Abstract
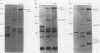
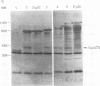
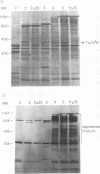
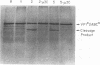
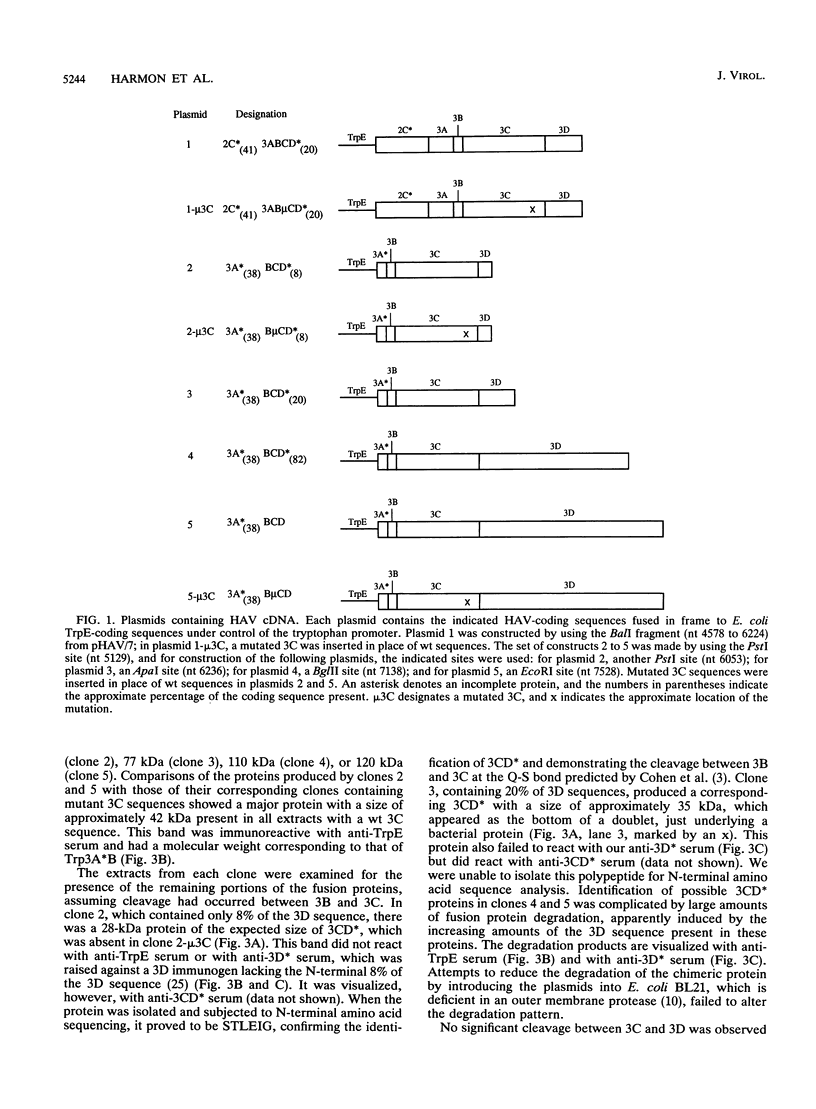
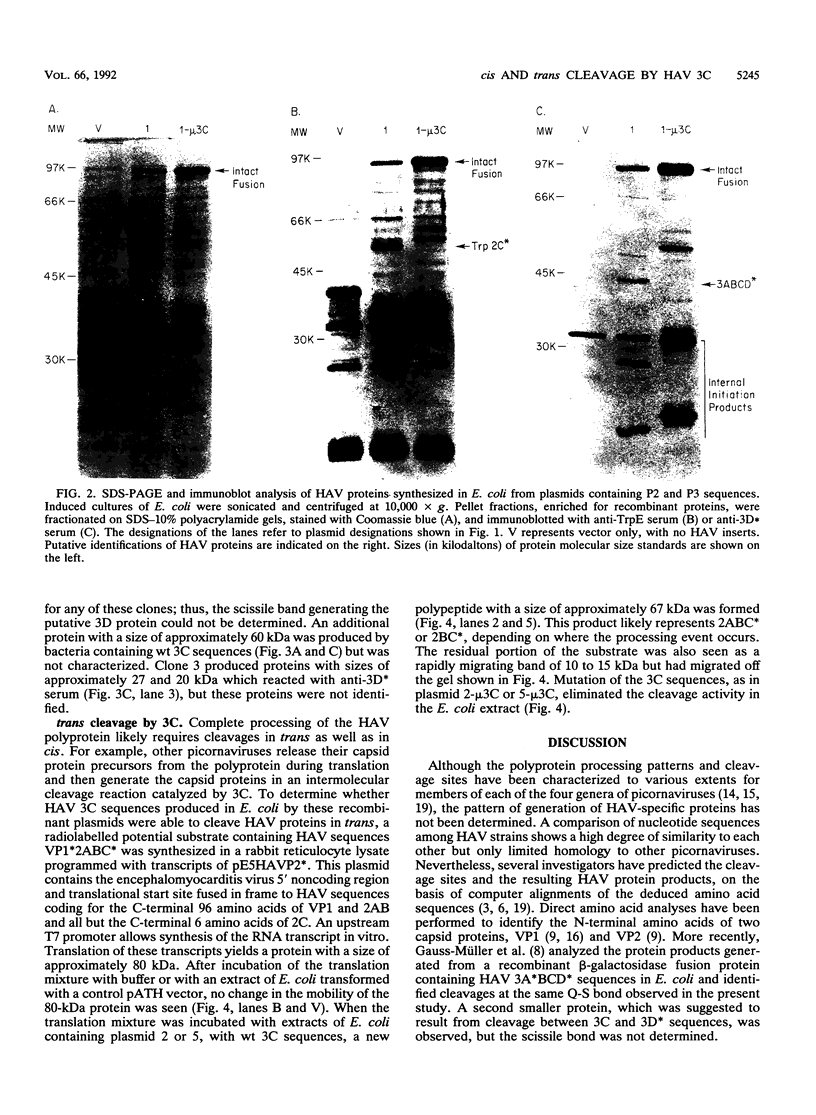
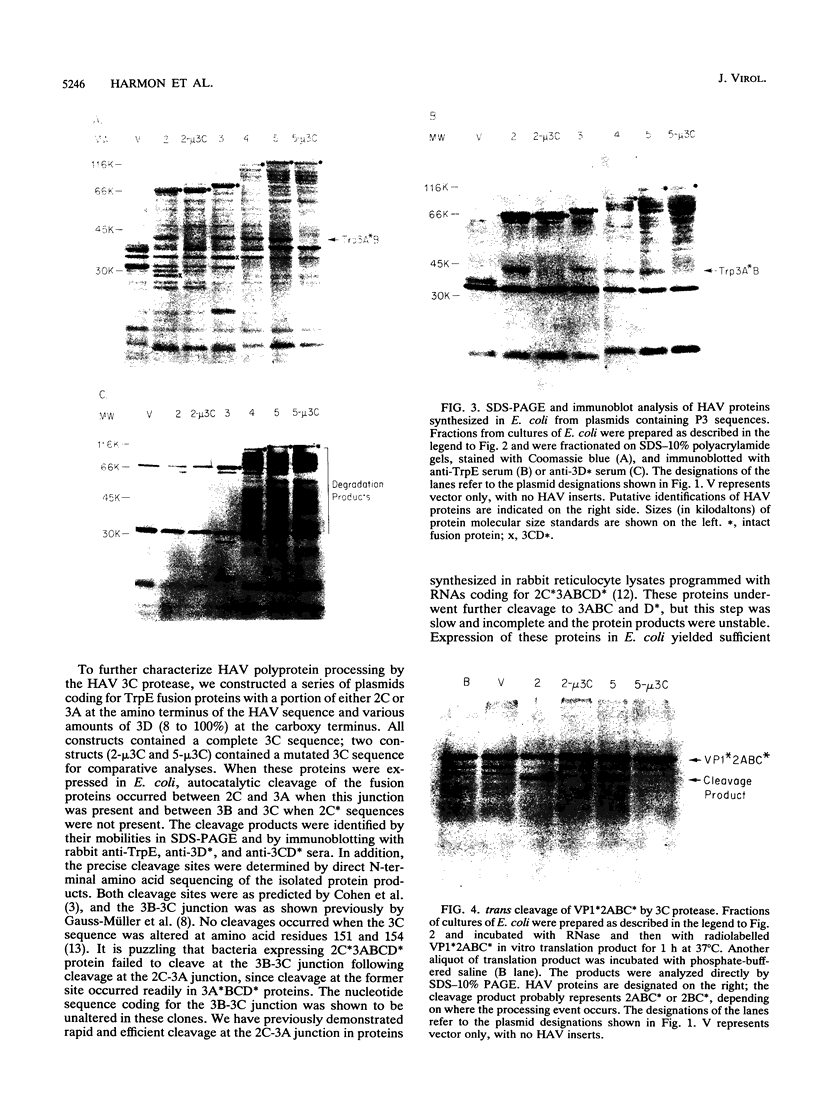
To determine the P3 region protein-processing sites cleaved by the hepatitis A virus 3C protease, a nested set of constructs containing a portion of 3A (3A* [the asterisk denotes an incomplete protein]), 3B and 3C and various amounts of 3D, fused in frame to Escherichia coli TrpE-coding sequences under control of the tryptophan promoter, was made. Additional plasmids that encoded a portion of 2C (2C*) and the P3 proteins, including complete or incomplete 3D sequences, were constructed. After induction, E. coli containing these recombinant plasmids produced high levels of fusion proteins as insoluble aggregates. 3C-mediated cleavage products were identified by comparison of expression with a matching set of plasmids, containing an engineered mutation in 3C. Cleavage products were detected by immunoblot analyses by using antisera against the TrpE protein, against 3D*, and against 3CD*. Scissile bonds were determined by N-terminal amino acid sequencing of the proteins formed by cleavage. The results showed that when a portion of 2C was present, the primary cleavage by the 3C protease was between 2C and 3A, and the cleavage site was QG, as predicted by J. I. Cohen, J. R. Ticehurst, R. H. Purcell, A. Buckler-White, and B. M. Baroudy, J. Virol. 61:50-59, 1987. Very little further cleavage of the released P3 protein was detected. When the fusion protein contained no 2C and included only 3A*-to-3D sequences, efficient cleavage occurred between 3B and 3C, at the QS pair, also as predicted by Cohen et al. (J. Virol. 61:50-59, 1987). The latter proteins were also cleaved between 3C and 3D, but less efficiently than between 3B and 3C. Extracts of bacteria expressing proteins from 3A* to 3D also cleaved a radiolabelled hepatitis A virus substrate containing VP1*2ABC* sequences in trans.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cho M. W., Ehrenfeld E. Rapid completion of the replication cycle of hepatitis A virus subsequent to reversal of guanidine inhibition. Virology. 1991 Feb;180(2):770–780. doi: 10.1016/0042-6822(91)90090-x. [DOI] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Feinstone S. M., Rosenblum B., Purcell R. H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987 Oct;61(10):3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Purcell R. H., Buckler-White A., Baroudy B. M. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987 Jan;61(1):50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulepis A. G., Locarnini S. A., Westaway E. G., Tannock G. A., Gust I. D. Biophysical and biochemical characterization of hepatitis A virus. Intervirology. 1982;18(3):107–127. doi: 10.1159/000149314. [DOI] [PubMed] [Google Scholar]
- De Chastonay J., Siegl G. Replicative events in hepatitis A virus-infected MRC-5 cells. Virology. 1987 Apr;157(2):268–275. doi: 10.1016/0042-6822(87)90269-8. [DOI] [PubMed] [Google Scholar]
- Diamond D. C., Wimmer E., von der Helm K., Deinhardt F. The genomic map of hepatitis A virus: an alternate analysis. Microb Pathog. 1986 Apr;1(2):217–219. doi: 10.1016/0882-4010(86)90023-9. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Gauss-Müller V., Jürgensen D., Deutzmann R. Autoproteolytic cleavage of recombinant 3C proteinase of hepatitis A virus. Virology. 1991 Jun;182(2):861–864. doi: 10.1016/0042-6822(91)90630-t. [DOI] [PubMed] [Google Scholar]
- Gauss-Müller V., Lottspeich F., Deinhardt F. Characterization of hepatitis A virus structural proteins. Virology. 1986 Dec;155(2):732–736. doi: 10.1016/0042-6822(86)90234-5. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon S. A., Summers D. F., Ehrenfeld E. Detection of hepatitis A virus RNA and capsid antigen in individual cells. Virus Res. 1989 Apr;12(4):361–369. doi: 10.1016/0168-1702(89)90093-2. [DOI] [PubMed] [Google Scholar]
- Jia X. Y., Ehrenfeld E., Summers D. F. Proteolytic activity of hepatitis A virus 3C protein. J Virol. 1991 May;65(5):2595–2600. doi: 10.1128/jvi.65.5.2595-2600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. Y., Scheper G., Brown D., Updike W., Harmon S., Richards O., Summers D., Ehrenfeld E. Translation of hepatitis A virus RNA in vitro: aberrant internal initiations influenced by 5' noncoding region. Virology. 1991 Jun;182(2):712–722. doi: 10.1016/0042-6822(91)90612-f. [DOI] [PubMed] [Google Scholar]
- Kräusslich H. G., Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- Lawson M. A., Semler B. L. Picornavirus protein processing--enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol. 1990;161:49–87. [PubMed] [Google Scholar]
- Linemeyer D. L., Menke J. G., Martin-Gallardo A., Hughes J. V., Young A., Mitra S. W. Molecular cloning and partial sequencing of hepatitis A viral cDNA. J Virol. 1985 May;54(2):247–255. doi: 10.1128/jvi.54.2.247-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J. L. Classification of hepatitis A virus as enterovirus type 72 and of hepatitis B virus as hepadnavirus type 1. Intervirology. 1982;18(3):105–106. doi: 10.1159/000149313. [DOI] [PubMed] [Google Scholar]
- Najarian R., Caput D., Gee W., Potter S. J., Renard A., Merryweather J., Van Nest G., Dina D. Primary structure and gene organization of human hepatitis A virus. Proc Natl Acad Sci U S A. 1985 May;82(9):2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- Parks G. D., Duke G. M., Palmenberg A. C. Encephalomyocarditis virus 3C protease: efficient cell-free expression from clones which link viral 5' noncoding sequences to the P3 region. J Virol. 1986 Nov;60(2):376–384. doi: 10.1128/jvi.60.2.376-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. V., Tada H., von der Helm K., Wissel T., Kiehn R., Wimmer E., Deinhardt F. The entire nucleotide sequence of the genome of human hepatitis A virus (isolate MBB). Virus Res. 1987 Aug;8(2):153–171. doi: 10.1016/0168-1702(87)90026-8. [DOI] [PubMed] [Google Scholar]
- Provost P. J., Hilleman M. R. Propagation of human hepatitis A virus in cell culture in vitro. Proc Soc Exp Biol Med. 1979 Feb;160(2):213–221. doi: 10.3181/00379727-160-40422. [DOI] [PubMed] [Google Scholar]
- Szewczyk B., Summers D. F. Preparative elution of proteins blotted to Immobilon membranes. Anal Biochem. 1988 Jan;168(1):48–53. doi: 10.1016/0003-2697(88)90008-5. [DOI] [PubMed] [Google Scholar]
- Updike W. S., Tesar M., Ehrenfeld E. Detection of hepatitis A virus proteins in infected BS-C-1 cells. Virology. 1991 Nov;185(1):411–418. doi: 10.1016/0042-6822(91)90789-e. [DOI] [PubMed] [Google Scholar]
- Wheeler C. M., Fields H. A., Schable C. A., Meinke W. J., Maynard J. E. Adsorption, purification, and growth characteristics of hepatitis A virus strain HAS-15 propagated in fetal rhesus monkey kidney cells. J Clin Microbiol. 1986 Mar;23(3):434–440. doi: 10.1128/jcm.23.3.434-440.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]