Abstract
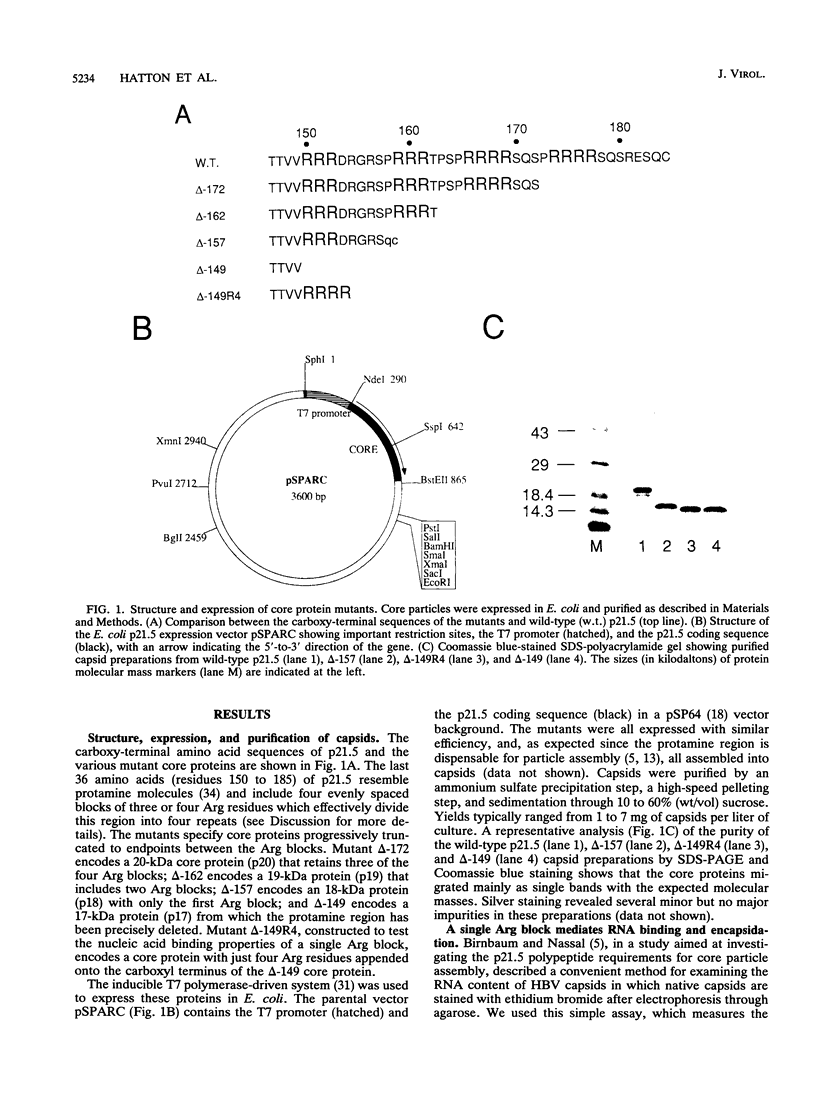
The hepatitis B virus capsid or core protein (p21.5) binds nucleic acid through a carboxy-terminal protamine region that contains nucleic acid-binding motifs organized into four repeats (I to IV). Using carboxy-terminally truncated proteins expressed in Escherichia coli, we detected both RNA- and DNA-binding activities within the repeats. RNA-binding and packaging activity, assessed by resolving purified E. coli capsids on agarose gels and disclosing their RNA content with ethidium bromide, required only the proximal repeat I (RRRDRGRS). Strikingly, a mutant in which four Arg residues replaced repeat I was competent to package RNA, demonstrating that Arg residues drive RNA binding. In contrast, probing immobilized core proteins with 32P-nucleic acid revealed an activity which (i) required more of the protamine region (repeats I and II), (ii) appeared to bind DNA better than RNA, and (iii) was apparently modulated by phosphorylation in p21.5 derived from Xenopus oocytes. Deletion analysis suggested that this activity may depend on an SPXX-type DNA-binding motif in repeat II. Similar motifs found in repeats III and IV may also function to bind DNA. On the basis of these observations, together with a reinterpretation of recent studies showing that capsid protein mutants cause defects in viral genome replication, we propose a model suggesting that hepadnavirus capsid proteins participate directly in the intracapsid reverse transcription of RNA into DNA. In this model, repeat I binds RNA whereas the distal repeats are progressively recruited to bind elongating DNA strands. The latter motifs may be required for replication to be energetically feasible.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albin C., Robinson W. S. Protein kinase activity in hepatitis B virus. J Virol. 1980 Apr;34(1):297–302. doi: 10.1128/jvi.34.1.297-302.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Fuller S. D. A model for the hepatitis B virus core protein: prediction of antigenic sites and relationship to RNA virus capsid proteins. EMBO J. 1988 Mar;7(3):819–824. doi: 10.1002/j.1460-2075.1988.tb02880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990 Nov;64(11):5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum F., Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990 Jul;64(7):3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Enders G. H., Ganem D., Varmus H. E. 5'-terminal sequences influence the segregation of ground squirrel hepatitis virus RNAs into polyribosomes and viral core particles. J Virol. 1987 Jan;61(1):35–41. doi: 10.1128/jvi.61.1.35-41.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson M. A., Marion P. L., Robinson W. S. Core particles of hepatitis B virus and ground squirrel hepatitis virus. II. Characterization of the protein kinase reaction associated with ground squirrel hepatitis virus and hepatitis B virus. J Virol. 1982 Aug;43(2):741–748. doi: 10.1128/jvi.43.2.741-748.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina A., Bonelli F., Zentilin L., Rindi G., Muttini M., Milanesi G. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J Virol. 1989 Nov;63(11):4645–4652. doi: 10.1128/jvi.63.11.4645-4652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich W. H., Goldmann U., Müller R., Stibbe W., Wolff W. Specificity and localization of the hepatitis B virus-associated protein kinase. J Virol. 1982 Jun;42(3):761–766. doi: 10.1128/jvi.42.3.761-766.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R. C., Lavine J. E., Chang L. J., Varmus H. E., Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990 Apr 5;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- Hirsch R. C., Loeb D. D., Pollack J. R., Ganem D. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J Virol. 1991 Jun;65(6):3309–3316. doi: 10.1128/jvi.65.6.3309-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker-Niepmann M., Bartenschlager R., Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990 Oct;9(10):3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Machida A., Ohnuma H., Tsuda F., Yoshikawa A., Hoshi Y., Tanaka T., Kishimoto S., Akahane Y., Miyakawa Y., Mayumi M. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J Virol. 1991 Nov;65(11):6024–6030. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Payne J. M., Laybourn P. J., Dahmus M. E. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989 Nov 25;264(33):19621–19629. [PubMed] [Google Scholar]
- Petit M. A., Pillot J. HBc and HBe antigenicity and DNA-binding activity of major core protein P22 in hepatitis B virus core particles isolated from the cytoplasm of human liver cells. J Virol. 1985 Feb;53(2):543–551. doi: 10.1128/jvi.53.2.543-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh J., Zweidler A., Summers J. Characterization of the major duck hepatitis B virus core particle protein. J Virol. 1989 Mar;63(3):1371–1376. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill G., Zentgraf H., Schaller H., Bosch V. The duck hepatitis B virus DNA polymerase is tightly associated with the viral core structure and unable to switch to an exogenous template. Virology. 1988 Mar;163(1):123–132. doi: 10.1016/0042-6822(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Roossinck M. J., Siddiqui A. In vivo phosphorylation and protein analysis of hepatitis B virus core antigen. J Virol. 1987 Apr;61(4):955–961. doi: 10.1128/jvi.61.4.955-961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht H. J., Bartenschlager R., Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989 Jul;63(7):2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring D. N., Ou J. H., Masiarz F. R., Rutter W. J. A signal peptide encoded within the precore region of hepatitis B virus directs the secretion of a heterogeneous population of e antigens in Xenopus oocytes. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8405–8409. doi: 10.1073/pnas.85.22.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring D. N., Ou J. H., Rutter W. J. Assembly of viral particles in Xenopus oocytes: pre-surface-antigens regulate secretion of the hepatitis B viral surface envelope particle. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9338–9342. doi: 10.1073/pnas.83.24.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989 May 5;207(1):61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- Whitten T. M., Quets A. T., Schloemer R. H. Identification of the hepatitis B virus factor that inhibits expression of the beta interferon gene. J Virol. 1991 Sep;65(9):4699–4704. doi: 10.1128/jvi.65.9.4699-4704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. Q., Walter M., Standring D. N. Hepatitis B virus p25 precore protein accumulates in Xenopus oocytes as an untranslocated phosphoprotein with an uncleaved signal peptide. J Virol. 1992 Jan;66(1):37–45. doi: 10.1128/jvi.66.1.37-45.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- Yu M., Summers J. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. J Virol. 1991 May;65(5):2511–2517. doi: 10.1128/jvi.65.5.2511-2517.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. L., Standring D. N. Production of hepatitis B virus nucleocapsidlike core particles in Xenopus oocytes: assembly occurs mainly in the cytoplasm and does not require the nucleus. J Virol. 1991 Oct;65(10):5457–5464. doi: 10.1128/jvi.65.10.5457-5464.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Yang S. Q., Standring D. N. Characterization of hepatitis B virus capsid particle assembly in Xenopus oocytes. J Virol. 1992 May;66(5):3086–3092. doi: 10.1128/jvi.66.5.3086-3092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]