Abstract
Lipopolysaccharide (LPS) of Gram-negative bacteria is a common pathogen-associated molecular pattern (PAMP) that induces potent innate immune responses. The host immune response against LPS is triggered by myeloid differentiation factor 2 (MD-2) in association with Toll-like receptor 4 (TLR4) on the cell surface. The MD-2/TLR4-mediated LPS response is regulated by the evolutionarily related complex of MD-1 and Toll-like receptor homolog RP105. Here, we report crystallographic and biophysical data that demonstrate a previously unidentified direct interaction of MD-1 with LPS. The crystal structure of chicken MD-1 (cMD-1) at 2.0 Å resolution exhibits a β-cup-like fold, similar to MD-2, that encloses a hydrophobic cavity between the two β-sheets. A lipid-like moiety was observed inside the cavity, suggesting the possibility of a direct MD-1/LPS interaction. LPS was subsequently identified as an MD-1 ligand by native gel electrophoresis and gel filtration analyses. The crystal structure of cMD-1 with lipid IVa, an LPS precursor, at 2.4 Å resolution revealed that the lipid inserts into the deep hydrophobic cavity of the β-cup-like structure, but with some important differences compared with MD-2. These findings suggest that soluble MD-1 alone, in addition to its complex with RP105, can regulate host LPS sensitivity.
Keywords: lipopolysaccharide, MD-2, RP105, TLR4, innate immunity
Lipopolysaccharide (LPS, endotoxin) in the outer membrane of Gram-negative bacteria is a pathogen-associated molecular pattern (PAMP) that potently stimulates host innate immunity (1, 2). Delayed or insufficient immune response against LPS results in uncontrolled infection, but overamplified responses can lead to life-threatening septic shock manifested by hypotension, hypoperfusion, and multiple organ failure (3, 4). Immune responses to LPS are mediated by a complex composed of myeloid differentiation factor 2 (MD-2) and Toll-like receptor 4 (TLR4) (5, 6).
MD-2 is coexpressed and forms a stable heterodimeric complex with TLR4 on the cell surface (5, 7). MD-2 folds into a β-cup-like architecture, and a large hydrophobic cavity between the two β-sheets provides a major binding site for LPS (8, 9). The recently determined crystal structure of the LPS-bound TLR4/MD-2 complex has led to a proposed mechanism for LPS-induced TLR4 activation (9), where five of the six acyl chains of LPS are completely embedded inside the MD-2 cavity. The remaining acyl chain bulges out of the cavity, but is stabilized through hydrophobic interactions with a second TLR4/MD-2 heterodimer. This LPS-induced oligomerization would bring the two intracellular domains of TLR4 into close proximity, resulting in recruitment of intracellular adaptors to initiate signaling (10). MD-2 is also secreted in a soluble form that confers responsiveness against LPS (11).
MD-1 and MD-2 are both members of group I of the MD-2-related lipid-recognition (ML) family that is characterized by a lipid binding function (12). MD-1 shares ~20% sequence identity with MD-2. MD-1 is associated with and functions with radioprotective 105 (RP105), which is homologous to TLR4 with ~30% sequence identity (13, 14). MD-1 is indispensable for cell surface expression of RP105 (13–15). RP105 and TLR4 are both expressed on macrophages, dendritic cells, and B cells (16). These common features indicate evolutionary relatedness between MD-1/RP105 and MD-2/TLR4. Furthermore, these two receptor complexes are functionally related, as MD-1/RP105 regulates MD-2/TLR4-mediated LPS responses. In dendritic cells and macrophages, MD-1/RP105 down-regulates LPS responses by interacting with MD-2/TLR4 (16, 17). In B cells, the MD-1/RP105 complex promotes LPS-driven cell proliferation and antibody production (18, 19).
Despite the evolutionary and functional connection between MD-1 and MD-2, LPS binding to MD-1 has not yet been reported under assay conditions that were applied for detection of the MD-2/LPS interaction (20, 21). Thus, it has been generally accepted that MD-1 is not able to bind LPS. Interestingly, recent findings have shown that house dust mite allergens, such as Der p 2 and Der f 2, which are distant homologs of MD-2, also interact with LPS (22–24). Given that MD-1 is the closest homolog of MD-2, it seemed curious that MD-1 was reported not to interact with LPS (12).
Here, we present the crystal structure of chicken MD-1 (cMD-1) at 2.0 Å resolution. The cMD-1 structure reveals a β-cup-like fold, similar to that seen in MD-2, but with large structural variation in loops that might be involved in ligand binding. The MD-1 structure houses a large hydrophobic cavity that, unexpectedly, was found to contain additional long and continuous electron density, suggesting that MD-1 can accommodate a lipid-like molecule, such as LPS. This finding led to our reevaluation of a possible MD-1/LPS interaction by biophysical and crystallographic methods. Native electrophoresis and gel filtration analyses provided strong evidence for a physical interaction between MD-1 and LPS. Moreover, the interaction and mode of binding were visualized in the crystal structure of cMD-1 complexed with an LPS precursor, lipid IVa at 2.4 Å resolution. The structure indicates that the MD-1 cavity hosts one lipid IVa molecule, but in a different binding mode from MD-2.
Results
Overall Structure of cMD-1.
The cMD-1 structure was initially determined at 3.0 Å resolution by single isomorphous replacement with anomalous scattering (SIRAS) using a PIP derivative (Table S1). The final 2.0 Å structure (cMD-1 residues 21–158 with the two C-terminal residues 159–160 excluded due to poor electron density) contains two molecules in the asymmetric unit that exhibit essentially identical structures with an rmsd value of 0.08 Å for 138 Cα atoms.
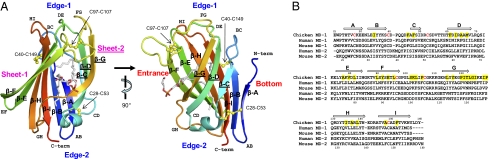
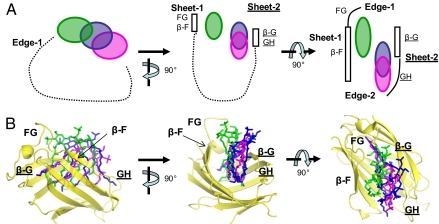
cMD-1 adopts a β-cup-like fold, consisting of nine β-strands that form two antiparallel β-sheets (Fig. 1). Six β-strands (β-A, β-B, β-I, β-H, β-E, and β-F) contribute to sheet-1, and the remaining three β-strands (β-C, β-D, and β-G) to sheet-2. The nine β-strands are interconnected by eight loops designated AB, BC, CD, DE, EF, FG, GH, and HI. Loops BC/DE/FG/HI and the N-terminal end form one edge (edge-1) of the β-cup, and loops AB/CD/EF/GH along with the C-terminal end contribute to the other (edge-2).
Fig. 1.
cMD-1 structure and sequence. (A) Overall structure of cMD-1 and its hydrophobic cavity. The cMD-1 structure is shown in ribbons that are colored from N terminus (blue) to C terminus (red). Labels for β-strands in sheet-2 are underlined. Disulfide bonds are shown in yellow ball-and-stick models, and a putative PGT molecule inside the MD-1 cavity is shown in sticks (carbon, gray; oxygen, red; phosphorus, orange). (B) MD-1 and MD-2 sequence alignment. cMD-1 cysteine residues that form disulfide bonds are colored in red, and cMD-1 residues that make contacts with lipid IVa are highlighted in yellow. cMD-1 β-strands are shown as arrows above the cMD-1 sequence.
cMD-1 contains three intramolecular disulfide bonds between C28 and C53; C40 and C149; and C97 and C107 (Fig. 1). The C40–C149 and C97–C107 disulfide bridges introduce structural constraints into the relatively flexible BC and FG loops by linking each loop to an ordered β-strand (β-I and β-F, respectively). The C28–C53 disulfide links β-A of sheet-1 to loop CD of sheet-2, and may stabilize the β-cup-like structure by connecting the two discontinuous β-sheets.
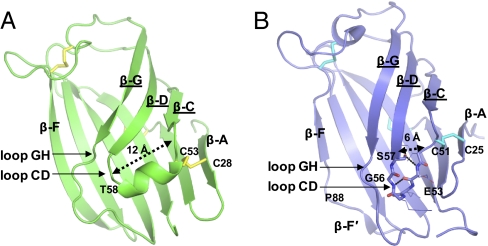
MD-1 and MD-2 of the ML family group I display similar overall structures with identical β-strand organization and disulfide bonds (Fig. 2). However, the rmsd of 2.8 Å between the MD-1 and MD-2 structures indicates some regions have substantial differences. The most pronounced structural deviations are found in the loops, particularly in the CD and GH loops of edge-2 that are involved in ligand interaction (see text following; Fig. 2). In addition, a noticeable change is found in β-strand F. MD-1 β-F is 11 residues long (Fig. 2A), but the corresponding region in MD-2 is broken into two short β-strands, β-F and β-F′, due to disruption of the main-chain H-bond network around Pro88 (Fig. 2B).
Fig. 2.
Structural comparison between cMD-1 (A) and hMD-2 (B; PDB ID code 2e56). Regions that differ between MD-1 and MD-2 are highlighted.
Hydrophobic Cavity of cMD-1 and Binding of a Lipid-Like Molecule.
The two β-sheets of cMD-1 form a cavity that is open to solvent through a channel enclosed by β-F, loop FG, β-G, and loop GH (Fig. 1 and Fig. S1A). The cMD-1 cavity (11 Å × 8 Å) is smaller than that of MD-2 (20 Å × 8 Å) due to partial closure of its entrance (Fig. S1). Accordingly, the internal volume of the cMD-1 cavity (~1,300 Å3, as calculated by CASTp server) (25) is smaller than that of MD-2 (1,700–1,900 Å3).
The partial closure of the cMD-1 cavity entrance mainly results from a pronounced inward shift of loop GH (residues 122–125) toward β-F, compared with the equivalent residues in MD-2 (Fig. 2 and Fig. S1). The neighboring residues 58–60 from loop CD follow a similar structural shift, suggesting a synchronized movement of loops GH and CD. Thus, the size of the cavity entrance is remotely governed by the structure of loop CD. In MD-1, a large bulge (~12 Å) is formed in the middle of loop CD by a six-residue 310 helix that pushes the GH loop toward β-F (Fig. 2A). In MD-2, a tight β-turn is found in loop CD that keeps its flanking regions close together (~6 Å; Fig. 2B). Formation of the β-turn in the MD-2 CD loop is driven by a main-chain H bond between Glu53 and Gly56, and further stabilized by another H bond between main chain of residue 53 and the side-chain hydroxyl of residue 57 [Ser57 in human MD-2 (hMD-2) and Thr57 in mouse MD-2 (mMD-2)]. Consequently, β-G and loop GH, proximal to the C-terminal end of loop CD, adopt a conformation that maintains a wider entrance to the cavity in MD-2. Thus, loops GH and CD appear to act as a single, structural dynamic unit that regulates access to the MD-1 and MD-2 cavities.
The internal surface of the cavity is highly hydrophobic with only two hydrophilic residues (Glu94 and Lys103) located at the cavity entrance. This observation suggests that any MD-1 ligand should be an amphipathic lipid-like molecule (i.e., a lipid molecule containing extended hydrophobic acyl chains with a more polar head group). Interestingly, the cMD-1 structure reveals additional long, continuous, two-pronged electron density that penetrates deep into the hydrophobic cavity with a connecting bridge between the prongs at the cavity entrance (Fig. S2). Above the bridge, additional weak electron density for a polar head group extends from the surface of cMD-1. Given the shape of the electron density and the nature of its environment, a phospholipid molecule, 1-myristoyl-2-palmitoyl-3-phosphatidylglycerol (PGT), was built into the electron density, although other abundant cellular lipids are certainly a possibility. The PGT head group exhibits weak electron density, and its structure is slightly different in the two molecules in the crystal asymmetric unit, presumably due to flexibility or heterogeneity. However, the glycerol backbone and almost all of the two acyl chains show clear and interpretable electron density. Thus, this crystallographic evidence suggests that cMD-1 binds a lipid-like molecule in its hydrophobic cavity.
Interaction of MD-1 with LPS.
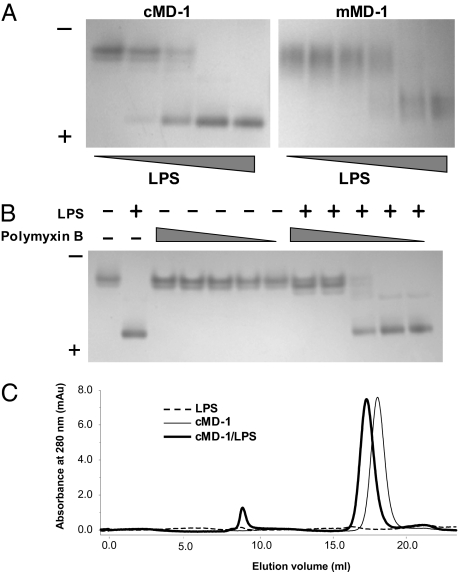
This structural observation led us to hypothesize that MD-1 is able to interact with LPS. To investigate a possible interaction between MD-1 and LPS, native PAGE was performed using purified cMD-1 as well as mouse MD-1 (mMD-1). As hypothesized, the mobility of MD-1 was enhanced by the negative charges of bound LPS in a dose-dependent manner (Fig. 3A). To test binding specificity, we examined whether cMD-1 competes for LPS with polymyxin B, a cyclic amphipathic peptide (Fig. 3B). Polymyxin B binds the lipid A moiety of LPS, resulting in neutralization of LPS bioactivity (26). Polymyxin B itself did not modulate the mobility of cMD-1 but did block the LPS-induced band shift of cMD-1, demonstrating that cMD-1 specifically binds the lipid A moiety (Fig. 3B). Furthermore, the cMD-1/LPS mixture eluted before cMD-1 in a gel filtration column (Fig. 3C), where the peak shift (0.7 mL) corresponds to the ~5-kDa difference in molecular weight of the LPS used in the experiment. Thus, we conclude that MD-1 directly interacts with LPS.
Fig. 3.
Interaction of MD-1 with LPS. (A) Native PAGE of cMD-1/LPS (Left) and mMD-1/LPS (Right) mixtures. (B) Native PAGE shows competition of cMD-1 with polymyxin B for LPS. (C) Gel filtration analysis of cMD-1, LPS, and their mixture.
Structure of cMD-1 Complexed with Lipid IVa.
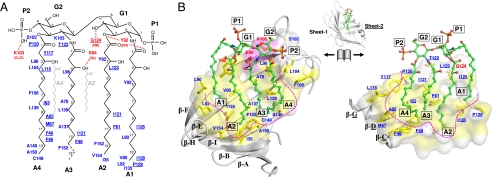
Given the hydrophobic interior, the cMD-1 cavity would provide a suitable binding site for the lipid A moiety of LPS. However, the cavity entrance is not spacious enough to house the two glucosamine moieties of LPS (Fig. S1A), suggesting that the cavity entrance must undergo substantial structural rearrangement upon LPS binding. To provide insights into the LPS binding mode, we determined the crystal structure of cMD-1 complexed with lipid IVa, an LPS precursor (cMD-1lipid IVa; Fig. 4). Lipid IVa represents the minimal LPS structure that binds MD-2, and does not have the A3 O3′- and A4 O3′-linked acyl chains (A3′ and A4’, respectively) of lipid A (Fig. 5A). Otherwise, lipid A and lipid IVa share an identical chemical structure composed of two phosphate groups (P1 and P2), two glucosamine moieties (G1 and G2), and four acyl chains (A1–A4). We crystallized the cMD-1/lipid IVa complex in the same crystal lattice as cMD-1 bound to the putative PGT (cMD-1PGT). The cMD-1lipid IVa structure was determined at 2.4 Å resolution by molecular replacement with two cMD-1/lipid IVa complexes, A and B, in the asymmetric unit. Complexes A and B contain cMD-1 residues 21–160 and 21–158, respectively. The two cMD-1 molecules are essentially identical with an rmsd of 0.08 Å for 138 Cα atoms, and the two lipid IVa molecules occupy similar positions. Thus, only the cMD-1/lipid IVa interaction in complex A is described herein.
Fig. 4.
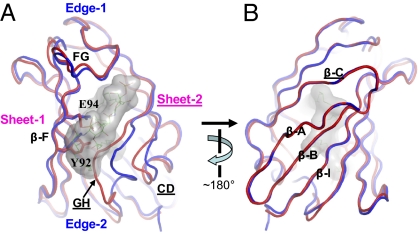
Conformational changes of cMD-1 upon lipid IVa binding. (A) Cavity entrance view. cMD-1PGT and cMD-1lipid IVa are displayed by red and blue coils, respectively. Lipid IVa bound to cMD-1 is shown as surface representation (gray) and also depicted in lines. (B) The view from the cavity bottom.
Fig. 5.
Interaction of cMD-1 with lipid IVa observed in the cMD-1/lipid IVa structure. (A) Schematic diagram of lipid IVa and interacting cMD-1 residues. Lipid IVa atoms that were not observed in the cMD-1/lipid IVa structure are represented by broken lines. Two extra lipid A acyl chains that are not present in lipid IVa are shown by light gray lines. cMD-1 residues that make H bonds (red dashed lines) with lipid IVa are labeled in red, and the remaining interacting residues are in blue. Residues of the sheet-2 side are underlined to discriminate those from sheet-1. (B) Open-book view of the cMD-1/lipid IVa interaction. To achieve the view, the cMD-1 structure (gray ribbons) shown in the middle was split between sheets-1 and -2, and then the sheet-1 (Left) and sheet-2 (Right) sides were rotated by 90° in the left and right directions, respectively, along the vertical axis. Lipid IVa is depicted in a ball-and-stick model (green). cMD-1 residues that form the internal cavity are color coded in surface representation (polar residues H/N/Q/S/T/C, gray; charged residues K/R/D/E, magenta; aliphatic and aromatic residues A/G/I/L/M/W/F/W/Y, yellow). cMD-1 residues that interact with lipid IVa are shown in sticks. The internal surface of the cavity is outlined in magenta broken lines.
One lipid IVa molecule binds inside the hydrophobic cMD-1 cavity (Fig. 4 and Fig. S3A). Lipid IVa makes contacts (≤4.5 Å) with 29 cMD-1 residues and buries ~940 Å2 of cMD-1 accessible surface area (total of 33 residues) that is provided by all β-strands, except β-A, and loops FG/GH (Fig. 5 and Fig. S4). The two phosphoglucosamine moieties of lipid IVa are located at or near the cavity entrance, and surrounded by β-F, loop FG, β-G, and loop GH. The four acyl chains fill almost the entire cMD-1 cavity. Glucosamine G2 and its connected acyl chains A3/A4 of lipid IVa are located in a similar position as PGT in the cMD-1PGT structure (Fig. S3B) and do not appear to induce any substantial structural rearrangement of cMD-1, in comparison with G1 and A1/A2 whose effect is described below.
Structural Rearrangement of cMD-1 upon Lipid IVa Binding.
Substantial structural deviations were identified between cMD-1PGT and cMD-1lipid IVa despite their essentially identical secondary structure (rmsd 1.3 Å; Fig. 4). Lipid IVa binding, especially the glucosamine G1 and acyl chains A1/A2, expands the MD-1 cavity and increases its volume by ~600 Å3 through large structural shifts of loops CD/GH and a slight separation of two β-sheets (Fig. 4A). Such structural changes gradually become pronounced from the cavity bottom toward the entrance. The β-strands A, B, I, and C at or near the cavity base do not show any significant structural changes (Fig. 4B), whereas substantial rearrangements are observed at and near the cavity entrance (Fig. 4A). Loop GH that surrounds the cavity entrance from sheet-2 shifts up to 6.8 Å away from β-F of sheet-1 and opens up the cavity entrance. Interestingly, loop GH, the most flexible of the MD-1 loops, is noticeably stabilized upon lipid IVa binding, as indicated by large decreases in B value (~34 Å2 for residues 128–129; Fig. S5). This loop GH shift is accompanied by movement in the C-terminal region of loop CD (Fig. 4A). This finding, combined with our previous structural comparisons of MD-1 and MD-2 (Fig. 2), confirms that loops GH and CD act a single, dynamic, structural unit. In addition, small shifts up to 1.2 Å were observed for β-F and loop FG along with rotamer changes of Glu94 and Tyr92 (Fig. 4A).
In comparison with cMD-1, binding of tetra-acylated lipid IVa did not significantly change the hMD-2 structure (rmsd for 144 Cα atoms, 0.25 Å), suggesting that the MD-2 cavity is already preconfigured for lipid IVa (8). However, the LPS-bound hMD-2/hsTLR4 structure showed that hexa-acylated LPS binding induces shifts in loops GH and CD of MD-2 (9). NPC2, a cholesterol-binding ML family protein, also experiences cholesterol-induced conformational changes in regions that correspond to loops GH and CD of MD-1, resulting in an enlarged lipid-binding cavity (27, 28). These findings suggest that ML family members use an intrinsic structural plasticity in loops GH and CD to bind their specific lipid ligands.
Lipid IVa Binding Mode of cMD-1.
The hydrophilic moiety of lipid IVa (P2-G2-G1-P1; Fig. 5A) extends over cMD-1 residues Lys103, Glu94, and Tyr92 and makes hydrophilic contacts with β-F and loop FG on sheet-1 (Fig. 5 and Fig. S6A Right). Lys103 is located next to P2 and G2, and forms main-chain H bonds with P2. Tyr92 and Glu94 reposition their side chains upon lipid IVa binding and H bond with G1 O3/A1 O1′ and A3 O3′, respectively. An additional H bond is observed between the Gly124 main chain and G1 O4 on β-sheet-2.
In contrast, three of the acyl chains, A2, A3, and A4, are completely embedded in the cavity and make extensive van der Waals contacts with hydrophobic residues lining the cavity (Fig. 5B and Fig. S3A). These three acyl chains extend into the cavity in an almost perpendicular orientation to the G1-G2 sugar backbone and are aligned side-by-side through hydrophobic interactions. A2 adopts an essentially fully extended conformation, whereas A3 and A4 adopt ~100° and ~120° kinks at their C8′ and C9′ atoms, respectively, due to space constraints within the cavity.
Acyl chain A1 is distinct in its conformation and exposure. The C1′–C3′ atoms are aligned parallel to the G1–G2 sugar backbone and perpendicular to the rest (C4′–C14′) of A1 (Fig. 5B). Consequently, A1 C7′–C14′ stacks with A2 C1′–C7′. Interestingly, only A1 C14′ is completely buried and makes extensive contacts with Val80, Pro129, and Ile135, whereas the other A1 C1′–C13′ atoms form limited hydrophobic contacts with Leu90, Tyr92, and Ile125. Thus, ~40% of A1 is exposed to solvent (Fig. S6A).
Lipid Binding Modes of MD-1 vs. MD-2.
Structural comparison of lipid IVa molecules bound to cMD-1 (lipid IVacMD-1) and hMD-2 (lipid IVahMD-2) demonstrates distinct lipid IVa binding modes (Fig. 6) (8). First, the hydrophilic phosphosugar backbone interaction of lipid IVa with MD-1 and MD-2 occurs on opposite sides of the cavity entrance. MD-1 uses β-F and loop FG on sheet-1 for interaction, whereas MD-2 employs β-G and loop GH, which are positioned below β-F and loop FG, on sheet-2. As a result, the lipid IVacMD-1 sugar backbone shifts by 8–15 Å toward the N-terminal end of loop FG and away from the cavity, compared with lipid IVahMD-2. Such a shift results in solvent exposure of the lipid IVacMD-1 A1 acyl chain (Fig. S6A). In contrast, MD-2 completely buries all of its acyl chains of lipid IVa inside the cavity (Fig. S6B).
Fig. 6.
Comparison of lipid IVacMD-1, lipid IVahMD-2 (PDB ID code 2e59), and LPShMD-2/hsTLR4 (PDB ID code 3fxi) structures. For clarity, only the lipid A portion of LPS is shown. (A) Schematic diagram of the relative ligand disposition in the hydrophobic cavities of MD-1 and MD-2. The phosphosugar backbone of ligand is represented by a circle (lipid IVacMD-1, green; lipid IVahMD-2, magenta; LPShMD-2/hsTLR4, blue). The internal surface of the cavity is schematically shown by broken lines. (B) The relative location and disposition of lipid IVacMD-1 (green), lipid IVahMD-2 (magenta), and LPShMD-2/hsTLR4 (blue) as observed in the superimposed MD-1 and MD-2 structures (yellow).
Unexpectedly, when LPS binds to hMD-2/hsTLR4 (LPShMD-2/hsTLR4), structural similarities and dissimilarities are exhibited (Fig. 6). LPShMD-2/hsTLR4 is distinct from lipid IVacMD-1 in that the LPShMD-2/hsTLR4 phosphosugar backbone interacts mainly with β-G and loop GH of MD-2, rather than β-F and loop FG as in cMD-1/lipid IVa. However, LPShMD-2/hsTLR4 and lipid IVacMD-1 are much more alike in their disposition, exposure, and orientation. LPS contains two extra acyl chains, and thus the LPS sugar backbone is displaced by ~5 Å from the cavity toward edge-1, with respect to lipid IVahMD-2, which then places LPShMD-2/hsTLR4 between lipid IVahMD-2 and lipid IVacMD-1 in the other structures. Second, the A1 acyl chain of LPShMD-2/hsTLR4 is exposed, much like that of lipid IVacMD-1 (Fig. S6 A and C). Last, the phosphosugar backbones of LPShMD-2/hsTLR4 and lipid IVacMD-1 are oriented in the same relative order of P2-G2-G1-P1 in the direction of β-G, whereas that of lipid IVahMD-2 is positioned in the reverse orientation, P1-G1-G2-P2 (Fig. S6). Taken together, MD-1 and MD-2 use different surfaces for ligand recognition, but lipid IVacMD-1 more closely resembles LPShMD-2/hsTLR4 in lipid orientation and exposure.
Discussion
Here, we present the crystal structure of MD-1 where the hydrophobic cavity of the β-cup fold contains a lipid-like molecule (Fig. 1). Similarly, other ML family proteins have been found to contain additional, elongated electron density of various lengths and orientations inside their hydrophobic cavities in their crystal structures (8, 29, 30). For example, MD-2 has three long tubes of electron density, each of which was modeled as a myristoyl acid molecule (Fig. S1B) (8). Der f 2 harbored an elongated U-shaped molecule (30). GM2AP, a ganglioside GM2 binding protein, was purified with a mixture of lipids and a PGT-like molecule was observed in the crystal structure (29). These lipid-like pseudoligands may be either cellular components encountered during recombinant protein synthesis inside cells or medium components that were captured after protein expression. Likewise, it is highly probable that MD-1 in vivo also harbors a lipid molecule derived from cells or from serum. Whereas the exact biological significance of these pseudoligands is unclear, they appear to have a key structural role. Such lipids fill the empty space within the hydrophobic cavity, preventing cavity collapse or disintegration. Although they likely play an indispensable role for structural stabilization of the β-cup fold, we assume that they can be easily replaced by bona fide ligands. Many other cell-surface receptors, such as CD1, which also binds lipids and glycolipids, and MHC class I and II molecules, are not found in an unliganded state and are normally occupied with endogenous self-ligands until displaced by foreign antigens (31, 32).
The observation of a lipid-like molecule in the cMD-1PGT structure inspired us to reevaluate the interaction between MD-1 and LPS. Indeed, we were able to identify direct interaction by native gel electrophoresis and gel filtration analyses (Fig. 3). Furthermore, our crystal structure of the cMD-1/lipid IVa complex strongly supports a direct LPS/MD-1 interaction and provides insights into the MD-1/LPS binding mode (Fig. 5). Tetra-acylated lipid IVa induces a conformational change at loop GH in MD-1 (Fig. 4). Nevertheless, one of the lipid IVa acyl chains is not completely buried inside the MD-1 cavity, suggesting that the cavity is not spacious enough to fully enclose an entire lipid IVa molecule. Thus, hexa-acylated LPS would be expected to induce further structural changes, potentially through the flexible MD-1 loop GH to expand the cavity, but it is also expected that the additional acyl chains in LPS would be somewhat exposed. Moreover, the LPS sugar backbone might be displaced further from the cavity, as seen for LPS bound to MD-2.
Two structural distinctions between lipid IVa-bound MD-1 and MD-2 imply that MD-1 has a lower affinity for LPS than MD-2. First, lipid IVa binding induces substantial conformational changes in loops GH and CD of MD-1, whereas these changes are not seen in MD-2. The lipid IVa-induced structural rearrangement in MD-1 would likely consume energy and, therefore, contribute to an affinity decrease. Second, the nonpolar A1 acyl chain of lipid IVa is partially exposed to solvent and would not be in as energetically favorable an environment as its counterpart in hMD-2, which is completely buried in the hydrophobic cavity. The lower affinity of MD-1 for LPS may provide an explanation why previous reports failed to detect the LPS/MD-1 interaction by coimmunoprecipitation analysis (20, 21).
It is well documented that RP105-tethered MD-1 regulates an MD-2/TLR4-mediated LPS response (16–19). In contrast, such studies on soluble MD-1 have been scarce. It is apparent that MD-1 circulates as a soluble form in the serum of skin-grafted mice at a concentration comparable to that of MD-2 in healthy human donors (33, 34). However, the biological role of soluble MD-1 has not been addressed. Our study on the MD-1/LPS interaction allows us to speculate that soluble MD-1 may function as a modulator of host sensitivity to LPS. One possible scenario for the modulation is that soluble MD-1 can sequester LPS from LPS receptors, such as LPS-binding protein (LBP) and CD14, as well as MD-2, to inhibit cell activation by LPS. Another interesting model for the function of soluble MD-1 is based on the role of soluble MD-2 in opsonizing Gram-negative bacteria and facilitating their phagocytosis and intracellular removal via TLR4 signaling (33, 35). By analogy with soluble MD-2, soluble MD-1 might dock to LPS in the outer membrane of Gram-negative bacteria and stimulate TLR4-mediated phagocytosis of the bacteria. This model is supported by the observation that MD-1 is immunoprecipitated with TLR4 in HEK293 cells that coexpress TLR4 and MD-1 (36).
In addition to the soluble form, MD-1 also exists as a complex with RP105 on the cell surface (13, 14), and the complex exerts a regulatory activity on MD-2/TLR4-mediated LPS responses via direct interaction of MD-1/RP105 with MD-2/TLR4 (16). To address whether MD-1 interacts with RP105 in a similar manner as MD-2 and TLR4, tertiary structures as well as primary sequences were compared between MD-1 and MD-2. MD-2 uses two opposite surfaces, corresponding to the primary and homodimerization interfaces, to interact with TLR4 (7, 9) (Fig S7A). The primary interface drives formation of the 1:1 TLR4/MD-2 complex and is located mainly at MD-2 loops DE and FG on edge-1. The homodimerization interface is formed between two 1:1 MD-2/TLR4 complexes upon LPS-induced TLR4/MD-2 homodimerization and includes MD-2 residues from loops EF and GH on edge-2. MD-1 shares relatively high sequence identity (28–39%) with MD-2 in the primary interface residues, despite low overall sequence identity (~20%; Fig. S7B). Moreover, the MD-1 and MD-2 primary interface residues exhibit relatively small structural deviations (~1.9 Å in average), compared with the overall rmsd value of 2.8 Å (Fig. S7C). The similarities in sequences and structures suggest that MD-1 uses the same DE/FG loops to interact with RP105 as MD-2 does with TLR4. In contrast, the homodimerization interface residues exhibit substantial sequence and structure dissimilarities between MD-1 and MD-2, where the sequence identity is very low (0–20%; Fig. S7D) and structural deviation is substantial (~4.3 Å in average), in particular for loop GH (~6.8 Å in average; Fig. S7E). Thus, MD-1 is unlikely to use loops EF/GH for RP105 interaction. Taken together, these observations suggest that MD-1/RP105 forms a similar structural architecture as the 1:1 MD-2/TLR4 complex, but does not resemble that of the 2:2 MD-2/TLR4 complex.
Loop GH, which exhibits pronounced sequence and structure dissimilarities between MD-1 and MD-2, appears to provide MD-1 and MD-2 with specific binding capabilities via sequence variation and structure plasticity. MD-1 loop GH changes its conformation and dynamics to accommodate lipid IVa, and MD-2 loop GH undergoes substantial structural rearrangement (3.3 Å on average) upon LPS binding, including a drastic movement of Phe126 into the MD-2 cavity, to recruit another 1:1 MD-2/TLR4 complex for signaling (7, 9). Moreover, sequence differences in residues 122 and 125 of loop GH between hMD-2 and mMD-2 determines species specificity in lipid IVa response (37), presumably due to differential positioning of lipid IVa in the MD-2 cavity that then affects the ability of lipid IVa to recruit another 1:1 complex (38). These observations imply that the sequence and structure diversity of loop GH is critical for MD-1- and MD-2-specific functions. Thus, it will be interesting to see if loop GH is also involved in MD-1/RP105’s specific function concerning regulation of MD-2/TLR4-mediated LPS response.
In conclusion, our crystal structures of cMD-1lipid IVa and cMD-1PGT, together with additional biophysical data, demonstrate a direct interaction of MD-1 with LPS. Through its LPS binding ability, soluble MD-1 might then play a crucial role in modulating the host immune response against bacterial infection.
Materials and Methods
For details of protein expression and purification, protein/ligand interaction, crystallization, and structure determination, see SI Materials and Methods. Briefly, cMD-1 and mMD-1 were expressed in insect cells by a baculovirus expression system and purified to homogeneity by a series of chromatography. MD-1/LPS interaction was analyzed by native PAGE and gel filtration. cMD-1PGT and cMD-1lipid IVa were crystallized by sitting-drop, vapor diffusion. X-ray diffraction data were collected at synchrotron sources. The cMD-1PGT and cMD-1lipid IVa structures were determined by SIRAS and molecular replacement methods, respectively. Data collection, phasing, and refinement statistics are summarized in Tables S1 and S2.
Supplementary Material
Acknowledgments
We thank X. Dai (The Scripps Research Institute) for assistance with X-ray data collection, and R. L. Stanfield (The Scripps Research Institute) and Y. S. Choo (Sanford-Burnham Medical Research Institute) for critical discussions. This work was supported by National Institutes of Health Grant AI042266 (to I.A.W.). X-ray diffraction datasets were collected at the SSRL beamline 11-1, the ALS beamline 8.2.2, and the APS beamline 23ID-B.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors for cMD-1PGT and cMD-1lipid IVa have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3mtx and 3mu3, respectively).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004153107/-/DCSupplemental.
References
- 1.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gangloff M, Gay NJ. MD-2: The Toll ‘gatekeeper’ in endotoxin signalling. Trends Biochem Sci. 2004;29:294–300. doi: 10.1016/j.tibs.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 4.Beutler B, Rietschel ET. Innate immune sensing and its roots: The story of endotoxin. Nat Rev Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 5.Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagai Y, et al. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 7.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 9.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 10.Bryant CE, Spring DR, Gangloff M, Gay NJ. The molecular basis of the host response to lipopolysaccharide. Nat Rev Microbiol. 2010;8:8–14. doi: 10.1038/nrmicro2266. [DOI] [PubMed] [Google Scholar]
- 11.Visintin A, Mazzoni A, Spitzer JA, Segal DM. Secreted MD-2 is a large polymeric protein that efficiently confers lipopolysaccharide sensitivity to Toll-like receptor 4. Proc Natl Acad Sci USA. 2001;98:12156–12161. doi: 10.1073/pnas.211445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inohara N, Nuñez G. ML—a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]
- 13.Miura Y, et al. RP105 is associated with MD-1 and transmits an activation signal in human B cells. Blood. 1998;92:2815–2822. [PubMed] [Google Scholar]
- 14.Miyake K, et al. Mouse MD-1, a molecule that is physically associated with RP105 and positively regulates its expression. J Immunol. 1998;161:1348–1353. [PubMed] [Google Scholar]
- 15.Nagai Y, et al. Requirement for MD-1 in cell surface expression of RP105/CD180 and B-cell responsiveness to lipopolysaccharide. Blood. 2002;99:1699–1705. doi: 10.1182/blood.v99.5.1699. [DOI] [PubMed] [Google Scholar]
- 16.Divanovic S, et al. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat Immunol. 2005;6:571–578. doi: 10.1038/ni1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Divanovic S, et al. Regulation of TLR4 signaling and the host interface with pathogens and danger: The role of RP105. J Leukoc Biol. 2007;82:265–271. doi: 10.1189/jlb.0107021. [DOI] [PubMed] [Google Scholar]
- 18.Nagai Y, et al. The radioprotective 105/MD-1 complex links TLR2 and TLR4/MD-2 in antibody response to microbial membranes. J Immunol. 2005;174:7043–7049. doi: 10.4049/jimmunol.174.11.7043. [DOI] [PubMed] [Google Scholar]
- 19.Yazawa N, et al. CD19 regulates innate immunity by the toll-like receptor RP105 signaling in B lymphocytes. Blood. 2003;102:1374–1380. doi: 10.1182/blood-2002-11-3573. [DOI] [PubMed] [Google Scholar]
- 20.Tsuneyoshi N, et al. The functional and structural properties of MD-2 required for lipopolysaccharide binding are absent in MD-1. J Immunol. 2005;174:340–344. doi: 10.4049/jimmunol.174.1.340. [DOI] [PubMed] [Google Scholar]
- 21.Visintin A, Latz E, Monks BG, Espevik T, Golenbock DT. Lysines 128 and 132 enable lipopolysaccharide binding to MD-2, leading to Toll-like receptor-4 aggregation and signal transduction. J Biol Chem. 2003;278:48313–48320. doi: 10.1074/jbc.M306802200. [DOI] [PubMed] [Google Scholar]
- 22.Trompette A, et al. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature. 2009;457:585–588. doi: 10.1038/nature07548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichikawa S, et al. Lipopolysaccharide binding of the mite allergen Der f 2. Genes Cells. 2009;14:1055–1065. doi: 10.1111/j.1365-2443.2009.01334.x. [DOI] [PubMed] [Google Scholar]
- 24.Keber MM, Gradisar H, Jerala R. MD-2 and Der p 2—a tale of two cousins or distant relatives? J Endotoxin Res. 2005;11:186–192. doi: 10.1179/096805105X35206. [DOI] [PubMed] [Google Scholar]
- 25.Dundas J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(Web Server issue):W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhor VM, Thomas CJ, Surolia N, Surolia A. Polymyxin B: An ode to an old antidote for endotoxic shock. Mol Biosyst. 2005;1:213–222. doi: 10.1039/b500756a. [DOI] [PubMed] [Google Scholar]
- 27.Xu S, Benoff B, Liou HL, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J Biol Chem. 2007;282:23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedland N, Liou HL, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc Natl Acad Sci USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright CS, Zhao Q, Rastinejad F. Structural analysis of lipid complexes of GM2-activator protein. J Mol Biol. 2003;331:951–964. doi: 10.1016/s0022-2836(03)00794-0. [DOI] [PubMed] [Google Scholar]
- 30.Johannessen BR, et al. Structure of the house dust mite allergen Der f 2: Implications for function and molecular basis of IgE cross-reactivity. FEBS Lett. 2005;579:1208–1212. doi: 10.1016/j.febslet.2004.11.115. [DOI] [PubMed] [Google Scholar]
- 31.Bjorkman PJ, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 32.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 33.Jain V, et al. Phagocytosis and intracellular killing of MD-2 opsonized gram-negative bacteria depend on TLR4 signaling. Blood. 2008;111:4637–4645. doi: 10.1182/blood-2007-11-126862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadidi S, Chen Z, Phillips J, Yu K, Gorczynski RM. Antisense deoxyoligonucleotides or antibodies to murine MD-1 inhibit rejection of allogeneic and xenogeneic skin grafts in C3H mice. Transplantation. 2002;73:1771–1779. doi: 10.1097/00007890-200206150-00013. [DOI] [PubMed] [Google Scholar]
- 35.Tissières P, et al. Soluble MD-2 is an acute-phase protein and an opsonin for Gram-negative bacteria. Blood. 2008;111:2122–2131. doi: 10.1182/blood-2007-06-097782. [DOI] [PubMed] [Google Scholar]
- 36.Acosta C, Davies A. Bacterial lipopolysaccharide regulates nociceptin expression in sensory neurons. J Neurosci Res. 2008;86:1077–1086. doi: 10.1002/jnr.21565. [DOI] [PubMed] [Google Scholar]
- 37.Vasl J, Oblak A, Gioannini TL, Weiss JP, Jerala R. Novel roles of lysines 122, 125, and 58 in functional differences between human and murine MD-2. J Immunol. 2009;183:5138–5145. doi: 10.4049/jimmunol.0901544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng J, Lien E, Golenbock DT. MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J Biol Chem. 2010;285:8695–8702. doi: 10.1074/jbc.M109.075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.