Abstract
The active center of RNA polymerase can hydrolyze phosphodiester bonds in nascent RNA, a reaction thought to be important for proofreading of transcription. The reaction proceeds via a general two Mg2+ mechanism and is assisted by the 3′ end nucleotide of the transcript. Here, by using Thermus aquaticus RNA polymerase, we show that the reaction also requires the flexible domain of the active center, the trigger loop (TL). We show that the invariant histidine (β′ His1242) of the TL is essential for hydrolysis/proofreading and participates in the reaction in two distinct ways: by positioning the 3′ end nucleotide of the transcript that assists catalysis and/or by directly participating in the reaction as a general base. We also show that participation of the β′ His1242 of the TL in phosphodiester bond hydrolysis does not depend on the extent of elongation complex backtracking. We obtained similar results with Escherichia coli RNA polymerase, indicating that the function of the TL in phosphodiester bond hydrolysis is conserved among bacteria.
Catalysis by the active center of RNA polymerase (RNAP) is thought to proceed through a general two Mg2+ ion mechanism (1–4). Structural and biochemical studies, however, revealed that during phosphodiester bond synthesis a flexible domain of the active center, the trigger loop (TL), is also required for catalysis (5–10). The TL was proposed to act by orienting the triphosphate moiety of the incoming NTP for efficient attack by the 3′ hydroxyl of RNA (5–7, 10). In crystal structures of RNAP elongation complexes, the TL was observed in two distinct states, folded and unfolded. Only in the folded conformation are amino acids of the TL close enough to the catalytic center to be able to participate in the reaction, so the folded form of the TL is assumed to be the catalytically active state (6–9).
The active center of RNAP is also able to perform hydrolysis of the phosphodiester bond of the transcript (3, 11, 12). This reaction almost exclusively proceeds at the penultimate (second) phosphodiester bond at the 3′ end of RNA transcript (12, 13). Such preference is because of the fact that the rate of hydrolysis of the second phosphodiester bond is hugely increased by assistance from the 3′ end NMP of the RNA (transcript-assisted hydrolysis) (12). Transcript-assisted hydrolysis proceeds in a 1-bp backtracked conformation of the elongation complex, in which the 3′ end NMP of the transcript disengages from the complementary template base to assist with the second phosphodiester bond hydrolysis (12, 14). The 3′ end NMP was proposed to provide its chemical groups to the active center of RNAP to increase the affinity to the second Mg2+ ion (Mg2+II) and/or to position the attacking water molecule (12). Reaction is especially efficient when RNAP incorporates an erroneous nucleotide, given that the misincorporated elongation complex is stabilized in the 1-bp backtracked state. Hydrolysis of the second phosphodiester bond, therefore, is thought to be important for proofreading of transcription (12).
The data on participation of the TL in the cleavage reaction are controversial. The TL was proposed to be required for RNA hydrolysis on the basis of the fact that RNA cleavage by Thermus aquaticus RNAP was inhibited by the antibiotic Streptolydigin (Stl) (6). According to crystallographic data, Stl binds to the TL and freezes it in the unfolded, inactive, conformation (6, 15). This proposal, however, was inconsistent with the results of a recent crystallographic study of several backtracked elongation complexes formed by yeast RNAP II (backtracked by 1, 2, 3, 4, 7, and 13 bp) (14). The TL residues in these complexes were too far from the catalytic Mg2+ and the reacting groups, suggesting that the TL may not participate in RNA cleavage. A recent biochemical study on Escherichia coli RNAP also suggested that the TL is not required for intrinsic hydrolysis (10). The authors used RNAP that lacked the TL (ΔTL RNAP) and showed that WT and ΔTL RNAPs cleaved RNA in 2-bp backtracked elongation complexes at similar rates. Notably, these rates were 1–2 orders of magnitude slower than those observed in earlier studies with T. aquaticus RNAP (12). These results suggested that the mechanisms of RNA hydrolysis used by E. coli and T. aquaticus RNAPs may differ. It was also suggested that the features of the structure of elongation complexes (extent of complementarity of the DNA strands and/or length of the RNA/DNA hybrid) may influence the mechanism of hydrolysis (10). Taken together, the role of the TL in RNA hydrolysis, and consequently in proofreading of transcription, remains unclear.
Results
H1242 of the TL Is Essential for Phosphodiester Bond Hydrolysis.
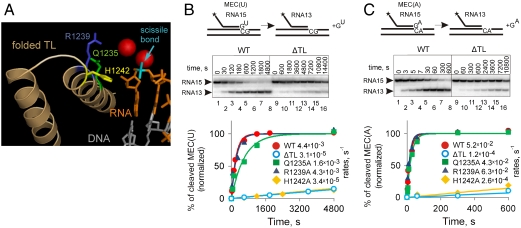
We investigated the mechanism whereby the TL participates in phosphodiester bond hydrolysis and the impact of this on proofreading of transcription. We used T. aquaticus RNAP, for which several crystal structures have been solved (16–18). Its active center is identical in amino acid sequence to that of Thermus thermophilus RNAP, for which several crystal structures are also available (6, 7, 15, 19–22). We constructed mutant RNAPs either lacking the whole TL (ΔTL RNAP) (5, 6) or bearing single alanine substitutions of the TL amino acids, R1239, H1242, and Q1235 (5). In the crystal structure of the T. thermophilus RNAP elongation complex with NTP bound in the active center, these amino acids are in the close proximity to the reacting groups (7). During phosphodiester bond formation, R1239 and H1242 were proposed to stabilize the transition state of the reaction (5, 7, 10), whereas Q1235 was suggested to properly orient R1239 and H1242 (5). These amino acids may potentially occupy similar positions close to the reactants during phosphodiester bond hydrolysis and thus participate in the reaction (Fig. 1A). We used elongation complexes in which the 3′ end NMP of the RNA was noncomplementary to the corresponding template base, but with fully complementary template and nontemplate strands (Fig. S1). Such complexes are stabilized in the 1-bp backtracked conformation, thus eliminating possible translocational effects on the rate of second phosphodiester bond hydrolysis. Given that the properties of the reactions depend on the identity of the 3′ end NMP of the transcript, we used elongation complexes bearing either AMP or UMP at the 3′ end [MEC(A) and MEC(U), respectively; Fig. S1]. The 3′OH and the phosphate group of UMP, and the 3′OH, the phosphate, and the N7 atom of AMP have been proposed to participate in chelation of the second catalytic Mg2+ ion, as inferred from the effects of substitutions of these groups on the Mg2+ dependence of the second phosphodiester bond hydrolysis in these complexes (12). The pH profiles of hydrolysis in these complexes were shown to be different, suggesting that the base moieties of the AMP and UMP may influence the reaction (12).
Fig. 1.
H1242 of the TL is required for hydrolysis of phosphodiester bonds. (A) Structural alignment of the yeast RNAP II elongation complex in a pretranslocated state [Protein Data Bank (PDB) ID code 1I6H (29)] and the T. thermophilus elongation complex with folded TL [PDB ID code 2O5J (7)] suggests that Q1242 (Green), R1239 (Blue), and H1242 (Yellow) of the folded TL (Light Brown) may come in close proximity to the scissile phosphodiester bond (Cyan) of RNA in the active center. RNA is shown in red and template DNA in gray. Mg2+ ions of the active center are shown as spheres. (B and C) Kinetics of phosphodiester bond hydrolysis in MEC(U) and MEC(A) (Fig. S1), respectively, for WT, ΔTL, Q1242A, R1239A, and H1242A RNAPs. Schematics above the gels show the reactions; the asterisk indicates that RNA is labeled at the 5′ end. Representative gels for WT and ΔTL are shown in the middle of each panel (gels for other mutants are shown in Fig. S2). Fractions of cleaved RNA15 against time were fitted to a single exponential equation (Solid Lines) and were normalized to the predicted amplitude. Reaction rates (s-1) are shown in the graphs legends.
Deletion of the TL drastically decreased the rate of second phosphodiester bond hydrolysis in both MEC(A) and MEC(U) (Fig. 1 B and C). This result is consistent with data on inhibition of the cleavage reaction by the antibiotic Stl, which inhibits TL folding (6), and indicates that the TL is indeed required for phosphodiester bond hydrolysis. Next, we tested single amino acid substitutions in these reactions. Substitutions R1239A and Q1235A had no or minor effects on the reactions (Fig. 1 B and C). Interestingly, alanine substitution of H1242 decreased the hydrolysis rates in both MEC(U) and MEC(A) by ∼130- and ∼200-fold, respectively, effects close to those of the whole TL deletion (∼140- and ∼430-fold, respectively; see also Fig. 1 B and C). These results thus indicate that the function of the TL in hydrolysis is determined by H1242. Moreover, if during phosphodiester bond hydrolysis and phosphodiester bond synthesis the TL folds in a similar way, H1242 may be positioned in close proximity to the scissile phosphodiester bond (Fig. 1A).
Two Roles of H1242 in Phosphodiester Bond Hydrolysis.
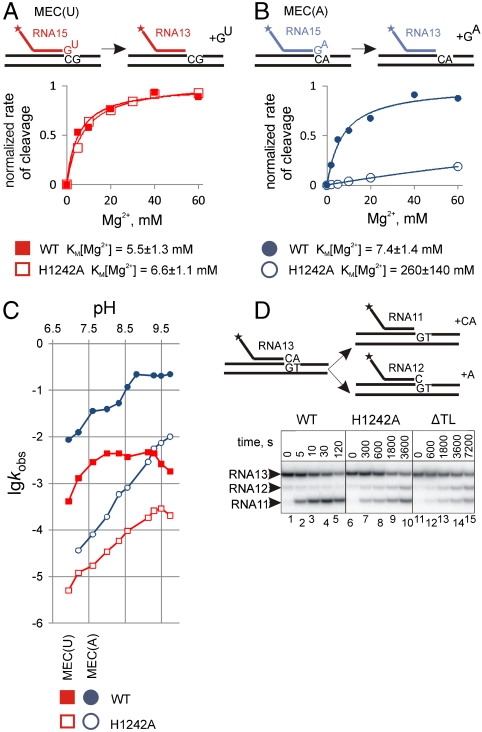
To investigate the role that H1242 plays during hydrolysis, we examined the Mg2+ and pH dependences of phosphodiester bond hydrolysis catalyzed by WT and H1242A RNAPs in MEC(U) and MEC(A) (Fig. 2 A–C). As seen from Fig. 2A, the H1242A substitution did not change the KM[Mg2+] during hydrolysis in MEC(U). The result therefore suggests that H1242 does not participate in Mg2+ chelation and must assist the reaction via another mechanism. In contrast, the pH profile of hydrolysis in MEC(U) was strongly altered by the H1242A mutation (Fig. 2C), suggesting that H1242 directly participates in the reaction. The pH profile of hydrolysis in MEC(U) by WT RNAP reveals that the apparent pKa of the reaction is around 7–8 (Fig. 2C), which is in the pKa range for a histidine residue. This result suggests that H1242 participates in hydrolysis as a general base, presumably by deprotonating the attacking water molecule. Note, however, that the slope of the log-log curve is less than 1, indicating that more than one pKa is being titrated even in the absence of H1242, and some other groups, possibly UMP, still contribute to general acid/base catalysis of phosphodiester bond hydrolysis.
Fig. 2.
H1242 of the TL directly participates in phosphodiester bond cleavage. (A and B) Mg2+ dependences of the hydrolysis of the second phosphodiester bond in MEC(U) and MEC(A), respectively, by WT and H1242A RNAPs. Schematics above the plots show the reactions; the asterisk indicates that RNA is labeled at the 5′ end. The fits of data to the Michaelis–Menten equation are shown as solid lines. The kcat (reaction rate in saturating Mg2+) of the reactions were taken as 1 for clarity. KM[Mg2+] values are shown below the plots. (C) pH profiles of second phosphodiester bond hydrolysis in MEC(U) (Red) and MEC(A) (Blue) complexes (colors correspond to reactions shown in A and B) by WT (Solid Squares and Circles, respectively) and H1242A (Empty Squares and Circles, respectively) RNAPs. (D) The first and the second phosphodiester bond hydrolysis in the elongation complex with the correctly paired 3′ end AMP of the RNA (schematically shown above the gel; EC1 in Fig. S1) by WT, H1242A, and ΔTL RNAPs.
Unexpectedly, the KM[Mg2+] for hydrolysis in MEC(A) by H1242A RNAP was ∼35 times higher than that by WT RNAP (Fig. 2B). This effect of the H1242A substitution on the KM[Mg2+] was of similar magnitude to that of a carbon substitution of N7 of the 3′ end AMP (12) N7 being proposed to participate in the chelation of Mg2+II (12, 14). The above result therefore suggests that, in the absence of H1242, the 3′ end AMP of the RNA cannot be oriented properly in the active center to assist RNAP in chelating the Mg2+II ion via its N7 and, possibly, via its 3′OH groups.
The pH profile of hydrolysis by H1242 RNAP in MEC(A) was strongly altered as compared to that of WT RNAP (Fig. 2C). However, unlike MEC(U), the slope of the pH profile of hydrolysis by H1242A RNAP in MEC(A) was close to 1, indicating that only one pKa is being titrated (pKa of Mg2+-OH- complex) and that the reaction is no longer assisted by either H1242 or the 3′ end AMP. This result is consistent with the above proposal that H1242 is responsible for proper orientation of the AMP moiety in the active center. We cannot distinguish if H1242 acts as a general base during hydrolysis in MEC(A), as it does in MEC(U), or if the altered pH profile is because of the loss of assistance from the 3′ AMP. However, disappearance of the inflection point at pH ∼ 7.5 in the pH profile of H1242A RNAP argues in favor of the former possibility. From the above data, we conclude that during phosphodiester bond hydrolysis H1242 of the TL directly participates in the reaction and that it does so in two ways: (i) as a general base and (ii) by positioning the 3′ NMP moiety of the RNA for transcript-assisted catalysis. It is also possible that the 3′ end AMP and the TL may mutually affect their conformations in the active center and, as a result, their modes of participation in the reaction.
Previously, we have proposed that in the correct elongation complexes (with a correctly paired 3′ end of the RNA) cleavage still proceeds almost exclusively at the second phosphodiester bond, because of assistance from the 3′ end NMP of the RNA, which makes slow unassisted cleavage of the ultimate phosphodiester bond at the 3′ end of the RNA undetectable (12). Following from the above, H1242 of the TL is essential to orient the 3′ end AMP of the RNA for AMP-assisted hydrolysis of the second phosphodiester bond. Therefore, in an elongation complex with the 3′ end AMP of the RNA correctly paired, the H1242A and ΔTL RNAPs should lose their preference for second phosphodiester bond cleavage. In agreement with this prediction, the H1242A mutation reduced, and deletion of the whole TL eliminated the preference for second phosphodiester bond hydrolysis (Fig. 2D). The differences in the magnitude of the effects of the H1242A substitution versus deletion of the TL were also consistent with their effects on the rate of AMP-assisted hydrolysis (Fig. 1C).
Role of the TL in Proofreading of Transcription.
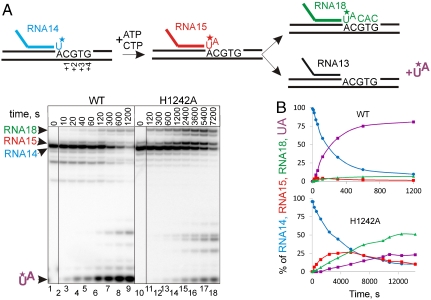
Proofreading via RNA cleavage was proposed to be an important property of transcription (12), because misincorporation leads to the production of erroneous RNA messages and results in stalling of elongation (5, 12, 13, 23). Given that the TL is required for efficient transcript-assisted cleavage, the intrinsic way of correcting errors, we expected that mutation of the catalytic H1242 would strongly diminish proofreading activity. To test that, we supplied elongation complexes with NTPs complementary for incorporation at positions +2, +3, and +4 but not +1, thus promoting a misincorporation event followed by extension of the erroneous transcript (see the scheme in Fig. 3A). Transcripts in the initial elongation complexes were labeled at the 3′ end, allowing us to simultaneously observe misincorporation, erroneous transcript extension, and transcript-assisted proofreading via dinucleotide cleavage. As seen from Fig. 3 A and B, H1242A RNAP readily extended about half of the transcripts beyond the misincorporated nucleotide, whereas WT RNAP removed almost all (∼90%) of the wrong NMP from the RNA without transcribing through the erroneous position. Note also that the rate of incorporation by H1242A RNAP was ∼20 times slower than that of WT RNAP, which gives H1242A RNAP more time for proofreading. Therefore, the actual contribution of the TL to proofreading is expected to be an order of magnitude stronger than observed in the experiment. From this we propose that the TL plays a critical role in transcript-assisted proofreading.
Fig. 3.
Impact of TL catalyzed hydrolysis on the proofreading of transcription. (A) The scheme above the gels shows the experimental setup. RNA in the elongation complex (EC2 in Fig. S1) is labeled at the 3′ end by incorporation of [α32P]UTP (asterisk in the scheme). The 3′ end labeling of RNA enables the monitoring of misincorporation, readthrough, and proofreading simultaneously. Misincorporation of 1 mM ATP, readthrough in the presence of 100 μM CTP, and proofreading (cleavage of pUpA) by WT and H1242A RNAPs are shown. The cleavage products, migrating slower than a dinucleotide, originate from hydrolytic cleavage in readthrough complexes (with RNA longer than 15 nucleotides) after backtracking (see Fig. 4 and Fig. S3) (30). Products are designated in colors the same as those used in the scheme of the reaction shown above the gels. A black vertical line separates lanes originating from the same gel that were brought together. (B) Graphic representation of the data from A. The color code for the plots is the same as in A.
The TL Is Required for RNA Hydrolysis in Complexes Backtracked by > 1 bp.
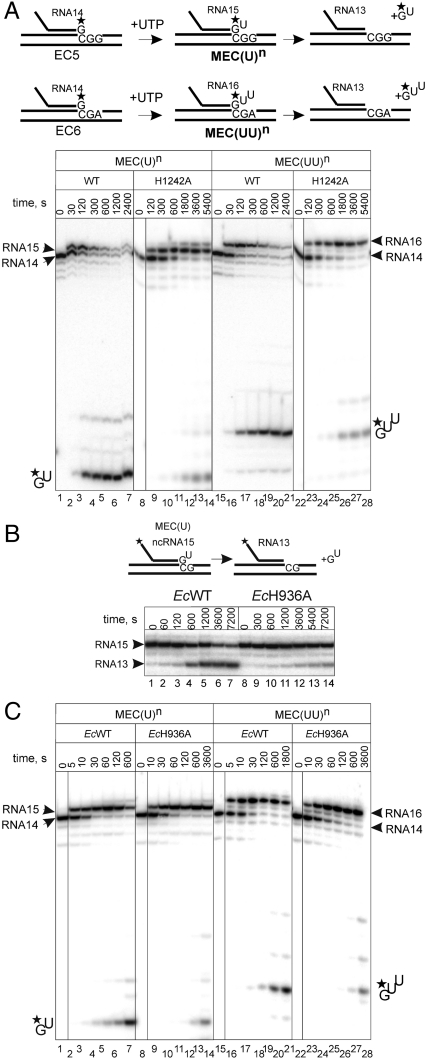
In the course of carrying out the above experiments, we uncovered complexes that had undergone efficient cleavage at the third phosphodiester bond. Formation of such complexes required misincorporation of NMP that was complementary to position +2. In this case, misincorporation was followed by efficient extension by one more NMP, to position +2 (Fig. 4A and Fig. S3A). Almost no proofreading of the misincorporation event was detected, but instead efficient cleavage occurred at the third phosphodiester bond (Fig. 4A and Fig. S3A). This result indicates that, after extension to +2 position, the elongation complex backtracks by 2 bp. To investigate the role of H1242 in hydrolysis of the third phosphodiester bond, we made use of such an elongation complex, MEC(UU)n (“n” for naturally backtracked), in which RNAP is backtracked by 2 bp and hydrolyzes the third phosphodiester bond with release of the 3′ terminal trimer pGpUpU (cartoon in Fig. 4A). We used MEC(U)n obtained through misincorporation of UMP [rather than via assembly with mismatched RNA like MEC(U)] as a control. As expected, hydrolysis by H1242A RNAP in MEC(U)n was strongly diminished as compared to WT RNAP (Fig. 4A; compare lanes 1–7 and 8–14). The third phosphodiester bond in MEC(UU)n was also cleaved much more slowly by H1242A RNAP than by WT RNAP (Fig. 4A; compare lanes 15–21 and 22–28). Therefore, our results suggest that participation of H1242 of the TL in phosphodiester bond hydrolysis is independent of the extent of elongation complex backtracking. Whether or not the hydrolysis in complexes backtracked by more than 1 bp is assisted by the transcript requires further investigation. However, the comparable rates of hydrolysis of the second and the third phosphodiester bonds (compare lanes 1–7 and 15–21 in Fig. 4A and lanes 1–7 and 8–14 in Fig. S3A) argues in favor of participation of the transcript in the reaction.
Fig. 4.
Histidine of the TL is required for RNA hydrolysis irrespective of the extent of elongation complex backtracking, and this function of the TL is conserved among bacteria. (A) Schematics at the top of the panel shows formation of elongation complexes MEC(U)n backtracked by 1 bp [formed from elongation complex EC6 (Fig. S1)] and MEC(UU)n backtracked by 2 bp [formed from elongation complex EC5 (Fig. S1)]. Below: Kinetics of hydrolysis of the second and the third phosphodiester bonds in MEC(U)n and MEC(UU)n, respectively, formed via 1 mM UTP misincorporation by WT and H1242A RNAPs. A black vertical line separates lanes originating from the same gel that were brought together. (B) Hydrolysis of the second phosphodiester bond in assembled MEC(U) by E. coli EcWT and EcH936A RNAPs. (C) Kinetics of hydrolysis of the second and the third phosphodiester bonds in MEC(U)n and MEC(UU)n, respectively, formed via 1 mM UTP misincorporation by E. coli EcWT and EcH936A RNAPs (see scheme of the reactions in A). A black vertical line separates lanes originating from the same gel that were brought together.
The Role of the TL in RNA Cleavage Is Conserved Among Bacteria.
To test if the role of H1242 of the TL in RNA hydrolysis is conserved among bacterial RNAPs, we used E. coli RNAP, which is only distantly related to T. aquaticus. We constructed mutant E. coli RNAP (EcH936A RNAP) bearing an alanine substitution of H936, the homologue of H1242 in T. aquaticus. First, we analyzed cleavage by EcWT and EcH936A RNAPs in an assembled elongation complex in which the 3′ end UMP of RNA was mispaired, MEC(U). As seen from Fig. 4B, hydrolysis was much slower for EcH936A RNAP than for EcWT RNAP, paralleling our results with T. aquaticus RNAP. We also analyzed hydrolysis of the second and the third phosphodiester bonds in “naturally” backtracked elongation complexes that had undergone misincorporation events, MEC(U)n and MEC(UU)n. As seen from Fig. 4C, EcH936A RNAP was slower in cleavage of either the second (compare lanes 1–7 and 8–14) or the third (compare lanes 15–21 and 22–28) phosphodiester bonds than EcWT RNAP. These results indicate that H936 of E. coli RNAP participates in hydrolysis of phosphodiester bonds of the transcript and does so irrespective of the extent of complex backtracking. Taken together, our results suggest that participation of the TL in RNA cleavage is conserved among bacteria.
Note that the TL histidine substitution had less effect on hydrolysis by E. coli RNAP than by T. aquaticus RNAP [∼18- and ∼130-fold effects in MEC(U), respectively]. This difference suggests that H936 of E. coli RNAP may catalyze RNA cleavage less efficiently than H1242 of T. aquaticus RNAP, which would be consistent with the observation that H936 of E. coli RNAP also makes a more modest contribution to phosphodiester bond formation than its equivalent in T. aquaticus RNAP (5, 10). Such differences between T. aquaticus and E. coli RNAPs can be caused by the presence of a large insertion in the TL of E. coli RNAP (24), which may influence catalysis.
Discussion
Hydrolysis of the second phosphodiester bond of the transcript is assisted by the 3′ end NMP of the transcript, which was proposed to contribute to chelation of Mg2+II (12). The principal finding of the present study is that phosphodiester bond hydrolysis requires the TL of the RNAP active center, which participates in the reaction in two distinct ways. First, the invariant histidine residue of the TL directly participates in catalysis of RNA cleavage as a general base, presumably via deprotonating the attacking water molecule. Second, TL serves to orient the NMP of the transcript that assists hydrolysis. This function seems to be similar to the one proposed for the TL in phosphodiester bond synthesis on the basis of biochemical and crystallographic studies: The TL was suggested to orient the triphosphate moiety of the NTP for the reaction to proceed (5, 7, 10). Therefore, TL-assisted hydrolysis cooperates with the ribozyme-like transcript-assisted hydrolysis to increase the efficiency of the reaction. This situation resembles modern ribozymes, whose activities are frequently assisted by protein components.
Earlier we proposed that the dependence of the pH profiles of transcript-assisted hydrolysis on the identity of the RNA 3′end NMP may be because of participation of the RNA 3′NMP in the reaction as a general acid/base (12). The discovery of the new player in the reaction, the TL, suggests that the 3′ end NMP may also change the microenvironment for TL-assisted hydrolysis or influence the orientation of the catalytic histidine of the TL. However, the different pH profiles of hydrolysis in the MEC(A) and MEC(U) complexes, even in the absence of the TL (Fig. S4), argue that the 3′ end NMP may still directly participate in the reaction.
Our discovery of the important role of the TL in phosphodiester bond hydrolysis contradicts the conclusions of the recent study on E. coli RNAP (10). The authors showed that deletion of the whole TL did not influence the rate of the reaction. However, they noted that, because of extensive noncomplementarity of the template and nontemplate strands and a short RNA-DNA hybrid, the elongation complexes they used might be “locked” in a configuration that precludes the TL from functioning (10). This idea is supported by the fact that the rates of hydrolysis by both E. coli and T. aquaticus WT RNAPs observed in our study (Figs. 1B and 4B) were about an order of magnitude faster than those observed by Zhang et al. (10), even though we used a lower pH (pH 8.0 versus 9.0) and lower Mg2+ concentration (10 versus 20 mM). In addition to using complexes assembled with mispaired 3′ end of RNA, we also used elongation complexes that had undergone misincorporation prior to cleavage [MEC(U)n, MEC(G)n, MEC(UU)n, and MEC(GG)n], a more physiological way to bring and stabilize complexes in 1- and 2-bp backtracked states (Fig. 4 A and C).
Notably, in the crystal structures of yeast RNAP II in the backtracked state the invariant histidine residue of the TL, H1085 (homologue of H1242 of T. aquaticus), is far from the scissile phosphodiester bond, making it unlikely that it participates in the cleavage reaction (14). However, the rate of phosphodiester bond hydrolysis by yeast RNAP II is about 2 orders of magnitude slower than that of T. aquaticus RNAP (12, 14). This rate difference is close to the effect of TL deletion in T. aquaticus RNAP on hydrolysis (see above). Taken together, these data suggest that RNAP II may not use its TL for hydrolysis. Nevertheless, it remains unclear whether the eukaryotic TL has lost its function in RNA hydrolysis or the bacterial TL has acquired it after these multisubunit RNAPs diverged early in evolution.
Recently, it was suggested that misincorporation at template position +1 occurs faster if the noncognate NTP is complementary to the template base in position +2 (25). The authors proposed that in this situation the +2 template base may shift into the active center (template misalignment) thus leading to recognition of the noncognate NTP as a cognate one. We did not observe any dependency of the rate of misincorporation of a noncognate NTP in position +1 on the identity of the +2 position of the template for either T. aquaticus or E. coli RNAPs (Fig. S3B). We, however, observed the following propensity. When there is no complementarity of the +1 misincorporated NMP and +2 position of the template, the misincorporated complex is stabilized in a 1-bp backtracked state and thus can only slowly be extended. In contrast, when incoming NTP is noncomplementary to position +1 but complementary to position +2 of the template, after misincorporation at +1, the mismatched RNA is quickly extended to position +2. These observations suggest that the noncognate NMP at the 3′ end of the RNA in such a misincorporated complex is recognized by RNAP as a correct one, thus allowing its efficient extension. Though disagreeing with Kashkina et al. (25) on the mechanism of the phenomenon, our results agree with their proposal that misincorporation of the NTP complementary to the +2 base of the template may indeed be the major pathway of acquisition of errors in RNA (25).
Material and Methods
Mutant RNAP Construction and Purification.
T. aquaticus RNAP lacking the TL (β′ residues 1238–1254 substituted by glycine residue) was constructed as described (5, 6). T. aquaticus mutant RNAPs bearing single alanine substitutions of the β′ Q1235, R1239, and H1242 (Q1235A, R1239A, and H1242A RNAPs, respectively) were obtained by site-directed mutagenesis in a coexpression system for recombinant RNAP (5, 26). WT and mutant core RNA polymerases were purified as described (26). To obtain E. coli EcH936A RNAP, the mutation was introduced in the β′ subunit coded under inducible promoter in plasmid pRL663. WT and mutant RNAPs production was performed as described (27) and their purification as described (28).
Transcription Assays.
Elongation complexes were assembled and immobilized on Ni-nitrilotriacetate beads (Qiagen) as described (5, 12), in transcription buffer containing 40 mM KCl and 20 mM Tris pH 8.0, unless otherwise specified. Sequences of the oligos used for the elongation complexes assembly are shown in Fig. S1. RNA was either kinased at the 5′ end (12) or labeled at the 3′ end after elongation complex assembly by incorporation of [α-32P]GTP or [α-32P]UTP (PerkinElmer), with subsequent removal of unincorporated nucleotide through beads washing with transcription buffer. Cleavage reactions were initiated by addition of 10 mM MgCl2 (final concentration), unless otherwise specified. Misincorporation was initiated by addition of a mixture 10 mM MgCl2, 1 mM noncognate NTP, and, where specified, 100 μM NTPs complementary to the following positions (see Fig. 3A) (final concentrations). Reactions were incubated at 40 °C for times indicated in the figures and were stopped by addition of loading buffer containing formamide. Products were resolved by denaturing (8 M Urea) 23% PAGE, revealed by PhosphorImaging (GE Healthcare), and analyzed by using ImageQuant software (GE Healthcare). To determine the rate of phosphodiester bond hydrolysis, a proportion of the cleaved RNA was plotted against time and fitted to a single exponential equation by using nonlinear regression (5, 12). To determine the rate of misincorporation, the proportion of complexes that had undergone misincorporation (and subsequent cleavage) was plotted against time and fitted as above. To determine the Km[Mg2+] for cleavage in MEC(A) and MEC(U), the reaction rates obtained in various MgCl2 concentrations were fitted to the Michaelis–Menten equation (5, 12).
Supplementary Material
Acknowledgments.
We thank Jeff Errington for his critical reading of the manuscript. The work was supported by research grants from the United Kingdom Biotechnology and Biological Sciences Research Council, the Royal Society, and the European Research Council (ERC-2007-StG 202994-MTP) (to N.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914424107/-/DCSupplemental.
References
- 1.Steitz TA. A mechanism for all polymerases. Nature. 1998;391(6664):231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 2.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90(14):6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sosunov V, et al. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22(9):2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosunov V, et al. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33(13):4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuzenkova Y, et al. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8(1):54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Temiakov D, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19(5):655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448(7150):163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30(5):547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127(5):941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2009;17(1):99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92(10):4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313(5786):518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 13.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93(4):627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 14.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuske S, et al. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122(4):541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296(5571):1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 17.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme-DNA complex. Science. 2002;296(5571):1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 19.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417(6890):712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 20.Vassylyev DG, et al. Structural basis for transcription inhibition by tagetitoxin. Nat Struct Mol Biol. 2005;12(12):1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artsimovitch I, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122(3):351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Perederina A, et al. Regulation through the secondary channel—Structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118(3):297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: Control of the fidelity of transcription. Science. 1993;262(5135):867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 24.Chlenov M, et al. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase beta’ subunit. J Mol Biol. 2005;353(1):138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 25.Kashkina E, et al. Template misalignment in multisubunit RNA polymerases and transcription fidelity. Mol Cell. 2006;24(2):257–266. doi: 10.1016/j.molcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Kuznedelov K, Minakhin L, Severinov K. Preparation and characterization of recombinant Thermus aquaticus RNA polymerase. Methods Enzymol. 2003;370:94–108. doi: 10.1016/S0076-6879(03)70009-3. [DOI] [PubMed] [Google Scholar]
- 27.Yuzenkova J, et al. Mutations of bacterial RNA polymerase leading to resistance to microcin j25. J Biol Chem. 2002;277(52):50867–50875. doi: 10.1074/jbc.M209425200. [DOI] [PubMed] [Google Scholar]
- 28.Kashlev M, et al. Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol. 1996;274:326–334. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- 29.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292(5523):1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 30.Laptenko O, Borukhov S. Biochemical assays of Gre factors of Thermus thermophilus. Methods Enzymol. 2003;371:219–232. doi: 10.1016/S0076-6879(03)71016-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.