Abstract
The RNA-dependent RNA polymerase core complex formed upon infection of Escherichia coli by the bacteriophage Qβ is composed of the viral catalytic β-subunit as well as the host translation elongation factors EF-Tu and EF-Ts, which are required for initiation of RNA replication. We have determined the crystal structure of the complex between the β-subunit and the two host proteins to 2.5-Å resolution. Whereas the basic catalytic machinery in the viral subunit appears similar to other RNA-dependent RNA polymerases, a unique C-terminal region of the β-subunit engages in extensive interactions with EF-Tu and may contribute to the separation of the transient duplex formed between the template and the nascent product to allow exponential amplification of the phage genome. The evolution of resistance by the host appears to be impaired because of the interactions of the β-subunit with parts of EF-Tu essential in recognition of aminoacyl-tRNA.
Keywords: protein biosynthesis, virus
Many RNA viruses encode an RNA-dependent RNA polymerase (RdRp) responsible for the replication of their genome. Because the viral genomes are limited in size, host-encoded proteins play important roles during most steps of viral infection (1). Often, the virally encoded RdRp interacts with host-encoded proteins during infection to form a complex, which ensures template specificity and polymerase activity.
Several viral RdRps recruit host proteins with a canonical role in protein biosynthesis. A classical example is the Qβ replicase complex consisting of the catalytic subunit encoded by the virus, the β-subunit, which hijacks the host translation elongation factors EF-Tu and EF-Ts as well as ribosomal protein S1 to form the Qβ replicase holoenzyme. Once assembled, the holoenzyme is capable of replicating the plus-stranded genome and generating the complementary minus strand. An additional host protein, Hfq, is required for efficient replication in vivo and in vitro (2). Contrary to the Qβ plus strand, the minus strand, as well as certain synthetic polyribonucleotides and small replicating RNAs (RQ RNAs) (3) are productive templates in the absence of Hfq and even with a Qβ replicase core complex lacking the S1 protein (4). The Qβ replicase amplifies the Qβ genome 104-fold in less than 1 hour during infection (5). Whereas the Qβ replicase exhibits extraordinary specificity toward its templates that prevents it from replicating host RNA, the template recognition elements remain poorly defined. Yet both the Qβ plus and minus strands contain the motif 5′…CCCAOH-3′, which is recognized by the replicase during initiation of replication (6).
In an uninfected bacterium, EF-Tu mediates the binding of amino-acylated tRNA (aa-tRNA) to the ribosomal A site via formation of a ternary complex, EF-Tu:GTP:aa-tRNA, and is released from the ribosome upon GTP hydrolysis in the form of EF-Tu:GDP. Subsequently, EF-Ts catalyzes the exchange of GDP for GTP on EF-Tu. In the Qβ replicase, the EF-Tu:EF-Ts subcomplex is necessary for initiation of replication of both the minus and plus strands (7) with EF-Tu being involved in template binding and recognition (8). EF-Ts may stabilize EF-Tu in a conformation with affinity for Qβ RNA templates instead of aa-tRNAs (9). Ribosomal protein S1 mediates recognition of the Qβ plus strand (8), whereas the RNA chaperone Hfq is believed to mediate the access of the β-subunit to the 3′ end of the plus strand (10).
The eukaryotic counterpart of EF-Tu, eEF1A, is present in RdRp complexes formed upon infection with bovine viral diarrhea virus (11), polio virus (12), and tobacco mosaic virus (13). Structures of a variety of RdRps have been determined in the apo state or in complex with nucleotides and/or RNA and thereby provided insight into the molecular mechanisms of initiation and elongation (14). However, no structures of RdRp complexes consisting of virus- and host-encoded proteins have been described. Here we report the structure of the Qβ replicase core complex consisting of the viral β-subunit in complex with elongation factors EF-Tu and EF-Ts. The structure reveals how the virus elegantly takes advantage of the normal function of EF-Tu in translation to replicate its own genome.
Results
Functional Relevance of the Crystallized Protein.
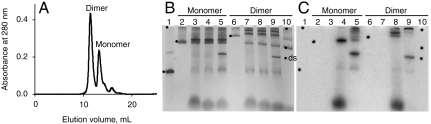
For crystallization, we used a fusion protein (consisting of EF-Ts, EF-Tu, and His6-tagged β-subunit), whose activity is similar to that of the noncovalent complex of the same proteins (15). A sequence encoding the cleavage site for tobacco etch virus (TEV) protease (16) was introduced between the genes encoding EF-Tu and the β-subunit to avoid aggregation of the fusion protein. Gel filtration in a high salt buffer resolved TEV-digested preparations of both the core enzyme (EF-Ts:EF-Tu:β-subunit) and the holoenzyme (EF-Ts:EF-Tu:β-subunit:S1), into two peaks corresponding to a monomer and dimer of the Qβ replicase (Fig. 1A) of the same subunit composition (Fig. S1A). Of these, only the dimer of the core replicase was crystallizable. The core dimer was virtually inconvertible to the monomer and vice versa, and no dimer formation was observed with the wild-type Qβ replicase core (Fig. S1 B and C). The specific activity of the dimeric and monomeric preparations as assayed in a poly(C)-directed reaction was about 10,000 units/mg (Fig. S1D). The dimer and monomer showed similar thermal stabilities and temperature optima (Fig. S1 E and F) as the wild-type core replicase. Also, the dimer and monomer were equally efficient in the amplification of RQ RNA (Fig. S1G).
Fig. 1.
Dimer and monomer of the Qβ replicase core. (A) Resolution of the TEV-digested fusion protein into dimer and monomer by gel filtration in a buffer containing 500 mM NaCl. (B) Silver stained gel and (C) its radioactivity pattern after nondenaturing PAGE of RQ135 RNA (lane 1), the monomer (lane 2), the dimer (lane 6), and complexes formed by the monomer and dimer in the presence of the RNA and GTP (lanes 3 and 7), plus CTP and [α-32P]UTP (lanes 4 and 8) or CTP, [α-32P]UTP, and ATP (lanes 5 and 9), and in the presence of GTP and no RNA (lane 10). The dots on the gel are radioactive markers identifying the positions of nonradioactive bands in C. Dot “ds” marks the position of the double-stranded RNA product.
Upon RNA binding, the monomer produced one slower migrating band during nondenaturing PAGE, whereas the dimer produced two bands, presumably containing one and two template molecules, respectively (Fig. 1B). Each of the RNA-bound bands become labeled in the presence of GTP, CTP, and [α-32P]UTP (Fig. 1C, lanes 4 and 8), when only a 5-nt-long product strand might have been synthesized on the 3′-terminal template sequence, 5′-…UAGCCC-3′ (3). The fact that such a short product remained tightly bound during the long electrophoresis procedure suggests that each of the RNA-bound bands represents the closed conformation of the replicative complex (17). When all four NTPs were present providing for the synthesis of full length product, the label was separated into a double-stranded product and high molecular weight complexes, which tend to be larger in the case of the dimer (Fig. 1C, lanes 5 and 9). Thus, the dimer complex retains its integrity during RNA synthesis, although it partially dissociates into monomers in the presence of GTP. This dissociation does not relate to RNA synthesis, because it was also observed in the absence of template (Fig. 1B, lane 10) and no label migrated ahead of the dimer band upon initiation (Fig. 1C, lane 8). Hence, the crystallizable dimeric Qβ replicase core represents a fully active complex indistinguishable from either the monomer or the wild-type core complex by a number of functional criteria demonstrating the functional relevance of the structure.
Structure of the Core Replicase.
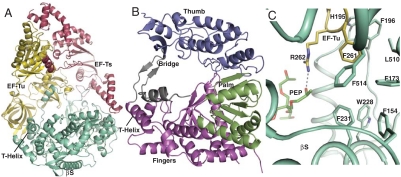
X-ray diffraction data extending to a maximum resolution of 2.5 Å (Table S1) were collected and the structure of the Qβ core replicase was determined by molecular replacement. In the asymmetric unit of the crystal, two monomers (Fig. 2A) of the core replicase related by a twofold rotation symmetry axis are present (Fig. S2C) and may correspond to the dimer found in solution. We have traced the same 1,219 residues per monomeric core replicase. In both copies, disordered are the EF-Ts residues 1–2, 283, and the histidine connecting it with EF-Tu; EF-Tu residues 1–8 and 42–63; and β-subunit residues 1–6, 114–118, 520–532, and 574–589. The β-subunit (Fig. 2B and Fig. S2D) has the appearance of a cupped right hand known from other polymerase structures (Fig. S3) (18) and can be divided into three domains: the palm, the thumb, and the fingers. Five conserved motifs, A–E (Fig. S4), with established functions in the catalytic mechanism are located in the palm domain (19). This domain is composed of two α-helices packed against one face of a four-stranded antiparallel β-sheet containing motifs A, C, and D (Fig. S3F). These secondary structure elements are organized with the topology βαββαβ also found in the RNA-recognition motif. A third helix is located on the opposite face of the four-stranded sheet, and this helix also packs against a small two-stranded, antiparallel β-sheet at the end of the palm domain containing the E motif. This sheet also forms the interface of the palm domain toward the thumb domain.
Fig. 2.
Structure of the Qβ replicase core complex. (A) Cartoon representation of the monomeric core replicase containing β-subunit (Green, labeled “βS”) in complex with EF-Tu (Yellow) and EF-Ts (Dark Red). (B) The domains of the β-subunit. The thumb domain is colored blue, the palm domain green, the fingers domain magenta, and the bridge region gray. (C) The PEP (Green Sticks) binding site on the β-subunit; a hydrogen bond with Arg262 of EF-Tu (Yellow) is shown as a dashed line. Binding of PEP possibly stabilizes the interaction of EF-Tu Phe261 with the hydrophobic cluster in the β-subunit.
The thumb domain (Fig. 2B and Figs. S2D and S4) consists of three segments: The first (residues 7–87) precedes the fingers domain and is α-helical, whereas the second (residues 408–494) follows the palm domain. The third, C-terminal segment (residues 560–573) contributes to the three-stranded β-sheet at the tip of the thumb domain, whereas the remaining 16 residues are disordered in agreement with mutational analysis showing that residues beyond 573 are dispensable (Table S2). The fingers domain (Fig. 2B and Fig. S2D) of the β-subunit consists of a four-stranded, antiparallel β-sheets and nine α-helices, and contains three segments: (i) residues 88–253 preceding the palm domain, (ii) residues 282–327 inserted between motifs A and B of the palm domain, and finally (iii) a single α-helix containing residues 505–518, which we will refer to as the T helix. The fingers domain contains a structurally conserved subdomain (residues 88–143, 208–253, and 282–327) that packs against the palm domain and contains secondary structure elements with a connectivity found in other RdRps as well (Fig. S3 G–L). In contrast, the residues 144–207 and the T helix appear to be structurally unique compared to other RdRps.
In other RdRps, the F motif is found in the N terminal of their sequences, where it connects the thumb domain with the fingers domain, and the motif is possibly involved in template unwinding (19). This motif cannot be identified in the β-subunit by sequence alignment. In the β-subunit, the function of the F motif is structurally fulfilled by the highly conserved loop 213–218 in the fingers domain with assistance from the loop 77–90 that connects the thumb and the fingers domain. The fingers domain of the β-subunit is also connected to the thumb domain through what we will refer to as the “bridge” (Fig. 2B and Fig. S2D) region crossing from the thumb to the fingers domain and back again. The bridge region consists of two flexible segments, residues 497–504 and 520–559, which flank the T helix in the fingers domain (Fig. S4). The region 520–532 is disordered in our structure. A small, two-stranded antiparallel β-sheet in the bridge is suspended between the thumb and fingers domains approximately 30 Å above the catalytic core structure of the palm domain. The single α-helix in the bridge region is located between the T helix and the second β-strand of the bridge (Fig. 2B). Two other RdRp structures contain elements resembling the bridge region. In the RdRp from hepatitis C virus, a β-hairpin extends from the thumb domain towards the palm domain (20). It has been suggested to undergo a conformational change to accommodate the RNA between the thumb and the fingers domain, which could position the hairpin in a location similar to that of the bridge β-sheet in the Qβ replicase. The second case is the λ3 reovirus RdRp, which has a C-terminal annular α-helical structure, known as the bracelet domain (21). Intriguingly, the β-subunit bridge together with EF-Tu domains 2–3 also forms an annular structure with a hole of similar size and located in the same orientation (see below).
Alignment of RdRp sequences from the virus family Leviviridiae suggests two subclasses corresponding to the Allolevivirus genus, to which the Qβ phage belongs, and the Levivirus genus (Figs. S4 and S5 A–F). In Levivirus, the polymerases are 30–50 residues shorter than in Allolevivirus because of deletions in especially the thumb domain (Figs. S4 and S5 E and F). Comparison of the structure of the β-subunit with this alignment suggests that all Leviviridiae polymerases have a structure similar to that of the β-subunit from the Qβ phage. In particular, they will all have the bridge region, although the Qβ protein has a longer bridge than all other Allolevivirus RdRps, which may be lacking the bridge α-helix.
Intermolecular Contacts in the Monomeric and Dimeric Qβ Replicase Core Complexes.
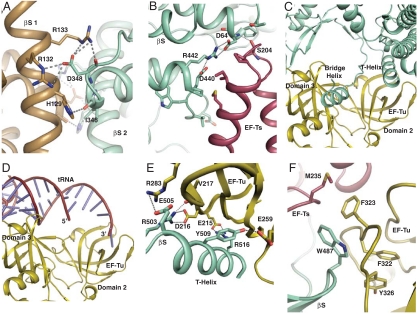
The interface between the two β-subunits within the putative dimer of the core enzyme is substantial with a buried surface area of 1,770 Å2 (Table S3). Residues 121–135 and 345–350 contribute to the dimer interface between the β-subunits (Fig. S6F), and these are highly conserved in Allolevivirus. In addition to van der Waals interactions between the two β-subunits, residues Arg132 and Arg133 of each β-subunit form salt bridges with Asp348 from the opposing β-subunit (Fig. 3A). The dimer is further stabilized by minor EF-Tu:EF-Tu contacts involving residues 38–40 (Fig. S6G) located immediately prior to the switch I region.
Fig. 3.
Intermolecular contacts within the Qβ replicase. Hydrogen bonds and electrostatic interactions are shown as dashed lines. (A) Details of the β-subunit (βS): β-subunit interface. (B) Contacts between the EF-Ts (Dark Red) coiled-coil motif and the β-subunit (Green) thumb domain. (C) The interface between the β-subunit and EF-Tu (Yellow) involving the T helix and bridge helix from the β-subunit and domains 1 and 3 from EF-Tu. (D). The complex of EF-Tu and aa-tRNA in the same orientation as in C. (E) Close-up of the β-subunit T-helix interaction with EF-Tu domain 2. (F) Insertion of the β-subunit thumb domain between EF-Tu domain 3 and EF-Ts.
The EF-Tu:EF-Ts:β-subunit complex forms a chair-shaped structure of maximum dimensions 116 × 79 × 80 Å, where the viral subunit forms the seat and host proteins the back of the chair (Fig. 2A). The chair has one armrest formed by the EF-Ts coiled-coil region (residues 187–226), which is in contact with the regions 62–64 and 436–442 of the β-subunit thumb domain (Fig 3B and Fig. S6 D and E), with the latter being completely conserved. EF-Tu:EF-Ts in the Qβ replicase complex superimpose with a rmsd of 1 Å over 592 Cα atoms onto the free complex from Research Collaboratory for Structural Bioinformatics (RCSB) entry 1EFU (22). Only the EF-Ts coiled-coil motif is significantly affected, and the interaction causes EF-Ts residues 190–217 in the coiled coil to rotate by 18° compared to EF-Tu:EF-Ts. The interface between EF-Ts and the β-subunit is substantial with a buried surface area of 1,522 Å2 (Table S3).
Even more extensive contacts are formed between the β-subunit and EF-Tu with a buried surface area of 3,766 Å2 (Table S3 and Fig. S6 A and B). The most prominent interactions are formed between EF-Tu domain 2 and the fingers domain, which includes the insertion of the T helix into the binding pocket for the CCA-aminoacyl group of aa-tRNA bound to EF-Tu:GTP (23) (Fig 3 C–E). In particular, Tyr509 of the β-subunit forms both hydrophobic interactions and a hydrogen bond with EF-Tu Glu259, and Arg516 of the β-subunit engages in a salt bridge with EF-Tu Glu215. The distant end of the β-subunit thumb domain is inserted between a region in EF-Tu domain 3 that also forms contacts with the aa-tRNA T stem (23) and the EF-Ts loop 233–239 (Fig. 3F). On the basis of the number of interactions, the region 481–491, not present in Levivirus, appears to play an important role in this interface involving all three proteins (Fig. 3F). A second conserved segment of the thumb domain, residues 566–573 neighboring the 481–491 region structurally, also interacts with both EF-Ts and EF-Tu. Finally, the α-helix of the β-subunit bridge region packs between EF-Tu domains 1 and 3 in the proximity of the switch regions I and II (residues 80–100) (Fig S6H). The β-subunit:EF-Tu interface is additionally stabilized by a molecule of the precipitant pentaerythritol propoxylate 5/4 PO/OH (PEP), which is trapped in a pocket formed by four helices in the fingers domain and the side chain of EF-Tu Arg262 that forms a hydrogen bond with PEP (Fig. 2C). The fixation of the EF-Tu arginine by PEP most likely results in increased hydrophobic interactions of the neighboring EF-Tu Phe261 with a hydrophobic cluster of six phenylalanine and one leucine side chains from the fingers domain (Fig. 2C), which keep the T helix in place for its interaction with the EF-Tu CCA pocket. A stabilizing effect of PEP is in agreement with its positive effect on replicase activity (Fig. S1D).
Regions of the β-Subunit with a Potential of Interacting with Substrates and Product.
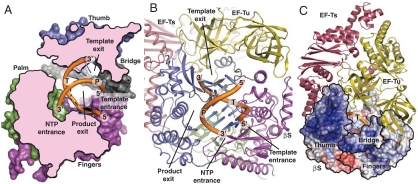
The locations of residues with a catalytic function during addition of NTP to the 3′ end of the product strand and residues creating the binding site for the transient template-product duplex are known from structures of initiating or elongating RdRps in complex with template, product, NTP, and metal ions (18). We have compared our apo structure with the structure of the Norwalk virus RdRp bound to CTP and product-template duplex (24) to identify regions of the β-subunit that are likely to interact with RNA, NTP, and metal ions during catalysis. On the basis of this comparison, the template is likely to enter through a channel formed within the fingers domain. It is bounded by a large loop containing residues 156–165, as well as one face of the four-stranded antiparallel β-sheet in the fingers domain (Fig. 4 A and B and Fig. S7). The β-subunit template channel is similar to those in the RdRps from Norwalk virus and hepatitis C virus (25). The putative NTP substrate entrance channel is located between residues 383–386 (from motif D) and 275–277 (in motif A) from the palm domain together with the segments 214–218 and 95–99 in the fingers domain (Fig. 4 A and B and Fig. S7). The substrate channel is flanked by five conserved lysine/arginine side chains that are capable of coordinating the incoming NTP. At the catalytic site, Asp274 in motif A and Asp359-Asp360 in motif C are expected to coordinate two Mg2+ ions that can mediate catalysis according to the two-metal-ion mechanism (18, 26). The strictly conserved Lys214 and Arg220 are likely to form electrostatic interactions with the NTP phosphates as well. Nucleic acid polymerases have been suggested to use as a general acid to protonate the pyrophosphate-leaving group, and in the polio virus RdRp Lys359 in motif D has been assigned this function (27). On the basis of our structure, we suggest that the strictly conserved Lys386 in motif D of the β-subunit can fulfill a similar role.
Fig. 4.
RNA binding and surface properties of the β-subunit. The double-stranded RNA in the internal cavity of the β-subunit (colored as in Fig. 2B) was docked by comparison with the structure of the Norwalk virus RdRp, PDB ID code 3BSN. Template and product strand are labeled “T” and “P,” respectively, and 3′ and 5′ ends are labeled as well. (A) Cross-section through the β-subunit displaying the suggested template and NTP entrance channels together with the template exit channel. (B) Cartoon representation of the core replicase indicating the four putative channels. (C) Surface representation displaying the electrostatic potential of the β-subunit colored according to charge (Blue 10 kt/e, White 0, Red -10 kt/e) with EF-Tu and EF-Ts shown in cartoon mode. The positively charged blue surface patch containing elements of the bridge and thumb domains of the β-subunit is a possible binding site for the S1 protein, template, and product. The thumb, bridge, and fingers domains of the β-subunit are indicated on the surface representation with a black outline.
A large internal cavity in the β-subunit appears to be available for binding a product-template A-form RNA duplex as previously observed in RdRps from Picornaviridae and Caliciviridae (18). The base of this cavity is formed by the palm domain but flanked on each side by the thumb and the fingers domain (Fig. 4 A and B and Fig. S7). The bridge limits the cleft above the catalytic center and is located at the periphery of the predicted path of the duplex, suggesting an important function of the bridge as a strand separator. Coarse modeling indicates that the 5′ end of the product strand would collide with the β-sheet of the bridge after synthesis of 5–6 phosphodiester bonds. The product strand may thereby become single-stranded and exits the internal cavity before a duplex length of 6–7 base pairs is reached. One option is that it exits through the opening between the bridge, thumb, and fingers domains, but this already harbors the template entrance channel. Much more space is available in an opening between the palm and thumb domains, EF-Tu domain 3, and the EF-Ts coiled coil (Fig. 4 A and B). In contrast, the 3′ end of the template strand could maintain an A-form RNA conformation even after unwinding and pass through the annular hole between the bridge region, the thumb domain β-sheet, and EF-Tu domains 2–3 (Fig. 4 and Fig. S7). An exit path through an annular hole in the λ3 bracelet has also been suggested for the reovirus RdRp and suggested to be accessible to both single- and double-stranded RNA (21), but, unless significant conformational changes occur between the apo state of our structure and the actual replicating state, the hole in the Qβ replicase can accommodate only single-stranded RNA.
Putative Binding Sites for RNA and the S1 Protein.
Mapping of the electrostatic potential onto the surface of the β-subunit identifies a rather large, positively charged and partly conserved patch covering the distant end of the thumb domain and the outside of the bridge β-sheet loop (Fig. 4C and Fig. S5 A–D) with potential for interacting with a negatively charged macromolecule. Prime candidates are template/product RNA or the strongly acidic protein S1. The Qβ replicase holoenzyme can be dissociated into two enzymatically inactive complexes (28), S1:β-subunit and EF-Tu:EF-Ts, indicating that the main association of S1 with the Qβ replicase complex occurs via the β-subunit. Binding of S1 at this positively charged patch would place S1 in close proximity to the suggested template entrance and exit holes.
Discussion
Maintenance of High Amplification Efficiency.
The Qβ genome is amplified with extraordinary efficiency both in vivo and in vitro, which is partly achieved because the Qβ replicase is able to utilize both the minus and the plus strands as a template, thus rendering exponential amplification possible (29, 30). In contrast, double-stranded RNA is rejected as a template (31). Thus, for an RNA strand to serve as a template in multiple rounds of RNA synthesis, extensive base pairing between template and product strands should be prevented. Qβ replicase is the only RdRp with a proven ability to prevent the unwound template and product strands from reannealing (31), and this capacity enables exponential RNA amplification (Fig. S1G). The explanation has remained an enigma, but our structure suggests two structural features that provide a credible solution to the problem. First, the bridge region is probably involved in unwinding the duplex between the template and the nascent part of the product when the duplex reaches a maximum size of 6–7 base pairs. Second, the strong, intrastrand secondary structures typical of efficient Qβ replicase templates (32) may assist the Qβ replicase in obtaining the single-stranded state of template for subsequent cycles of replication. The separation between the proposed exit channels for template and product seems to favor formation of such intrastrand secondary structure relative to formation of extensive template-product duplex. The suggested encounter between RNA and the bridge region may in addition cause a conformational change that potentially could affect the interaction between the β-subunit and EF-Tu. Docking of the RNA duplex in the active sites and the suggested locations of the RNA entry/exit sites within the replicase dimer (Fig. S7F) does not support channeling of RNA between the two β-subunits in agreement with our functional studies (Fig. 1 and Fig. S1).
Functional Significance of Interactions Between the β-Subunit and Host Proteins.
Our structure provides a unique example of a viral RdRp complex containing host proteins, and features of this complex may apply to other cases, in particular to those viral RdRp complexes containing the eukaryotic homologue of EF-Tu, eEF1A. The basic catalytic machinery of the β-subunit is not in direct contact with the host proteins in agreement with the ability of the viral subunit to function in the elongation phase of RNA replication in their absence (33). Quantitatively, the interface between the β-subunit and EF-Ts is relatively small compared to the β-subunit:EF-Tu interface. Thus, it is interesting to consider why the β-subunit has evolved to depend on the EF-Tu:EF-Ts complex rather than on EF-Tu alone. In the bacterial cell, all EF-Ts is expected to be in complex with EF-Tu, whereas only 10–20% of the EF-Tu is found in complex with EF-Ts (34). The majority of the remaining EF-Tu is likely to be in complex with abundant aa-tRNA. Hence, if the β-subunit depended only on EF-Tu, it would encounter a strong, direct competition from aa-tRNA for the binding to EF-Tu. However, the vital importance of the coiled-coil domain of EF-Ts for formation of the Qβ replicase complex (9) and the observed interaction of the β-subunit with the coiled coil shows that virus also takes direct advantage of the presence of EF-Ts.
The existence of a correlation between the ability of EF-Tu for binding tRNA and its function within the Qβ replicase has been a matter of dispute. Our structure revives this debate, because the CCA-binding pocket of EF-Tu interacts directly with the β-subunit T helix. Originally, the replacement of endogenous EF-Tu with chemically or enzymatically modified variants incapable of binding tRNA was found not to affect the functionality of EF-Tu in the Qβ replicase complex (7). More recent studies of Qβ replicase complexes containing mutant species of EF-Tu (Table S2) with defects in tRNA binding showed a direct correlation between the tRNA-binding capacity of the mutants and their ability to sustain replication when part of a Qβ replicase complex (35), which is in agreement with our structure showing the binding of the β-subunit T helix to the EF-Tu CCA-binding pocket. A striking consequence of this interaction is that the host is unlikely to evolve resistance towards Qβ by mutations in the aminoacyl-binding pocket, because this could interfere with its normal function. A close parallel is the recognition of elongation factor 2 by pseudomonas exotoxin A at a site involved in contacts with the decoding center of the ribosome (36). Another intriguing question raised by the structure, and in particular the proposed location of the template exit channel, is whether the template CCA-3′ end at some point competes with the β-subunit T helix for binding to the CCA-aminoacyl pocket of EF-Tu.
Concluding Remarks.
The elucidation of the function of the host proteins in viral RdRp complexes is of potential importance in drug development against pathogenic viruses. Here we provide a description at the atomic level of a viral RdRp complex containing host proteins. In our case, the basic catalytic machinery is well separated from the host proteins, which seem to provide a stabilizing scaffold for the β-subunit and perhaps provide interaction sites for the template both before and after initiation of replication. Qβ replicase is the most efficient in vitro system for nucleic acid amplification, far more efficient than PCR or isothermal amplification systems (17). However, the utilization of Qβ replicase for amplification of desired sequences has yet to be realized because of its puzzling and extraordinary template specificity requiring neither promoters nor primers (7, 8). Although the described structure does not provide an immediate solution to this problem, it paves a way for a rational approach towards reaching this goal.
Materials and Methods
Detailed experimental procedures for all techniques used are given in SI Methods.
Expression and Purification.
The expression vector pBAD33Ts-Tu-β-3 (15) was modified to insert a TEV protease cleavage site in the fusion protein EF-Ts–EF-Tu–TEV–βS–6xHis. Escherichia coli BL21 (DE3) transformed with the resulting plasmid was grown in LB medium and induced with L-arabinose. Harvested cells were lysed by sonication. The fusion protein was purified by Ni2+ chelate chromatography and subsequently digested by TEV protease. The cleaved fusion protein was further purified by hydrophobic interaction chromatography and gel filtration.
Formation of Replicative Complexes.
Formation of replicative complexes was detected by a gel shift assay. The monomeric or dimeric form of Qβ replicase was incubated with a 139-nt-long derivative (17) of the minus strand of RQ135-1 RNA (3) in reaction buffer containing GTP and, where indicated, CTP and α-32P UTP, with or without ATP. The reaction conditions were adjusted to keep the molar amount of RNA product lower than the amount of enzyme. Subsequently, the samples were subjected to nondenaturing electrophoresis through an 8% polyacrylamide gel for 2 hours at 10–12 °C. Subsequently, the gel was silver stained and dried. 32P-labeled bands were revealed with a phosphor storage system and quantified by measuring intensities of the resulting bands.
Structure Determination.
Crystals were grown by vapor diffusion at 4 °C with a reservoir buffer containing 0.2 M KCl, 0.05 M Hepes-NaOH pH 7.5, and 27–30% vol/vol PEP. Reservoir buffer was mixed 1∶1 with dimeric core replicase (36 mg/mL). Crystals were soaked prior to flash freezing in a buffer containing 0.2 M KCl, 0.05 M Hepes-NaOH pH 7.5, and 35% PEP. Diffraction data were processed with XDS (37). Two copies of the EF-Tu:EF-Ts complex were located with PHASER (38), and RESOLVE located significant parts of the β-subunits after density modification. In an iterative manner, the remaining parts were build in COOT (39) and the resulting model refined with PHENIX.REFINE (40).
Supplementary Material
Acknowledgments.
We are grateful to K.M. Nielsen and G. Hartvigsen for excellent technical assistance, the staffs at European Synchrotron Radiation Facility and Swiss Light Source beam lines for help with data collection, M. Behrens for small angle X-ray scattering analysis, and M. Blaise and L. Sanderson for discussions. G.R.A. was supported by the Danish Science Research Council, Danscatt, the Vilhelm Petersen foundation, the Danish National Research Foundation, and a Hallas-Møller stipend from the Novo-Nordisk foundation. A.B.C. was supported by the Russian Foundation for Basic Research and the Molecular and Cell Biology Program of the Russian Academy of Sciences. C.R.K. was supported by the Danish Science Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors are deposited at the RCSB protein data bank as entry 3MMP.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003015107/-/DCSupplemental.
References
- 1.Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su Q, Schuppli D, Tsui HCT, Winkler ME, Weber H. Strongly reduced phage Qβ replication, but normal phage MS2 replication in an Escherichia coli K12 mutant with inactivated Qβ host factor (hfq) gene. Virology. 1997;227:211–214. doi: 10.1006/viro.1996.8302. [DOI] [PubMed] [Google Scholar]
- 3.Munishkin AV, et al. Efficient templates for Qβ replicase are formed by recombination from heterologous sequences. J Mol Biol. 1991;221:463–472. doi: 10.1016/0022-2836(91)80067-5. [DOI] [PubMed] [Google Scholar]
- 4.Kamen R, Kondo M, Römer W, Weissmann C. Reconstitution of Qβ replicase lacking subunit with protein-synthesis-interference factor i. Eur J Biochem. 1972;31:44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 5.Weissmann C. The making of a phage. FEBS Lett. 1974;40:S10–18. doi: 10.1016/0014-5793(74)80684-8. [DOI] [PubMed] [Google Scholar]
- 6.Tretheway DM, Yoshinari S, Dreher TW. Autonomous role of 3′-terminal CCCA in directing transcription of RNAs by Qβ replicase. J Virol. 2001;75:11373–11383. doi: 10.1128/JVI.75.23.11373-11383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenthal T, Carmichael GG. RNA replication: Function and structure of Qβ replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 8.Brown D, Gold L. RNA replication by Qβ replicase: A working model. Proc Natl Acad Sci USA. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karring H, et al. Qβ-phage resistance by deletion of the coiled-coil motif in elongation factor Ts. J Biol Chem. 2004;279:1878–1884. doi: 10.1074/jbc.M306605200. [DOI] [PubMed] [Google Scholar]
- 10.Schuppli D, et al. Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc Natl Acad Sci USA. 1997;94:10239–10242. doi: 10.1073/pnas.94.19.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CM, Perez DR, French R, Merrick WC, Donis RO. The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J Gen Virol. 2001;82:2935–2943. doi: 10.1099/0022-1317-82-12-2935. [DOI] [PubMed] [Google Scholar]
- 12.Harris KS, et al. Interaction ofpoliovirus ppolypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 13.Yamaji Y, et al. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 2006;347:100–108. doi: 10.1016/j.virol.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 14.Ortin J, Parra F. Structure and function of RNA replication. Annu Rev Microbiol. 2006;60:305–326. doi: 10.1146/annurev.micro.60.080805.142248. [DOI] [PubMed] [Google Scholar]
- 15.Kita H, et al. Functional Qβ replicase genetically fusing essential subunits EF-Ts and EF-Tu with beta-subunit. J Biosci Bioeng. 2006;101:421–426. doi: 10.1263/jbb.101.421. [DOI] [PubMed] [Google Scholar]
- 16.Carrington JC, Dougherty WG. A viral cleavage site cassette: Identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc Natl Acad Sci USA. 1988;85:3391–3395. doi: 10.1073/pnas.85.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ugarov VI, Demidenko AA, Chetverin AB. Qβ replicase discriminates between legitimate and illegitimate templates by having different mechanisms of initiation. J Biol Chem. 2003;278:44139–44146. doi: 10.1074/jbc.M305992200. [DOI] [PubMed] [Google Scholar]
- 18.Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bressanelli S, et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage—Structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 22.Kawashima T, Berthet-Colomunas C, Wulff M, Cusack S, Leberman R. The structure of the Escherichia coli EF-Tu·EF-Ts complex at 2.5 Å resolution. Nature. 1996;379:511–518. doi: 10.1038/379511a0. [DOI] [PubMed] [Google Scholar]
- 23.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 24.Zamyatkin DF, et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283:7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 25.Bressanelli S, Tomei L, Rey FA, De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 27.Castro C, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamen R. Characterization of the subunits of Qβ replicase. Nature. 1970;228:527–533. doi: 10.1038/228527a0. [DOI] [PubMed] [Google Scholar]
- 29.Dobkin C, Mills DR, Kramer FR, Spiegelman S. RNA replication: Required intermediates and the dissociation of template, product, and Qβ replicase. Biochemistry. 1979;18:2038–2044. doi: 10.1021/bi00577a030. [DOI] [PubMed] [Google Scholar]
- 30.Chetverin AB. Replicable and recombinogenic RNAs. FEBS Lett. 2004;567:35–41. doi: 10.1016/j.febslet.2004.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissmann C, Feix G, Slor H. In vitro synthesis of phage RNA: The nature of the intermediates. Cold SH Q B. 1968;33:83–100. doi: 10.1101/sqb.1968.033.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Brown D, Gold L. Selection and characterization of RNAs replicated by Qβ replicase. Biochemistry. 1995;34:14775–14782. doi: 10.1021/bi00045a019. [DOI] [PubMed] [Google Scholar]
- 33.Landers TA, Blumenthal T, Weber K. Function and structure in ribonucleic acid phage Qβ ribonucleic acid replicase. J Biol Chem. 1974;249:5801–5808. [PubMed] [Google Scholar]
- 34.Neuhard J, Nygaard P. Purines and pyrimidines. In: Ingraham JL, et al., editors. Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Vol 1. Washington, DC: Am Soc Microbiology; 1987. pp. 445–473. [Google Scholar]
- 35.Mathu SGJ, Knudsen CR, van Duin J, Kraal B. Isolation of Qβ polymerase complexes containing mutant species of elongation factor Tu. J Chromatogr B. 2003;786:279–286. doi: 10.1016/s1570-0232(02)00811-5. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen R, et al. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436(7053):979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. XDS. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. Vol F. Dordrecht: Kluwer Academic; 2001. [Google Scholar]
- 38.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.