Abstract
An important question in taste research is how 25 receptors of the human TAS2R family detect thousands of structurally diverse compounds. An answer to this question may arise from the observation that TAS2Rs in general are broadly tuned to interact with numerous substances. Ultimately, interaction with chemically diverse agonists requires architectures of binding pockets tailored to combine flexibility with selectivity. The present study determines the structure of hTAS2R binding pockets. We focused on a subfamily of closely related hTAS2Rs exhibiting pronounced amino acid sequence identities but unique agonist activation spectra. The generation of chimeric and mutant receptors followed by calcium imaging analyses identified receptor regions and amino acid residues critical for activation of hTAS2R46, -R43, and -R31. We found that the carboxyl-terminal regions of the investigated receptors are crucial for agonist selectivity. Intriguingly, exchanging two residues located in transmembrane domain seven between hTAS2R46, activated by strychnine, and hTAS2R31, activated by aristolochic acid, was sufficient to invert agonist selectivity. Further mutagenesis revealed additional positions involved in agonist interaction. The transfer of functionally relevant amino acids identified in hTAS2R46 to the corresponding positions of hTAS2R43 and -R31 resulted in pharmacological properties indistinguishable from the parental hTAS2R46. In silico modeling of hTAS2R46 allowed us to visualize the putative mode of interaction between agonists and hTAS2Rs. Detailed structure-function analyses of hTAS2Rs may ultimately pave the way for the development of specific antagonists urgently needed for more sophisticated analyses of human bitter taste perception.
Keywords: calcium imaging, G protein-coupled receptors, hTAS2R
The mammalian sense of taste is mediated by receptor proteins located in the oral cavity. Each of the five basic taste qualities, sour, salty, sweet, umami, and bitter, serves a specific function in the identification of food components occurring in an animal's diet (1). Because many toxic plant metabolites taste bitter, bitter taste receptors are thought to protect the organism against the ingestion of poisonous food compounds. The human bitter taste receptor gene family (hTAS2R) consists of ∼25 members belonging to the superfamily of G protein-coupled receptors (GPCR) (2–5). On the tongue, the expression of hTAS2Rs is confined to bitter taste-receptor cells, which coexpress specific signal transduction components (6). A main question in taste research is how such few receptors are capable of detecting thousands of structurally diverse bitter compounds.
The pharmacological properties of hTAS2Rs are characterized by two important features: (i) a rather broad tuning, exemplified by the fact that, based on current information on 20 deorphaned receptors, some hTAS2Rs responded to up to one-third of all bitter compounds tested (7); and (ii) although highly variable, the average affinity for bitter agonists is rather low compared with other GPCR-ligand interactions (7). Nevertheless, hTAS2Rs can discriminate even among chemically very similar bitter compounds with high accuracy (8). The combination of these two features results in the manifestation of agonist spectra, which are unique for every single hTAS2R, although some overlaps for individual bitter compounds are evident (9). Therefore, it seems likely that the ligand-binding pockets of hTAS2Rs differ considerably among each other.
Despite recent progress in structure determination of GPCRs, (reviewed in refs. 10–12), structural data on GPCRs are scarce and crystallized receptor proteins exhibit only low amino acid sequence similarity with TAS2Rs. Therefore, a detailed characterization of the structure-function relationship for this GPCR family is necessary to understand the molecular basis underlying the broad tuning and selectivity of its members.
Mammalian TAS2Rs exhibit considerable variability among species. Comparison of human and rodent TAS2R genes reveal, on the one hand, receptors that can be considered as one-to-one orthologs, and on the other hand, receptor subfamilies that underwent species-specific expansions and losses (13). One such subfamily consisting of 8 hTAS2Rs [hTAS2R19 (hTAS2R48), hTAS2R20 (hTAS2R49), hTAS2R30 (hTAS2R47), hTAS2R31 (hTAS2R44), hTAS2R43, hTAS2R45, hTAS2R46, hTAS2R50 (recently withdrawn former gene symbols in parenthesis)] has been identified previously (5) and several family members have been characterized (8, 14–17). This characterization demonstrated that all members of this group exhibit unique agonist activation profiles with rather few common activators.
For the present article, we focused on three members of this subfamily, hTAS2R46 (8), -R43, and -R31 (16), for structure-function analyses, taking advantage of the fact that relatively few different amino acid residues should determine their inherent agonist selectivity. By the generation of receptor chimeras and point-mutagenesis in combination with functional calcium imaging experiments, we identified receptor positions critically involved in agonist interaction.
Results
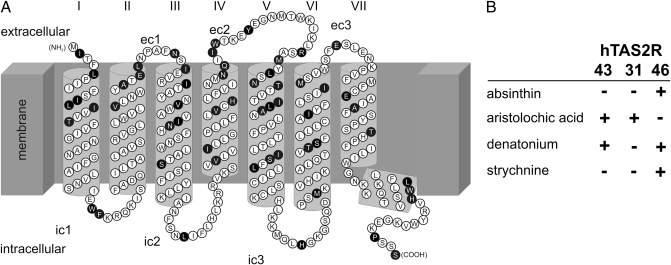
The amino acid sequences of hTAS2R46 and the related receptors, hTAS2R43 and -R31, share 87 and 85% identity, respectively. The amino acid identity of hTAS2R43 and -R31 is 89% (Fig. S1). The transmembrane topology of hTAS2R46 (Fig. 1A) is based on an alignment of rhodopsin family GPCRs. The majority of amino acid residues are identical in all three receptors (Fig. 1A, white circles). Variant positions reside predominantly in receptor parts oriented toward the extracellular side (Fig. 1A, black circles). Each receptor has a distinct set of bitter agonists (Fig. 1B). It appears reasonable to speculate that amino acid sequence differences observed among the compared hTAS2Rs must include agonist-selective residues.
Fig. 1.
Snake plot and pharmacological properties of hTAS2R46. (A) The positions of hTAS2R46 amino acid residues are based on multiple sequence alignments. Extracellular side, intracellular side, and membrane are labeled. Transmembrane domains are indicated by Roman numerals. The N terminus (NH2), extracellular loops (ec1–ec3), intracellular loops (ic1–ic3), and C terminus (COOH) are labeled. Positions in the hTAS2R46 sequence differing from either one of the receptors hTAS2R43 and -R31 are printed in black, invariant positions are shown in white. (B) Pharmacological response profiles of hTAS2R43, -R31, and -R46. Receptor activation (+) and absence of activation (−) by cognate bitter compounds (Left) is indicated. Note that each receptor possesses a unique agonist activation spectrum.
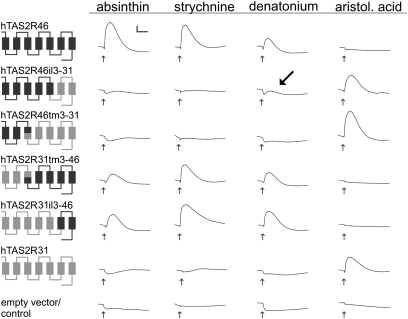
For identification of receptor regions involved in agonist-specific activation, chimeric receptors between hTAS2R46 and -R31 were constructed and challenged with the corresponding selective agonists (Fig. 2). Four chimeras, linked within the highly conserved third transmembrane domains (TM) (hTAS2R46tm3-31, hTAS2R31tm3-46) and third intracellular loops (hTAS2R46il3-31, hTAS2R31il3-46), respectively, were tested for function. Whereas hTAS2R46 responded to its agonists, absinthin, strychnine, and denatonium, hTAS2R31 was activated only by its specific agonist aristolochic acid. Analyses of the chimeric constructs revealed the importance of the C-terminal portions of both receptors for agonist selectivity. Even if the N-terminal two-thirds of receptor chimeras consist of hTAS2R31 residues (hTAS2R31tm3-46, hTAS2R31il3-46), they exhibit an hTAS2R46-like agonist profile and vice versa (hTAS2R46tm3-31, hTAS2R46il3-31). A low-amplitude signal (Fig. 2, arrow) obtained by denatonium stimulation of the construct hTAS2R46il3-31, but not hTAS2R46tm3-31, indicates that the central portion of hTAS2R46 contains residues important for denatonium activation.
Fig. 2.
Functional characterization of hTAS2R46-hTAS2R31 chimeras. The receptor chimeras used for functional calcium imaging experiments are indicated at the left. Parts originating from hTAS2R46 (black) and hTAS2R31 (gray) are shown. hTAS2R46 (row 1) and hTAS2R31 (row 6) refer to the native receptors. For each of the two receptors, two sets of chimeras were built, with junctures in intracellular loop 3 (hTAS2R46il3-31, row 2; hTAS2R31il3-46, row 5) and TM3 (hTAS2R46tm3-31, row 3; hTAS2R31tm3-46, row 4), respectively. HEK 293T Gα16gust44 cells transiently transfected with receptor constructs were stimulated with the bitter compounds indicated at the top. The compounds absinthin, strychnine, and denatonium are agonists of hTAS2R46, whereas hTAS2R31 is activated by aristolochic acid (aristol. acid). The applied agonist concentrations (100 μM absinthin, 30 μM strychnine, 1 mM denatonium benzoate, 3 μM aristolochic acid) were chosen to fully activate the native receptors. Changes in fluorescence after substance application (indicated by arrows under the corresponding traces) were monitored and are shown as traces. Residual denatonium responsiveness of the construct hTAS2R46il3-31 is indicated by a larger arrow. Scaling of traces is indicated by the inset at the top left of the figure (y axis, ΔF/F = 2,000 relative light units; x axis, time = 2 min).
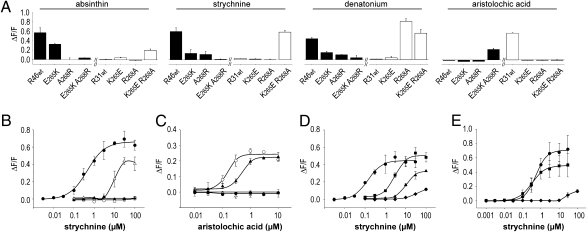
To identify specificity determining residues in the C-terminal receptor parts, we focused on TMs 6 and 7 of hTAS2R46 and -R31, where we observed two prominent differences located at the extracellularly oriented side of TM7. Whereas hTAS2R31 possesses two basic residues in positions 2657.39 (Lys) and 2687.42 (Arg) [indices refer to the positions according to Ballesteros-Weinstein numbering (18)], respectively, hTAS2R46 contains glutamate (E2657.39) and alanine (A2687.42) at the corresponding positions. Swapping of this amino acid pair generated the constructs hTAS2R31K265E R268A and hTAS2R46E265K A268R. Dose-response relationships determined with strychnine and aristolochic acid revealed a conversion of agonist specificities (Fig. 3). Whereas hTAS2R46E265K A268R was not activated by strychnine but responded to the hTAS2R31-agonist aristolochic acid, the corresponding mutant hTAS2R31K265E R268A exhibited a switch in agonist specificity in the opposite direction (Fig. 3A). Comparison of the EC50 values for strychnine between hTAS2R46 (0.43 ± 0.02 μM) and hTAS2R31K265E R268A (10.9 ± 1.8 μM) indicates that additional residues must contribute to full strychnine activation (Fig. 3B). However, because hTAS2R46E265K A268R is more sensitive to aristolochic acid stimulation (EC50 = 0.16 ± 0.02 μM) than hTAS2R31 itself (EC50 = 0.46 ± 0.01 μM), the residual peptide sequence of parental hTAS2R46 seems to be fully permissive for aristolochic acid activation (Fig. 3C).
Fig. 3.
Swapping of residues in TM7 causes switching of receptor specificities. For calcium imaging experiments, receptor constructs were expressed in HEK 293T Gα16gust44 cells and challenged with the indicated bitter compounds. Changes in fluorescence after substance application were monitored and plotted on the y axes (ΔF/F). (A) The applied agonist concentrations (100 μM absinthin, 30 μM strychnine, 1 mM denatonium benzoate, 3 μM aristolochic acid) were chosen to fully activate the native receptors. The amino acid sequence of hTAS2R46 (R46 wt) was changed either at position 265 (E265K), 268 (A268R), or at both positions (E265K A268R) into the corresponding hTAS2R31 amino acid residues (black bars). The amino acid sequence of hTAS2R31 (R31 wt) was mutated correspondingly resulting in the constructs hTAS2R31K265E, hTAS2R31R268A, and hTAS2R31K265E R268A (white bars). (B) Dose-response relationships of hTAS2R46 and –R31 mutants stimulated with strychnine. The receptor constructs hTAS2R46 (black circles), hTAS2R46E265K A268R (black triangles), hTAS2R31 (white circles), and hTAS2R31K265E R268A (white triangles) were transfected and subjected to calcium imaging analyses. Note that hTAS2R46E265K A268R is not activated by the hTAS2R46-selective agonist strychnine, whereas hTAS2R31K265E R268A gained strychnine responsiveness. (C) Same as B, stimulated with the hTAS2R31-selective agonist aristolochic acid. (D) Dependence of strychnine responsiveness on different residues at position 265 of the receptor hTAS2R46. The glutamate at position 265 (circles) was exchanged by aspartate (squares), glutamine (triangles), or lysine (diamonds) and the constructs subjected to calcium imaging. (E) Dependence of strychnine responsiveness on different residues at position 268 of the receptor hTAS2R46. The alanine at position 268 (circles) was exchanged by glycine (squares) or arginine (diamonds) and the constructs subjected to calcium imaging.
The switch in agonist specificity cannot be attributed to any of the two positions alone, as single mutations at these positions did not show a gain of responsiveness; instead, a strong reduction or loss-of-responsiveness toward the original agonist was evident (Fig. 3A). Whereas hTAS2R31 interaction with aristolochic acid appeared to be highly fragile, because both mutants, hTAS2R31K265E and hTAS2R31R268A, exhibited a loss-of-responsiveness and the corresponding mutants, hTAS2R46E265K and hTAS2R46A268R, showed no gain-of-responsiveness, activation by hTAS2R46 agonists was more gradual. The mutant hTAS2R46E265K demonstrated a limited effect on absinthin activation, with an EC50 of 27.3 ± 4.7 μM compared with hTAS2R46 with 9.9 ± 0.3 μM, an almost complete loss-of strychnine responsiveness, and a major reduction in denatonium activation. The mutant hTAS2R31K265E was not activated by any of the indicated compounds [however reduced responsiveness to acesulfame K and saccharin (Fig. S2), both known hTAS2R31 agonists (16), was evident]. The strong codependence of positions 2657.39 and 2687.42 is further substantiated by the observation that mutation of position 2687.42 alone resulted in a loss-of-responsiveness for absinthin or strychnine in the mutant hTAS2R46A268R, but not in a gain-of-responsiveness in the corresponding mutant, hTAS2R31R268A. Denatonium activation of both mutants is strongly affected (hTAS2R31R268A, EC50 = 299.9 ± 21.5 μM; hTAS2R46A268R, residual activation at 1 mM denatonium) indicating the importance of this position for denatonium stimulation.
Given the strong influence of positions 2657.39 and 2687.42 on receptor activation and agonist selectivity, we generated additional, nontemplate-derived mutations in these positions (Fig. 3D). Exchanging glutamate 2657.39 for aspartate reduced the EC50 value for strychnine activation (4.5 ± 0.48 μM); introduction of glutamine further reduced receptor responsiveness (threshold ∼10 μM), whereas lysine almost abolished receptor activation. This result indicated that the exact positioning of a negatively charged group is crucial for hTAS2R46’s sensitivity for strychnine. A minor elevation in strychnine sensitivity after exchanging alanine 2687.42 for glycine (Fig. 3E) (EC50= 0.34 ± 0.03 μM) may indicate a sterical influence of this position.
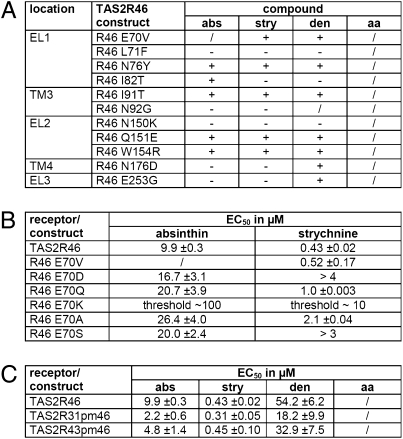
The different sensitivities of hTAS2R46 and hTAS2R31K265E R268A suggested the presence of additional strychnine-selective residues in hTAS2R46. We identified 11 additional amino acid differences in extracellular loops and upper parts of TMs of hTAS2R46 and -R31 and mutated the corresponding positions. The resulting mutants, hTAS2R46E70V, -L71F, -N76Y, -I82T, -I91T, -N92G, -N150K, -Q151E, -W154R, -N176D, and -E253G were functionally characterized with absinthin, strychnine, denatonium, and aristolochic acid (Fig. 4A). We challenged the mutated receptors with a saturating and a nonsaturating agonist concentration. Four of the mutants, hTAS2R46N76Y, -I91T, -Q151E, and -W154R, showed no obvious differences compared with hTAS2R46. Another three mutations, hTAS2R46L71F, -N92G, and -N150K, showed decreased receptor activation by all agonists. To rule out nonspecific effects, we confirmed that cellular expression levels of the constructs were comparable to reference constructs (Fig. S3). Four mutants, hTAS2R46E70V, -I82T, -N176D, and -E253G exhibited agonist-selective reductions of responses, the most pronounced seen for hTAS2R46E70V, which was not activated by absinthin. None of the mutations led to aristolochic acid sensitivity. In view of the results obtained with hTAS2R46/31-chimeras, a strong and agonist-selective effect of position 702.65 was not anticipated. We therefore analyzed additional mutations to investigate the apparent context-dependent influence of position 702.65 on receptor activation by absinthin and strychnine (Fig. 4B). Interestingly, only the exchange from glutamate to valine caused a selective loss of absinthin activation. We also identified two mutations (E70Q and E70A) resulting in a minor reduction in responsiveness, and one mutation (E70K) causing a strong reduction for both substances, whereas the majority of mutations decreased receptor responsiveness for either one of the agonists. Obviously, position 70, located at the base of extracellular loop 1, exerts an important function in the establishment of receptor conformation.
Fig. 4.
Pharmacological characterization of hTAS2R46 mutants. (A) Based on prominent amino acid sequence differences between hTAS2R46 and -R31, hTAS2R46 was mutated and subjected to calcium imaging experiments. HEK 293T-Gα16gust44 cells were transfected with the indicated hTAS2R46 constructs and stimulated with the hTAS2R46-selective agonists absinthin (abs), strychnine (stry), denatonium benzoate (den), and the hTAS2R31-selective agonist aristolochic acid (aa). The applied agonist concentrations (100 μM absinthin, 30 μM strychnine, 1 mM denatonium benzoate, 3 μM aristolochic acid) were chosen to fully activate the native receptors. Absence of activation (/), reduced responsiveness compared with hTAS2R46 (−), and largely unchanged responses (+) are indicated. (B) Mutagenesis of position 70 of hTAS2R46 selectively affected absinthin and strychnine responses. The replacement of glutamine 70 by different amino acid residues is indicated and the corresponding EC50 values for absinthin and strychnine are given. If no signal saturation was achieved, either an approximate EC50 was extrapolated or a threshold was determined. Only the E70V mutation caused a selective loss-of-response upon absinthin stimulation. (C) Functionally relevant hTAS2R46 residues identified in this study were collected in recipient receptors hTAS2R31 and -R43 resulting in the constructs hTAS2R31 PM46 (hTAS2R31 V70E F71L T82I G92N K150N D176N G253E K265E R268A) and hTAS2R43 PM46 (hTAS2R43 V70E T82I G253E K265E R268A), respectively.
To prove that we detected all positions that are different between hTAS2R31 and -R46 and contribute to selective activation of hTAS2R46, functionally relevant positions were assembled in the construct hTAS2R31 PM46 (hTAS2R31 V70E F71L T82I G92N K150N D176N G253E K265E R268A), and calcium imaging analyses were performed (Fig. 4C and Table S1). Comparison of the EC50 values of hTAS2R46 and hTAS2R31 PM46 demonstrated that these nine positions suffice to transfer the full pharmacological profile of hTAS2R46 onto hTAS2R31. Further evidence for this observation is provided by the fact that hTAS2R43 was also successfully engineered to exhibit an hTAS2R46-specific agonist activation profile. This time, because of higher amino acid sequence identity among hTAS2R46 and -R43, fewer exchanges were necessary to result in the mutant receptor hTAS2R43 PM46 (hTAS2R43 V70E T82I G253E K265E R268A), resembling the pharmacological features of hTAS2R46 (Fig. 4C and Table S1).
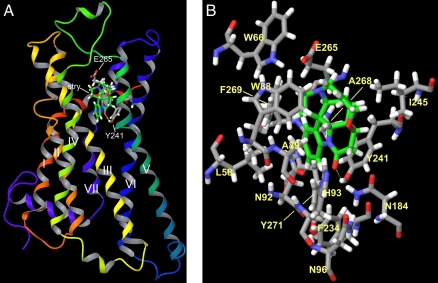
To investigate if the identified positions indeed form a binding pocket, we established an in silico model of hTAS2R46 (Fig. 5). Docking of strychnine into this model proposes the presence of a hydrogen-bond between Y2416.50 and the carbonyl-oxygen of strychnine. Intriguingly, the partially positive-charged N19 of the docked strychnine is positioned at a distance of ∼3Å from the negatively charged carboxylate anion of E2657.39. Moreover, one can congruently fit aristolochic acid onto the posed strychnine structure and observe that a hydrogen-acceptor site, a negatively charged carboxylate group, as well as an aromatic ring system is placed at largely identical positions, thus underscoring the result of the in silico docking approach and our considerations on a common pharmacophore structure (Fig. S4). Mutation of Y2416.50, which is present in hTAS2R46, -R43, and -R31, into phenylalanine resulted in a significant reduction in strychnine responsiveness corroborating the importance of the putative hydrogen-bond (Table S2). As many bitter compounds, including those activating hTAS2R46, are rather hydrophobic, it is not surprising that a number of residues located in the putative strychnine binding pocket are hydrophobic as well (L582.53, W662.61, W883.32, A893.33, F2346.43, I2456.54, A2687.42, F2697.43, Y2717.45). Of these, the highly conserved W883.32 was already shown to affect hTAS2R43 and -R30 (hTAS2R47) activation (14). Our mutagenesis on three additional positions, W662.61, A2687.42, F2697.43, rendered the resulting receptor almost nonfunctional (Fig. 3 and Table S2), supporting their importance. Another group of residues close to the docked strychnine consists of putative hydrogen-bond donors (N923.36, H933.37, N963.40, N1845.47, Y2717.45). As they are located in vicinity of Y2416.50 and the carbonyl oxygen of strychnine, one of them might surrogate for the residual strychnine responsiveness of hTAS2R46Y241F. Our data obtained for the mutant hTAS2R46N92G (Fig. 4) and a previous report on the importance of identical position in hTAS2R43 and -R31 (14) support this assumption.
Fig. 5.
In silico modeling of hTAS2R46 and its putative mode of interaction with strychnine. (A) Ribbon structure of the hTAS2R46 model with bound strychnine. The transmembrane domains of hTAS2R46 are labeled with Roman numerals. The extracellular side of the receptor is oriented to the top and the intracellular side is located at the bottom of the graphic. The carbon atoms of strychnine are shown in green. For better orientation, only two residues of the hTAS2R46 are shown. Tyrosine 241 is located in the upper part of TM6 forming a hydrogen-bond with strychnine (dashed yellow line) and glutamate 265 is at the top of TM7. (B) Close-up view of strychnine docked into the proposed binding pocket of hTAS2R46. Receptor residues located in close proximity to the docked strychnine (green carbon atoms) are indicated.
Discussion
A common feature of TAS2Rs is their ability to respond to many structurally different compounds. This ability could be facilitated by different mechanisms. (i) TAS2Rs may possess more than a single agonist binding pocket, and each pocket is tailored to accommodate subgroups of agonists sharing common structural details. (ii) There might be a single binding pocket providing access to multiple agonists establishing contacts with critical receptor residues. In this case, hTAS2R agonist selectivity may require that access of “wrong” agonists into the binding pocket is tightly restricted. (iii) Growing evidence for GPCR oligomerization (19) including hTAS2Rs (20) suggests that combinations of TAS2Rs might form oligomers acting as agonist binding units. However, at present, functional consequences of TAS2R oligomerization remain elusive (20).
Our data clearly support the existence of a single agonist binding pocket in hTAS2Rs. As demonstrated by the construction of receptor chimeras (Fig. 2), the resulting receptors changed their responsiveness to all tested agonists. Moreover, responsiveness of the chimeras, hTAS2R31tm3-46 and hTAS2R31il3-46 for hTAS2R46-specific agonists absinthin, strychnine, and denatonium was accompanied by a loss-of-responsiveness for the hTAS2R31-specific agonist aristolochic acid; vice versa, insensitivity for hTAS2R46 agonists of the chimeras hTAS2R46tm3-31 and hTAS2R46il3-31 was accompanied by aristolochic acid sensitivity. This observation indicates that the binding pockets in both receptors are located at similar, or at least, partially overlapping regions. Mutagenesis of hTAS2R46 further supports the presence of single binding pockets in hTAS2R46 and related receptors. With few exceptions, functionally relevant amino acid exchanges affected receptor responses for more than one agonist. The most convincing argument for a single binding pocket derives from the transfer of hTAS2R46-specific amino acid residues onto hTAS2R43 and -R31. Although the initial screening for functionally relevant exchanges in hTAS2R46 was done with strychnine, consistently the entire agonist-activation spectrum was transferred onto the recipient receptor (Fig. 4C and Table S1).
The construction of receptor chimeras demonstrated that agonist selectivity is predominantly determined by C-terminal receptor parts. Subsequent mutagenesis revealed two residues within TM7, which are highly divergent between hTAS2R44 and -R46, as being responsible for agonist selectivity. Exchanging 2657.39Glu and 2687.42Ala present in TM7 of hTAS2R46 for 2657.39Lys and 2687.42Arg, as found in hTAS2R31, not only led to aristolochic acid responsiveness of the mutant receptor hTAS2R46E265K A268R, but also to a loss-of-responsiveness for hTAS2R46-specific agonists. Further mutagenesis identified additional residues important for receptor-specific agonist activation, indicating that interaction with such a variety of compounds is governed by more than two amino acids in TM7 and may as well include residues that are identical in hTAS2R43, -R31, and -R46. Another component affecting agonist selectivity of hTAS2Rs could be a tight access control for “wrong” bitter compounds. A position to exemplify this principle is 702.65, at the interface between TM2 and EL1. Whereas hTAS2R46 has a glutamate in this position, the other receptors used in our study have valine instead. As seen for the receptor chimeras (Fig. 2), both residues appear fully compatible with receptor activation by hTAS2R46-specific as well as by hTAS2R31-specific agonists. However, exchanging glutamate for valine in the context of the receptor hTAS2R46 solely affects activation by absinthin (Fig. 4 A and B). On the other hand, exchanging glutamate to aspartate hardly affected the receptor's responsiveness to absinthin, but reduced activation by strychnine. Additional mutations in position 702.65 of hTAS2R46, introducing either glutamine, lysine, alanine, or serine did not point to a specific chemical group or a particular size of the side-chain required at this position, arguing against a direct contact between agonists and receptor at this position. Rather, it appears as if, perhaps based on intramolecular interaction between position 702.65 and a yet unknown part of the receptor, folding of extracellular regions affect access of agonists into the binding pocket.
The number and diversity of TAS2R agonists could indicate that the TAS2R binding pockets have a different architecture compared with other GPCRs. However, our data demonstrate that several of the residues contributing to agonist interaction are positionally conserved, compared with other GPCRs. Of the functionally analyzed positions (Figs. 3 and 4), nine affected receptor activation. Five of these positions were already shown to be involved in receptor-agonist interactions in other GPCRs [N923.36, compare with N92 in hTAS2R43/31 (14) and S159 in 5-HT2A receptor (21); Y2416.50, compare with F310 in α1B-adrenergic receptor (22); F2697.43, compare with in P2Y1 receptor (23)]. Most importantly, the positions in TM7, 2657.39 and 2687.42, were shown to be involved in receptor activation in a number of GPCRs. Position 7.39 has especially been associated with ligand interactions in 5-hydroxytryptamine (5-HT) receptors [5-HT1A (24, 25), 5-HT1B (26)], adrenergic receptors (27, 28), purinergic receptors [P2Y1 (23), P2Y2 (29)], and cholecystokinin-B (CCK-B)/gastrin receptor (30). Intriguingly, amino acid exchanges at this position often affected ligand selectivity (26, 28, 30), thus paralleling our observations. Tentatively, because of similarities in biological functions and pharmacological properties (broad but selective agonist spectra, relatively low affinities of olfactory receptors for their ligands), one may speculate that the binding site of olfactory receptors should share some characteristics with the binding pocket of hTAS2Rs. Indeed, the two positions in TM7 of hTAS2R46 were also predicted to be part of the odorant binding site in olfactory receptor proteins [Fig. S5; cf. Man et al. (31)]. However, a detailed structure-function analysis of an eugenol-responsive mouse OR [mOR-EG (32)] demonstrated that residues critical for agonist binding of this receptor are found in TM3, TM5, and TM6. A striking similarity between hTAS2R46 and mOR-EG is the high number of hydrophobic residues lining the presumed binding pockets.
In conclusion, our analysis of hTAS2R46 and related receptors showed that hTAS2Rs possess single binding pockets accommodating the various agonists via overlapping sets of amino acid residues. The majority of residues involved in agonist interactions are, although the types of amino acids differ, positionally conserved compared with other GPCRs. Exclusion of potential agonists from the hTAS2R binding pockets seems to contribute to selectivity. Future analyses might reveal additional residues important for receptor-ligand interaction and may ultimately pave the way toward the engineering of specific antagonists urgently needed for more sophisticated sensory and pharmacological characterization of bitter taste perception.
Materials and Methods
Taste Compounds.
Aristolochic acid, denatonium benzoate, strychnine, andrographolide, picrotoxinin, acesulfame K, saccharin (Sigma), and marrubiin (LGC Promochem) were purchased. Absinthin, parthenolide, santamarin, and sintenin were available from a previous study (8).
Bitter Taste Receptor Constructs.
We have used the hTAS2R variants hTAS2R46 NM_176887.2, hTAS2R31 NM_176885.2 (variant position L162M, compare with dbSNP rs# cluster id rs10743938), and hTAS2R43 NM_176884.2 (variant position W35S, compare with dbSNP rs# cluster id rs68157013; variant position H212R, compare with dbSNP rs# cluster id rs71443637). The coding regions fused with N-terminal sst3- and C-terminal hsv-tags (5) were cloned into pcDNA5/FRT (Invitrogen). For analyses of cellular expression levels, hTAS2Rs were generated as C-terminal GFP2 fusion proteins.
In Vitro Mutagenesis of hTAS2Rs.
Site-directed mutagenesis of hTAS2R cDNAs was done by PCR-mediated recombination (33), as described previously (34). For a list of oligonucleotides, see Table S3.
Functional Expression.
The functional calcium imaging analyses were done as described previously (8).
A detailed description of methods used for mutagenesis and functional expression is found in SI Materials and Methods.
In Silico Modeling of hTAS2R46.
The hTAS2R46 sequence was submitted to I-Tasser server (35). Docking of strychnine was done using the software module “induced fit” of the Schrodinger molecular modeling suite. Ballesteros-Weinstein numbering (18) was obtained using a combination of multiple sequence alignment of human bitter taste receptors with the structure-based alignment of rhodopsin family GPCRs using Expresso server (36).
Supplementary Material
Acknowledgments
We thank A. Levit for helpful discussions on molecular modeling aspects. This work has been supported by grants from the German Research Foundation, Deutsche Forschungsgemeinschaft (ME1024/2-3) (to W.M.) and Institute Danone (M.B.).
Footnotes
Conflict of interest statement: A.B., M.B., and W.M. have filed patents and patent applications on human bitter taste.
This article is a PNAS Direct Submission. B.K.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913862107/-/DCSupplemental.
References
- 1.Lindemann B. Taste reception. Physiol Rev. 1996;76:718–766. doi: 10.1152/physrev.1996.76.3.719. [DOI] [PubMed] [Google Scholar]
- 2.Adler E, et al. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 3.Chandrashekar J, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 4.Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 5.Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 6.Margolskee RF. Molecular mechanisms of bitter and sweet taste transduction. J Biol Chem. 2002;277:1–4. doi: 10.1074/jbc.R100054200. [DOI] [PubMed] [Google Scholar]
- 7.Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 8.Brockhoff A, Behrens M, Massarotti A, Appendino G, Meyerhof W. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J Agric Food Chem. 2007;55:6236–6243. doi: 10.1021/jf070503p. [DOI] [PubMed] [Google Scholar]
- 9.Behrens M, Meyerhof W. Bitter taste receptors and human bitter taste perception. Cell Mol Life Sci. 2006;63:1501–1509. doi: 10.1007/s00018-006-6113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson MA, Stevens RC. Discovery of new GPCR biology: One receptor structure at a time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 12.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- 14.Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2R receptors. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- 15.Pronin AN, et al. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn C, et al. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens M, et al. The human bitter taste receptor hTAS2R50 is activated by the two natural bitter terpenoids andrographolide and amarogentin. J Agric Food Chem. 2009;57:9860–9866. doi: 10.1021/jf9014334. [DOI] [PubMed] [Google Scholar]
- 18.Ballesteros J, Weinstein H. Integrated methods for the construction of three-dimensional models of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 19.Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn C, Bufe B, Batram C, Meyerhof W. Oligomerization of TAS2R Bitter Taste Receptors. Chem Senses. 2010;35:395–406. doi: 10.1093/chemse/bjq027. [DOI] [PubMed] [Google Scholar]
- 21.Almaula N, Ebersole BJ, Zhang D, Weinstein H, Sealfon SC. Mapping the binding site pocket of the serotonin 5-Hydroxytryptamine2A receptor. Ser3.36(159) provides a second interaction site for the protonated amine of serotonin but not of lysergic acid diethylamide or bufotenin. J Biol Chem. 1996;271:14672–14675. doi: 10.1074/jbc.271.25.14672. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, et al. Phe310 in transmembrane VI of the alpha1B-adrenergic receptor is a key switch residue involved in activation and catecholamine ring aromatic bonding. J Biol Chem. 1999;274:16320–16330. doi: 10.1074/jbc.274.23.16320. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Q, et al. A mutational analysis of residues essential for ligand recognition at the human P2Y1 receptor. Mol Pharmacol. 1997;52:499–507. doi: 10.1124/mol.52.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan XM, Peroutka SJ, Kobilka BK. Identification of a single amino acid residue responsible for the binding of a class of beta-adrenergic receptor antagonists to 5-hydroxytryptamine1A receptors. Mol Pharmacol. 1992;41:695–698. [PubMed] [Google Scholar]
- 25.Kuipers W, et al. Study of the interaction between aryloxypropanolamines and Asn386 in helix VII of the human 5-hydroxytryptamine1A receptor. Mol Pharmacol. 1997;51:889–896. doi: 10.1124/mol.51.5.889. [DOI] [PubMed] [Google Scholar]
- 26.Oksenberg D, et al. A single amino-acid difference confers major pharmacological variation between human and rodent 5-HT1B receptors. Nature. 1992;360:161–163. doi: 10.1038/360161a0. [DOI] [PubMed] [Google Scholar]
- 27.Suryanarayana S, Kobilka BK. Amino acid substitutions at position 312 in the seventh hydrophobic segment of the beta 2-adrenergic receptor modify ligand-binding specificity. Mol Pharmacol. 1993;44:111–114. [PubMed] [Google Scholar]
- 28.Suryanarayana S, Daunt DA, Von Zastrow M, Kobilka BK. A point mutation in the seventh hydrophobic domain of the alpha 2 adrenergic receptor increases its affinity for a family of beta receptor antagonists. J Biol Chem. 1991;266:15488–15492. [PubMed] [Google Scholar]
- 29.Erb L, et al. Site-directed mutagenesis of P2U purinoceptors. Positively charged amino acids in transmembrane helices 6 and 7 affect agonist potency and specificity. J Biol Chem. 1995;270:4185–4188. doi: 10.1074/jbc.270.9.4185. [DOI] [PubMed] [Google Scholar]
- 30.Kopin AS, McBride EW, Quinn SM, Kolakowski LF, Jr, Beinborn M. The role of the cholecystokinin-B/gastrin receptor transmembrane domains in determining affinity for subtype-selective ligands. J Biol Chem. 1995;270:5019–5023. doi: 10.1074/jbc.270.10.5019. [DOI] [PubMed] [Google Scholar]
- 31.Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004;13:240–254. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: Mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang G, Weiser B, Visosky A, Moran T, Burger H. PCR-mediated recombination: A general method applied to construct chimeric infectious molecular clones of plasma-derived HIV-1 RNA. Nat Med. 1999;5:239–242. doi: 10.1038/5607. [DOI] [PubMed] [Google Scholar]
- 34.Behrens M, et al. Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem. 2006;281:20650–20659. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armougom F, et al. Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34(Web Server issue):W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.