal tissue (10- to 12-day-old plants) were prepared using an Arabidopsis protocol with minor modifications (20). After SDS/PAGE, proteins were transferred onto a nitrocellulose membrane (Sartorious AG) using a blot system from Biorad and were detected with specific polyclonal antibodies produced in the laboratory.
Supplementary Material
Acknowledgments
We are grateful to Giulia Bonente (Università di Verona, Verona, Italy) for providing high light- and low light-adapted Chlamydomonas cells and to Fabien Nogué [Institut Jean-Pierre Bourgin (IJPB), Institut National de la Recherche Agronomique (INRA), Versailles, France] for suggestions on moss transformation. A doctoral grant was awarded to C.G. by Fondazione Cassa di Risparmio di Padova e Rovigo (CaRiPaRo). A.A. and R.B. received financial support from the Italian Ministry of Research (FIRB PAREALLELOMICS, Grant RBIP06CTBR). T.M. received financial support from the Università di Padova (Grant CPDA089403).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002873107/-/DCSupplemental.
References
- 1.Barber J, Andersson B. Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 3.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 4.Scott AC, Glasspool IJ. The diversification of Paleozoic fire systems and fluctuations in atmospheric oxygen concentration. Proc Natl Acad Sci USA. 2006;103:10861–10865. doi: 10.1073/pnas.0604090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters ER. Molecular adaptation and the origin of land plants. Mol Phylogenet Evol. 2003;29:456–463. doi: 10.1016/j.ympev.2003.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Genty B, Briantais J-M, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta. 1989;990:87–92. [Google Scholar]
- 7.Niyogi KK. Safety valves for photosynthesis. Curr Opin Plant Biol. 2000;3:455–460. doi: 10.1016/s1369-5266(00)00113-8. [DOI] [PubMed] [Google Scholar]
- 8.Külheim C, Agren J, Jansson S. Rapid regulation of light harvesting and plant fitness in the field. Science. 2002;297:91–93. doi: 10.1126/science.1072359. [DOI] [PubMed] [Google Scholar]
- 9.Nilkens M, et al. Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim Biophys Acta. 2010;1797:466–475. doi: 10.1016/j.bbabio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Li XP, et al. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- 11.Li XP, Phippard A, Pasari J, Niyogi KK. Structure-function analysis of photosystem II subunit S (PsbS) in vivo. Funct Plant Biol. 2002;29:1131–1139. doi: 10.1071/FP02065. [DOI] [PubMed] [Google Scholar]
- 12.Betterle N, et al. Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J Biol Chem. 2009;284:15255–15266. doi: 10.1074/jbc.M808625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonente G, Howes BD, Caffarri S, Smulevich G, Bassi R. Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J Biol Chem. 2008;283:8434–8445. doi: 10.1074/jbc.M708291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss AZ, Ruban AV, Horton P. The PsbS protein controls the organization of the photosystem II antenna in higher plant thylakoid membranes. J Biol Chem. 2008;283:3972–3978. doi: 10.1074/jbc.M707410200. [DOI] [PubMed] [Google Scholar]
- 15.Finazzi G, et al. Nonphotochemical quenching of chlorophyll fluorescence in Chlamydomonas reinhardtii. Biochemistry. 2006;45:1490–1498. doi: 10.1021/bi0521588. [DOI] [PubMed] [Google Scholar]
- 16.Bonente G, et al. The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem Photobiol. 2008;84:1359–1370. doi: 10.1111/j.1751-1097.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 17.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 18.Koziol AG, et al. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 2007;143:1802–1816. doi: 10.1104/pp.106.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anwaruzzaman M, et al. Genomic analysis of mutants affecting xanthophyll biosynthesis and regulation of photosynthetic light harvesting in Chlamydomonas reinhardtii. Photosynth Res. 2004;82:265–276. doi: 10.1007/s11120-004-2439-y. [DOI] [PubMed] [Google Scholar]
- 20.Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T. In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS ONE. 2008;3:e2033. doi: 10.1371/journal.pone.0002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovács L, et al. Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell. 2006;18:3106–3120. doi: 10.1105/tpc.106.045641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkum AWD, Vesk M. In: Photosynthesis in Algae. Larkum AWD, Douglas SE, Raven JA, editors. The Netherlands: Kluwer; 2003. pp. 11–28. [Google Scholar]
- 23.Gunning BES, Schwartz OM. Confocal microscopy of thylakoid autofluorescence in relation to origin of grana and phylogeny in the green algae. Aust J Plant Physiol. 1999;26:695–708. [Google Scholar]
- 24.Schaefer DG, Zrÿd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 25.Nishiyama T, et al. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: Implication for land plant evolution. Proc Natl Acad Sci USA. 2003;100:8007–8012. doi: 10.1073/pnas.0932694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murén E, Nilsson A, Ulfstedt M, Johansson M, Ronne H. Rescue and characterization of episomally replicating DNA from the moss Physcomitrella. Proc Natl Acad Sci USA. 2009;106:19444–19449. doi: 10.1073/pnas.0908037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamisugi Y, et al. The mechanism of gene targeting in Physcomitrella patens: Homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 2006;34:6205–6214. doi: 10.1093/nar/gkl832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XP, Muller-Moule P, Gilmore AM, Niyogi KK. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA. 2002;99:15222–15227. doi: 10.1073/pnas.232447699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballottari M, Dall'Osto L, Morosinotto T, Bassi R. Contrasting behavior of higher plant photosystem I and II antenna systems during acclimation. J Biol Chem. 2007;282:8947–8958. doi: 10.1074/jbc.M606417200. [DOI] [PubMed] [Google Scholar]
- 30.Sirikhachornkit A, Shin JW, Baroli I, Niyogi KK. Replacement of alpha-tocopherol by beta-tocopherol enhances resistance to photooxidative stress in a xanthophyll-deficient strain of Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8:1648–1657. doi: 10.1128/EC.00124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda H, DellaPenna D. Tocopherol functions in photosynthetic organisms. Curr Opin Plant Biol. 2007;10:260–265. doi: 10.1016/j.pbi.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Coesel S, et al. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009;10:655–661. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruban AV, et al. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature. 2007;450:575–578. doi: 10.1038/nature06262. [DOI] [PubMed] [Google Scholar]
- 34.Teardo E, et al. Evidences for interaction of PsbS with photosynthetic complexes in maize thylakoids. Biochim Biophys Acta. 2007;1767:703–711. doi: 10.1016/j.bbabio.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Ashton NW, Grimsley NH, Cove DJ. Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta. 1979;144:427–435. doi: 10.1007/BF00380118. [DOI] [PubMed] [Google Scholar]
- 36.Demmig-Adams B, et al. Using chlorophyll fluorescence to assess the fraction of absorbed light allocated to thermal dissipation of excess excitation. Physiol Plant. 1996;98:253–264. [Google Scholar]
- 37.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.