Abstract
Phytochromes are a collection of dimeric photoreceptors that direct a diverse array of responses in plants and microorganisms through photoconversion between a red light-absorbing ground state Pr, and a far-red light-absorbing photoactivated state Pfr. Photoconversion from Pr to Pfr is initiated by a light-driven rotation within the covalently attached bilin, which then triggers a series of protein conformational changes in the binding pocket. These movements ultimately affect an appended output module, which often has reversible protein kinase activity. Propagation of the light signal from the bilin to the output module likely depends on the dimerization interface but its architecture and response to phototransformation remain unclear. Here, we used single particle cryoelectron microscopy to determine the quaternary arrangement of the phytochrome dimer as Pr, using the bacteriophytochrome (BphP) from Deinococcus radiodurans. Contrary to the long-standing view that the two monomers are held together solely via their C-terminal region, we provide unambiguous evidence that the N-terminal bilin-binding region of BphP also provides a dimerization interface with the C-terminal kinase domain appearing as a more flexible appendage. The BphP monomers dimerize in parallel with the polypeptides intimately twisting around each other in a right-handed fashion. Based on this electron microscopic picture, we propose that the light-driven conformational changes transmitted from the chromophore to the output module along the spine of this extensive dimer interface is the central feature underpinning phytochrome signaling.
Keywords: dimerization, histidine kinase, light signaling, signaling helix
Phytochromes (Phys) encompass a diverse set of photochromic photoreceptors that enable the detection of red (R) and far-red light (FR). They were first discovered in higher plants based on their regulation of many aspects of photomorphogenesis and more recently have been found in numerous proteobacteria, actinobacteria, cyanobacteria, slim molds, and filamentous fungi where they play roles in photomovement, pigmentation, chromatic adaptation, and sexual reproduction (1–3). Phys are unique among photoreceptors by being able to assume two stable states, a Pr form that absorbs R and typically represents the ground state, and a Pfr form that absorbs FR and typically represents the photoactivated state. By photointerconverting between Pr and Pfr, Phys act as reversible switches in light perception.
Phys are homodimeric chromoproteins that share a common modular architecture across evolution (2–4). Light absorption occurs through a covalently attached bilin chromophore, which is cradled within a signature cGMP phosphodiesterase/adenyl cyclase/FhlA (GAF) domain. Often up and downstream of the GAF domain are Per/Arndt/Sim (PAS) and Phy-associated (PHY) domains, respectively, which together with the GAF domain comprise the complete photosensory module (Fig. 1A). All three domains have related core topological elements indicative of a common evolutionary ancestor. C-terminal to the photosensory module is an output (or effector) module that transduces the light signal to appropriate sensory responses. In many Phys, this output module contains a histidine kinase (HK) or related domain; their presence implicates these Phys in two-component phosphorelays that initiate their cascades by autophosphorylation of a conserved histidine with the bound phosphate then donated to a cognate response regulator to effect signal transmission (5). Accordingly, a number of Phys bearing such kinase domains have been shown to have reversible phosphotransferase activity in vitro [e.g., (6–9)]. Whereas the prevailing view is that autophosphorylation occurs in trans across the dimeric two-component receptor, recent studies indicate that cis autophosphorylation is also possible (10).
Fig. 1.
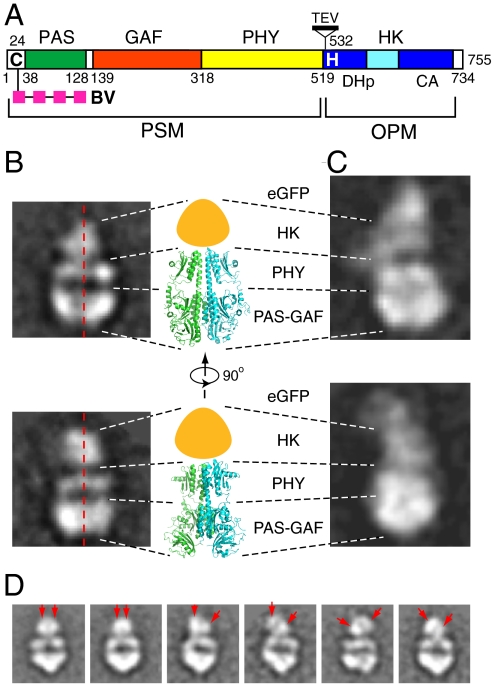
Domain organization and negative-staining EM image analysis of DrBphP. (A) The domain architecture of the DrBphP polypeptide revealing the linear arrangement of the PAS, GAF, PHY, and HK domains. Cys-24 (C) that covalently binds BV, and the DHp and CA subdomains and His532 (H) that serves as the phosphoacceptor site in the HK domain are indicated. TEV cleavage site introduced into DrBphP-TEV is shown. PSM, photosensory module. OPM, output module. (B, C) Two selected class averages of the negatively stained particles of DrBphP (B) and DrBphP-EGFP (C) in comparison with two side views of the PSM dimer from PaBphP as determined by X-ray crystallography [PDB ID 3C2W (15)]. The approximate position of the HK domains is indicated by the orange-colored shape. (D) Flexibility of the HK domains as observed by negative-staining EM. The right two pictures show DrBphP particles with symmetrically arranged HK domains (red arrows), whereas the remaining pictures show particles where the HK domains are variably displaced.
How light absorption by the bilin regulates signaling by the associated output module remains unclear. X-ray crystallographic structures of several Phys as Pr revealed that the bilin is buried deep within the GAF domain and anchored by contacts with surrounding amino acids (11–14). Interaction between the PAS and GAF domains is stabilized by a figure-of-eight knot formed by a GAF-domain loop lassoing a region N-terminal to the PAS domain. In addition to a long helical strand (46 residues spanning 12 turns) that connects the GAF and PHY domains, an unusually long hairpin emerging from the PHY domain reinforces their association by interacting electrostatically with the GAF domain near the bilin (11, 15). This hairpin interaction comprises 90% of the buried surface area between the GAF and PHY domains. Crystallographic studies showed that sister GAF domains also interact in parallel via congruent three-helix bundles, which may represent an important dimerization surface (13, 15).
The recently published NMR solution structures of a Phy GAF domain as Pr and Pfr have provided a possible model for photoactivation based on the modular Phy architecture (16, 17). Contrary to expectations that the D pyrrole ring of the bilin rotates during Pfr formation, the primary motion involves rotation of the A pyrrole ring. This rotation then triggers a series of rearrangements within the GAF domain, including a partial melting of one of the helices involved in GAF/GAF domain dimerization, and motion at the electrostatic contact between the PHY domain hairpin and the GAF domain. Taken together, such a cascade of events could modify both the intramolecular GAF/PHY and the intermolecular GAF/GAF contacts to ultimately alter the dimer interface and thus the spatial relationship between distal HK domains that may be critical to phosphotransfer.
Given the potential importance of dimerization to Phy signaling (13, 15, 18), a full appreciation of this contact along the entire homodimer is essential. Results from negative-staining and rotary-shadowing EM (19, 20) combined with limited proteolysis and crosslinking of plant Phys (20, 21) first proposed that these photoreceptors assume a roughly Y-shaped quaternary structure with the C-terminal output module providing the dimerization interface. This model was purportedly supported by small angle X-ray scattering of a bacteriophytochrome (BphP) from Rhodopseudomonas palustris (22). Unfortunately, this and the previous studies were potentially compromised by incorrect orientation of the particles. The crystallographic structures of the bilin-binding domains from Deinococcus radiodurans (Dr) and Pseudomonas aeruginosa (Pa) BphPs have since challenged this view by revealing that the dimer interface may also include the GAF and PHY domains binding in parallel (13, 15). However, given the absence of the output module in these structures and the fact that artifactual interfaces are sometimes generated under the harsh conditions used for crystallization, it remained unsettled as to how Phys dimerize. Evidence to the latter caveat was provided by the X-ray crystallographic analysis of the photosensory module from the cyanobacterial Phy Cph1 in Synechocystis PCC6803, which crystallized as an antiparallel and not parallel dimer (11).
As an alternative method to examine Phy dimerization, we exploited single particle cryoelectron microscopy to determine the quaternary structure of a Phy dimer, using full-length dimeric DrBphP as the example. Our 13 Å resolution model for Pr revealed a surprisingly long twisted dimerization contact that extends the length of the monomers from the GAF to the HK domains. Whereas the GAF and PHY domains appear to provide rigid interfaces, the HK domains interface appears more flexible, thus offering a model in which Phys photoregulate the activity for their output modules by adjusting the strength and/or orientation of this C-terminal contact.
Results and Discussion
Negative-Staining EM Image Analyses of DrBphP.
Prior studies showed that full-length DrBphP can be synthesized recombinantly and efficiently assembled with its native chromophore biliverdin IXα (BV) to generate a soluble dimeric R/FR photochromic biliprotein (6, 12, 23). To help orient the DrBphP particle in our EM density maps, we were also successful in expressing a soluble tagged version containing EGFP fused to the C-terminus (24). The appended EGFP (29 kDa) did not alter the photochemical properties of DrBphP except for the addition of GFP absorption to the absorption spectrum of DrBphP (Fig. S1 A and B). Maltose-binding protein, glutathione S-transferase (GST), and the T7 epitope combined with anti-T7 antibodies were also tested as both N- and C-terminal tags. Whereas the maltose-binding protein and GST tags substantially reduced DrBphP solubility, the DrBphP-T7 protein was soluble but the dimeric Y-shaped structure of the 150-kDa T7 antibody resembled too closely the dimeric DrBphP particles to be of help in orienting the raw EM images. We found that repurifying DrBphP and DrBphP-EGFP samples by size exclusion chromatography immediately before preparing the EM grids dramatically improved the homogeneity and contrast of the particles in the EM images, likely by excluding denatured chromoproteins.
The mass of the DrBphP dimer (166 kDa) is relatively small for single particle EM, which is generally applied to imaging protein complexes larger than 500 kDa (25). To overcome this limitation, we combined cryoEM with negative-staining EM. Despite its known problem with structural flattening by the stain salt, negative-staining offers high contrast that can reveal structural features of relatively small particles (26). These negative staining images allowed us to derive well-defined and consistent 2D class averages from raw particle images, which were then used to generate starting 3D models for further refinement of cryoEM images (Fig. 1 B–D and Fig. S1C). For cryoEM, the addition of a thin carbon film (approximately 20 Å) over the holey carbon film during EM grid preparation enabled formation of very thin vitreous ice. When combined with a relatively high underfocus value, this fixation method resulted in high-contrast EM images of DrBphP particles.
We recorded a set of high quality negative-staining micrographs and manually picked a dataset containing approximately 20,800 high-contrast DrBphP images that was computationally organized into 200 classes. Images belonging to the same class were then combined to generate average 2D particle views with improved signal-to-noise (Fig. S1C). Due to the elongated and somewhat flat shape of DrBphP generated by negative staining, many 2D averages were of various side views, a smaller number were in tilted and edge-on views, and a few cases represented top views. The dominant feature of the side views was the near mirror symmetry between the left and right halves of the particles (Fig. 1B), indicating that DrBphP is a 2-fold symmetric dimer generated by a parallel association of two monomers.
The orientations of the 2D views were then determined by negative-staining EM analysis of the DrBphP dimer bearing a C-terminal EGFP. As compared to DrBphP alone, we consistently observed an additional subparticle attached to one end of the side views and nowhere else, which likely represented two EGFP molecules based on its size and density (Fig. 1C). The EGFP moieties were not well-defined in the EM images, likely because the linker between the EGFP and DrBphP polypeptides allowed partial flexibility. Nevertheless, our localization of EGFP combined with the previous crystallographic structures of PAS-GAF and PAS-GAF-PHY domains (13, 15) allowed us to orient the DrBphP dimer in these side views (Figs. 1 B–C). This arrangement is roughly consistent with the parallel dimer arrangement predicted for the photosensory modules of DrBphP and PaBphP (13, 15), with the PAS-GAF, PHY, and HK domains predicted to stack on top of each other. The extensive contacts between dimers also implied that the photosensory and output HK modules of DrBphP (PAS-GAF-PHY-HK) both contribute to dimerization rather than the HK module being the sole interface (Fig. 1B). The densities in the side views reflecting the HK domains were the most variable as evidenced by their diffuse appearance and often slightly shifted locations relative to each other (Fig. 1D).
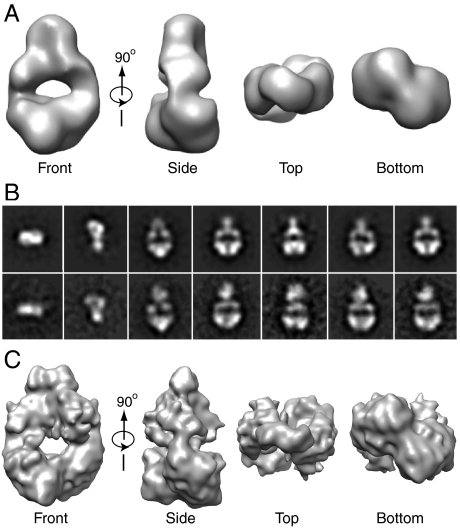
We determined by the common line technique an initial 3D map at 25 Å resolution from selected negative-staining 2D class averages and by applying 2-fold symmetry (Fig. 2A). The reprojections of the 3D map are mostly consistent with the reference-free 2D class averages, except minor inconsistence at the HK domain due to the above-noted flexibility (Fig. 2B). Notable features were a slightly twisted overall arrangement, an electron-dense region at the bottom that could be assigned to the PAS-GAF domains, and an electron transparent region at the center, (Fig. 2A). Based on the crystallographic structure of PaBphP (15), the hole likely represents the loose interface between the GAF and PHY domains in the dimer, which involves just the connecting helices and the PHY hairpin. Only part of the mass for the two HK domains was apparent with the missing density likely caused by their inherent flexibility.
Fig. 2.
3D reconstruction of the DrBphP dimer. (A) The 3D map presented in several surface views obtained from negative-staining EM images. (B) Several reference-free 2D class averages of the DrBphP in comparisons with the reprojections of the negative-staining 3D map. (C) The final 3D cryoEM map of Pr presented in several surface views.
CryoEM Image Analyses of DrBphP.
To obtain a more resolved 3D view of the DrBphP dimer devoid of potential stain artifacts, we performed cryoEM with the preparations frozen as Pr. In contrast to the abundance of the flat side views in the negatively stained images, the clearest views of DrBphP in vitreous ice were top and tilted views, due to the relatively high projection density of particles in these orientations. We recorded a cryoEM dataset of > 21,000 DrBphP particle projections, and refined the orientations of the images, first with the negative-staining 3D map and subsequently with the cryoEM-image derived map. After several rounds of iterative refinement, we finally obtained a stable cryoEM 3D map of the DrBphP dimer with the improved resolution of approximately 13 Å based on Fourier shell correlation of a 2-fold symmetric particle (Fig. 2C). The handedness of the cryoEM map, which was undefined by the common line technique, was deduced by docking the 3D map with the crystal structure of the DrBphP PAS-GAF region (12, 13)
Docking Crystal Structures for the PAS, GAF, PHY, and HK Domains into the DrBphP 3D Scaffold.
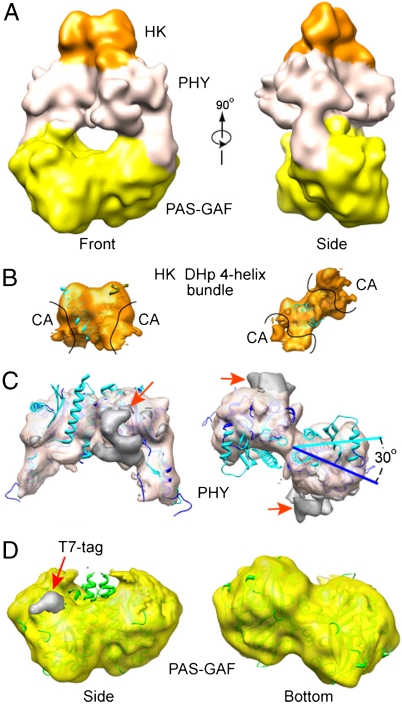
As with the negative-staining model, the cryoEM model depicted the DrBphP dimer as a twisted structure containing a central hole with only part of the HK domains evident (Fig. 2C). Support for this model was then provided by docking it with the X-ray crystallographic structures available for the major domains (Fig. 3A). The 1.45 Å resolution crystal structure of the DrBphP PAS-GAF dimer [PDB ID 2O9C (13)] fit extremely well into the bottom portion of the cryoEM model (correlation coefficiency of 95%) and even faithfully captured the dimer interface between the sister GAF domains (Fig. 3D). Only two small regions of density in the EM map corresponding to the PAS-GAF dimers were not occupied by the crystal structure (Fig. 3D, red arrow). These extra densities, located immediately outside the N-terminus of the crystal structure, appeared to be generated by the 17 residues upstream of Ala-4 that include the appended 14-residue T7 tag and spacer. This segment was disordered in the DrBphP PAS-GAF crystal structure (13).
Fig. 3.
Docking domain crystal structures into the 3D cryoEM map of the DrBphP dimer in the Pr state. (A) Two orthogonal surface views of the density map colored according to the location of the PAS-GAF (yellow), PHY (white), and HK domains (orange). (B–D) Docking of crystal structures for individual domains into the cryoEM map. The cryoEM densities for individual domains are segmented and shown separately in the semitransparent surface views. (B) Docking of the four-helix bundle from the DHp subdomain extracted from the T. maritima HK853 structure [PDB ID 3DGE (10, 27)]. The crystal structure of the CA subdomain was omitted due to the lack of density for this region in the cryoEM model. (C) Docking of the PHY domains extracted from the PaBphP photosensory module structure [PDB ID 3C2W (15)]. The PHY domains in the DrBphP cryoEM map (blue) are rotated approximately 30% from their positions in the PaBphP crystal structure (cyan). The largest region of unoccupied density is located by the red arrows. (D) Docking of the PAS-GAF domains from DrBphP [PDB ID 2O9C (13)]. The extra density in the cryoEM map marked by the red arrow is attributed to the disordered N-terminal 17 residues bearing the T7 tag.
In the absence of a crystal structure of the DrBphP PHY domain, we attempted to dock a comparable 23-kDa PHY domain structure, determined as part of a larger 2.9 Å resolution structure encompassing the photosensory module of PaBphP [PDB ID 3C2W (15)]. Although the two paired PaBphP PHY structures could be readily fit into the DrBphP cryoEM map, an exact alignment was not possible, suggesting that the conformation of the PHY domains determined by cryoEM differs subtly from that determined by X-ray crystallography. For example, fitting of the entire PaBphP photosensory module (PAS-GAF-PHY) as a rigid body into the cryoEM map revealed an approximately 30° rotation of the PHY domains (Fig. 3C). This discrepancy was not completely solved by docking the dimeric PHY domains of PaBphP alone as a single rigid body, nor by docking the two monomeric PHY domains as two separate rigid bodies (Fig. 3C). Given the low amino acid sequenced identity (21%) of their PHY domains, this structural difference could reflect inherent conformational differences between the photosensory modules of DrBphP and PaBphP. Alternatively, it could reflect natural flexibility in the GAF-PHY domain contacts that may be allowed by the extended connections that scaffold these two regions (long helical strand and PHY hairpin). In fact, such quaternary plasticity in the cross-domain helical strands has been seen by comparing the structures of the PaBphP1 and SynCph1 photosensory modules and by structural variations in the asymmetric crystal units of PaBphP (11, 15).
Both our negative staining and cryoEM maps contained much less density than expected for the two C-terminal 25 kDa HK domains regions (Fig. 2 A and C). Combined with structural heterogeneity observed in the 2D image averages for this region (Fig. 1C), this lack of density suggested that the HK domains are highly mobile in the DrBphP dimer. To predict how the HK domains are arranged and to assess how much of their density is missing in the cryoEM map, we attempted to dock a crystallographic structure of an HK domain. In the absence of a 3D structure for a Phy HK domain, we used the next closest available structure based on domain organization, that being the class I HK domain from the Thermotoga maritima soluble cytoplasmic sensor HK853 (PDB ID 3DGE) (10, 27). Like predictions for DrBphP and many other microbial Phys (4, 28), the T. maritima HK853 HK domain is divided into an N-terminal DHp subdomain, containing a helix-turn-helix bundle that houses the dimerization and histidine phosphoacceptor sites, followed by a C-terminal globular (CA) subdomain that directs ATP-dependent phosphorylation of the histidine. Upon placing the T. maritima HK domain dimer structure into the top region of the DrBphP EM map, it was immediately clear that much of the CA subdomain densities was missing in the EM map. In fact, only a portion of the DHp helix-turn-helix bundle was apparent, strongly suggesting that the HK domains in DrBphP can also dimerize but that most of the domain is highly flexible (Fig. 3B).
Revised Architecture of the DrBphP Dimer and Implications to Signaling.
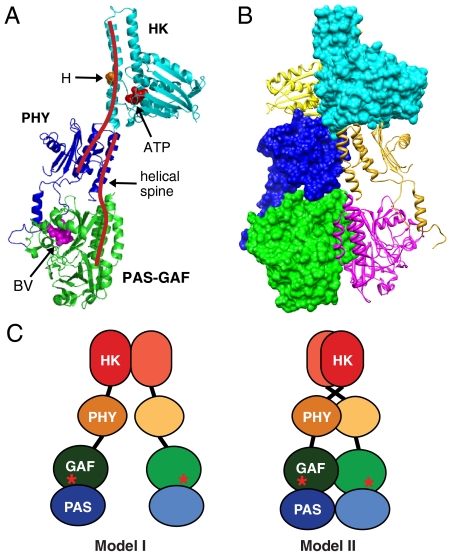
Based on our docking studies, we assembled a pseudoatomic model of the full-length DrBphP dimer (Fig. 4 A and B). In contrast to previous models, which predicted that Phys dimerize solely through their HK or HK-related domains (Fig. 4C, model I), our improved model shows that the PAS-GAF and PHY domains also contribute. In fact, each monomer of DrBphP appears to form an elongated and slightly bent structure that intimately twists in parallel around each other in a right-handed fashion, thus generating an extensive dimer contact involving most of each polypeptide (Fig. 4C, model II). Much of this contact involves an approximately 118 Å long spine comprised of two long overlapping helices, one extending from the GAF to the PHY domain (12 turns) and the other extending from PHY to the HK domain (14 turns). Based on EM density, the cryoEM structure also predicts flexibility in the GAF/PHY linkers and in the relative orientations of the sister HK domains. For T. maritima HK853, such flexibility in the HK domain is key to signaling in which a 20° rotation of the CA subdomain with respect to the DHp subdomain is likely required for differential autophosphorylation and signal output (10, 27).
Fig. 4.
Proposed model of the DrBphP dimer derived from both cryoEM and crystal structures. (A) Ribbon diagram of the DrBphP monomer assembled based on cryoEM from the crystal structures of the PAS-GAF domains (green) from DrBphP (13), the PHY domain (purple) from PaBphP (15), and the HK domain (cyan) from T. maritima HK853 (10). The positions of BV, the histidine (H) and ATP-binding site involved in autophosphorylation, and the helical spine are indicated. (B) Composite ribbon and space filling views of the DrBphP dimer showing the right-hand twisted association of the monomers. (C) The previous model (I) and our updated model (II) of Phy dimerization. Astericks indicate BV.
Combined with predictions generated from the paired Pr and Pfr structures of the bilin and GAF domain (16, 17), our cryoEM structure offers a plausible model for Phy phototransformation. Light absorption induces conformational changes within the bilin, especially with regard to the A pyrrole ring. Bilin motion then directs a set of local conformational changes within the GAF domain that affect the GAF/GAF dimerization interface and the GAF/PHY contact involving the PHY hairpin. These structural changes are then propagated along the interfaces between sister PHY domains and ultimately between the sister HK domains. Based on HK model from T. maritima HK853 (10, 27), activation of the Phy HK domain would then involve repositioning the phosphoacceptor histidine relative to the ATP-binding site to promote phosphotransfer in cis or trans within the HK dimer. Central to this model are the long helical bundles that connect the GAF, PHY, and HK domains in each Phy polypeptide and form the backbone of the dimeric structure. Such helical bundles are widely found in other signaling proteins where they connect modular domains that transmit signal information by sequential conformational perturbations (29). By transmitting motion from the bilin down this spine to the spatially distant output modules, it is conceivable that all Phys regardless of their output mechanism [e.g., HK, Ser/Thr kinase, methyl accepting, protein phosphatase 2C, diguanylate cyclase/phosphodiesterase (4, 7)] differentially photoregulate signaling simply by reorienting their sistered output regions. The ability to engineer novel photoswitches by attaching various output modules to a Phy photosensory core is testament to this principle (30).
Biochemical Support for the CryoEM Model for DrBphP Phototransformation.
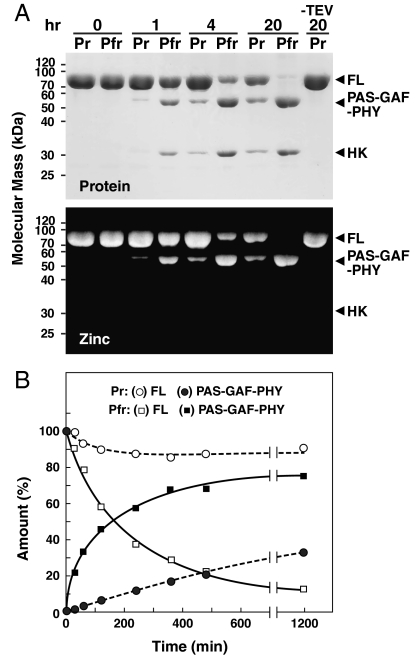
To help support our model of Phy photoactivation, we tested by proteolytic sensitivity whether conformational changes within the GAF domain of DrBphP are indeed propagated to the distal output module. Here, we introduced a tobacco etch virus (TEV) protease cleavage site immediately proximal (13 residues) to the phosphoacceptor histidine within the helical DHp subdomain (see Fig. 1A), and assayed if its exposure is affected by photoconversion. Recombinant DrBphP-TEV readily assembled with BV to generate a fully R/FR photochromic photoreceptor (Fig. S1B) and upon incubation with TEV protease was digested into the expected PAS-GAF-PHY and HK fragments (Fig. 5A). During native PAGE, the released PAS-GAF-PHY fragments migrated as the expected dimer (13), whereas the HK fragment migrated as a smear, suggesting that the released polypeptide formed a heterogeneous collection of dimeric and higher order complexes. Comparison of cleavage rates for Pr and an approximately 80% Pfr equilibrium mixture generated with saturating R revealed that the TEV site was substantially more exposed as Pfr (Fig. 5B). In fact, initial cleavage rates estimated that the region near the phosphoacceptor histidine is at least 20 times more solvent accessible as Pfr, strongly suggesting that a conformational change occurs surrounding this site as a result of long-range signaling from the chromophore.
Fig. 5.
Light-induced conformation changes transmitted from the bilin to the HK domain as determined by proteolytic susceptibility of DrBphP-TEV. See Fig. 1A for location of TEV cleavage site. (A) Proteolytic cleavage of DrBphP-TEV by TEV protease in the dark-adapted Pr state and following saturating R irradiation, which generates a mixture containing approximately 80% Pfr. Arrowheads locate the SDS-PAGE migration of the PAS-GAF-PHY and HK domains proteolytically released from the full-length chromoprotein (FL). (Upper) Detection of the protein products by staining with Coomassie Blue. (Lower) Detection of the BV-containing fragments by zinc-induced fluorescence following SDS-PAGE. (B) Rates of TEV digestion as Pr and Pfr. Open figures represent the rate of loss for full-length DrBphP-TEV. Closed figures represent the rate of appearance for the PAS-GAF-PHY fragment.
Final Perspectives.
Here, we present a detailed, unambiguous picture of a full-length Phy dimer that reveals an extensive intertwined dimerization interface between the two monomers, which may be central to signaling. Our work demonstrates further that a combination of negative-staining EM and cryoEM can provide important structural information for protein complexes previously considered too small for single particle EM analyses. The accuracy of the model was well supported by our ability to faithfully dock previously determined 3D structures of the various Phy subregions into the cryoEM model of the entire complex. With respect to Phys, this approach could overcome the substantial barriers inherent in other structure-based methods (NMR and X-ray crystallography) to provide testable models of action, including the 3D structure of full-length plant Phy dimers, and Phys in association with components of their immediate signal transduction chain(s).
At present, it is unclear whether our cryoEM model of DrBphP as Pr represents the inactive or active signaling state. Even though the HK domain of DrBphP contains all the necessary motifs for phosphotransfer including the positionally conserved histidine that should directly participate (31), we have not yet succeeded in detecting its kinase activity by standard ATP-dependent in vitro autophosphorylation assays. Extrapolation from other BphPs has been challenged in turn by the observations that some Phys are more active in phosphotransfer as Pr (8, 9), whereas others are more active as Pfr (6, 7). If our Pr model represents the more active form of DrBphP, then its twisting parallel dimer interface could encourage phosphotransfer within the dimer with photoconversion to Pfr then relaxing this twist and attenuating activity. If Pfr is more active, we might expect that photoconversion reorients the sister HK domains relative to each other to generate a new set of contacts that better promotes phosphotransfer. Clearly, cryoEM models of DrBphP as Pfr are now needed to assess the orientation of the sister HK domain relative to each other and the photosensory module and biochemical assays to determine which form (Pr or Pfr) is the active signaling conformer.
Materials and Methods
Preparation of D. radiodurans BphP Chromoproteins.
Full-length DrBphP [755 amino acids plus 22 for the T7 (MASMTGGQQMGRGS) and 6His tags (LEHHHHHH) (tag sequence underlined)] was expressed and assembled in Escherichia coli, and then purified by sequential nickel nitrilotriacetic acid and Superdex 200 size-exclusion chromatography as described (12, 13, 23). Previously frozen samples were repurified by Superdex-200 chromatography immediately before preparing the EM grids. DrBphP-EGFP was generated by appending the EGFP coding region (265 amino acids: Clontech) to the 3′ end of the last DrBphP codon using an XhoI site. DrBphP-TEV was created by introducing the TEV protease cleavage site (ENLYFQG) between residues 518 and 519 of a DrBphP flanked by an N-terminal 14-residue T7 tag and a C-terminal 19-residue Flag-6His tag [GGGDYKDDDDKLEHHHHHH (tag sequences underlined)]. Covalently attachment of the bilin and proper photoconversion of the chromoproteins were assessed by zinc-induced fluorescence of the bound bilin following SDS-PAGE and by absorption spectroscopy following saturating R and FR (12, 13). Protease sensitivity for DrBphP-TEV was determined by incubating the chromoprotein at 25 °C in the dark-adapted Pr state or during continuous irradiation with R (approximately 80% Pfr) in the presence of 100 units of the 27 kDa catalytic domain of TEV protease (Invitrogen AcTEV) per mg of DrBphP. Identity of the released fragments was confirmed by immunoblot analysis with anti-T7 (Novagen) and anti-Flag antibodies (Sigma).
Negative-Staining and CryoEM.
For negative-staining EM, the grids were prepared at room temperature in white light (mixed Pr/Pfr) with 50 μg/mL solutions and stained with two consecutive drops of 2% phosphotungstic acid (pH 7.4) for 30 sec. Images were recorded under low-dose conditions (10 e/ Å2) in a JEOL JEM-1200EX transmission electron microscope operated at 120 kV at a magnification of 50,000× and with an underfocus value of 1.5 μm. For cryoEM, the grids were prepared in the dark with the Pr state (confirmed by absorption spectroscopy) using a Vitrobot (FEI) with the specimen chamber set to 11 °C and 90% humidity. A 4.5-μL sample was applied to a quantifoil grid covered with a second layer of continuous thin carbon film, and after blotting off excess fluid, plunge frozen in liquid ethane. The frozen-hydrated samples were imaged in a JEOL JEM-2010F transmission electron microscope operated at 200 kV at the magnification of 50,000× with underfocus values varied from 2.0 to 4.5 μm. All cryoimages were recorded on Kodak SO-163 negative films, which were developed for 12 min in full-strength Kodak D-19 developer at 20 °C. Micrographs without sample drift and objective lens astigmatism were digitized with a Nikon Supercool scanner 8000ED at a step size of 7.3 μm, and binned for image processing to the pixel size of 12.7 μm, which corresponded to 2.54 Å at the sample level.
2D Image Classifications and 3D Image Reconstruction.
Image processing and 3D reconstruction of single particle images employed the software packages EMAN (32) and SPIDER (33). From the negative stained samples, 20,879 (DrBphP) and 7817 (DrBphP-EGFP) images were combined for the final reference-free 2D image classification and for the calculation of the starting 3D models. The cryoEM dataset combined 21,469 images. The defocus value was determined for each micrograph, and the contrast transfer function effect in the images was corrected by flipping the inverted phases. For 3D reconstruction, multiple starting models generated from selected 2D averages of the negatively stained particles by common line technique were used to refine the cryoEM dataset. All the models were first refined using 2 × 2 binned particle images (5.08 Å/pixel) and the derived structures were evaluated by examining consistency between their projections and a large number of reference-free 2D averages of both negative-staining and cryoEM data. Those 3D models that exhibited better consistency were further refined, with the derived 3D models examined further using the same method. Finally, the model that showed the best consistency was used as the starting model to refine the original cryoEM dataset. The resolution of the final map was estimated by Fourier shell correlation of two maps calculated separately from two halves of the dataset. The map was low-pass filtered to the estimated resolution of 13 Å for the cryoEM structure and 25 Å for the negative-staining EM structure. Surface rendering, docking of atomic structures, and density segmentation were performed in University of California-San Francisco (UCSF) Chimera (34).
Supplementary Material
Acknowledgments.
We thank Dr. Tao Wang for help with size-exclusion chromatography. This work was supported by grants from the Brookhaven National Laboratory Laboratory-Directed Research and Development Project 10-16 and National Institutes of Health Grant GM74985 (Huilin Li), and by National Science Foundation Grant MCB-07191530 and a grant from the University of Wisconsin-Madison College of Agricultural and Life Science (Hatch) (R.D.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001908107/-/DCSupplemental.
References
- 1.Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vierstra RD, Karniol B. Phytochromes in microorganisms. In: Briggs WR, Spudich JL, editors. Handbook of Photosensory Receptors. Weinheim, Germany: Wiley-VCH Press; 2005. pp. 171–196. [Google Scholar]
- 4.Karniol B, Wagner JR, Walker JM, Vierstra RD. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- 7.Yeh KC, Lagarias JC. Eukaryotic phytochromes—light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 9.Karniol B, Vierstra RD. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc Natl Acad Sci USA. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA. 2008;105:14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 13.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Stojkovic EA, Kuk J, Moffat K. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc Natl Acad Sci USA. 2007;104:12571–12576. doi: 10.1073/pnas.0701737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci USA. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornilescu G, Ulijasz AT, Cornilescu CC, Markley JL, Vierstra RD. Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. J Mol Biol. 2008;383:403–413. doi: 10.1016/j.jmb.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulijasz AT, et al. Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature. 2010;463:250–254. doi: 10.1038/nature08671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 19.Jones AM, Erickson HP. Domain structure of phytochrome from Avena sativa visualized by electron microscopy. Photochem Photobiol. 1989;49:479–483. doi: 10.1111/j.1751-1097.1989.tb09198.x. [DOI] [PubMed] [Google Scholar]
- 20.Tokutomi S, et al. A model for the dimeric molecular structure of phytochrome based on small-angle X-ray scattering. FEBS Lett. 1989;247:139–142. [Google Scholar]
- 21.Jones AM, Vierstra RD, Daniels SM, Quail PH. The role of separate molecular domains in the structure of phytochrome from Avena sativa. Planta. 1985;164:501–506. doi: 10.1007/BF00395966. [DOI] [PubMed] [Google Scholar]
- 22.Evans K, Grossmann JG, Fordham-Skelton AP, Papiz MZ. Small-angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state. J Mol Biol. 2006;364:655–666. doi: 10.1016/j.jmb.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 23.Wagner JR, et al. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J Biol Chem. 2008;283:12212–12226. doi: 10.1074/jbc.M709355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Gurtu V, Kain SR. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun. 1996;227:707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- 25.Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anantharaman V, Balaji S, Aravind L. The signaling helix: a common functional theme in diverse signaling proteins. Biol Direct. 2006;1:25. doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levskaya A, et al. Synthetic biology: Engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 31.Davis SJ, Vener AV, Vierstra RD. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- 32.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 33.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.