Abstract
A unique property of the mammalian embryo is that stem cells can be derived from its early tissue lineages. These lineages will give rise to the fetus as well as essential extraembryonic tissues. Understanding how chromatin regulation participates in establishment of these lineages in the embryo and their derived stem cells provides insight that will critically inform our understanding of embryogenesis and stem cell biology. Here, we compare the genomewide location of active and repressive histone modifications in embryonic stem cells, trophoblast stem cells, and extraembryonic endoderm stem cells from the mouse. Our results show that the active modification H3K4me3 has a similar role in the three stem cell types, but the repressive modification H3K27me3 varies in abundance and genomewide distribution. Thus, alternative mechanisms mediate transcriptional repression in stem cells from the embryo. In addition, using carrier chromatin immunoprecipitation we show that bivalent histone domains seen in embryonic stem cells exist in pluripotent cells of the early embryo. However, the epigenetic status of extraembryonic progenitor cells in the embryo did not entirely reflect the extraembryonic stem cell lines. These studies indicate that histone modification mechanisms may differ between early embryo lineages and emphasize the importance of examining in vivo and in vitro progenitor cells.
Keywords: epigenetics, methylation, embryogenesis, extraembryonic
Understanding the mechanisms by which tissues differentiate from each other during development is essential for understanding how to effectively use stem cells. During mouse development, the implanting blastocyst is composed of trophectoderm (TE), primitive endoderm (PE), and epiblast (EPI). At this stage in development, individual cells are already lineage restricted, but capable of differentiating into all of the more specialized cells within that lineage (1). Once established by lineage-specific transcription factor networks, the transcriptional memory of each progenitor cell may be maintained by epigenetic modifications (2). How genetic factors and epigenetic modifiers interact to regulate lineage decisions is therefore an important goal among stem cell researchers and developmental biologists alike. Targeted deletion of numerous epigenetic modifiers has been shown to result in ectopic expression of lineage-specific genes and periimplantation lethality, demonstrating that establishment and maintenance of an epigenetic program is essential for the earliest stages of development (3–5).
Permanent stem cell lines can be derived from the three tissue lineages present in the mouse blastocyst and are used as an in vitro model to study the regulation of early lineage progenitors. These cell lines include embryonic stem (ES) cells (6, 7), trophoblast stem (TS) cells (8), and extraembryonic endoderm stem (XEN) cells (9). All three stem cells can self-renew in vitro, but differentiate and contribute in a lineage-appropriate manner in vitro and in vivo (8–10). Epigenetic regulation of this process could provide an elegant mechanism for maintaining lineage identity following isolation of these progenitors from their embryonic environment during stem cell establishment. Lineage-specific differences in chromatin have been observed at the level of histone modifications (11, 12). Whether these lineage-specific differences in histone marks impact transcriptional regulation and are involved in the establishment or maintenance of lineage progenitors is not known. It is essential, therefore, to compare the histone methylation status of the three lineages derived from the mouse blastocyst.
Self-renewal in ES cells is dependent on maintaining appropriate epigenetic regulation, including nucleosome remodeling and modification of DNA and histones (2, 4, 13). In particular, bivalent domains that are composed of an unusual coexistence of active, such as histone H3 lysine 4 trimethylation (H3K4me3), and repressive, such as histone H3 lysine 27 trimethylation (H3K27me3), modifications have been shown to be overrepresented in putative regulatory loci of developmentally important genes in ES cells, as compared to other cell types (14–16). These findings led to the prediction that bivalent domains may contribute to pluripotency in ES cells by keeping developmental genes poised for activation during differentiation. However, Polycomb group (PcG) proteins, which mediate H3K27me3, are dispensable for maintaining pluripotency and lineage potential in ES cells, but are required for precise control of gene expression during differentiation (17–19). Together, these studies suggest that PcG-mediated epigenetic repression may regulate transitions in cell fate during development. However, extraembryonic lineages have been shown to differ from the pluripotent embryonic lineages in a number of epigenetic pathways. X-chromosome inactivation occurs in all lineages, but is paternally imprinted in extraembryonic cells and is specifically dependent on the PcG protein Eed to maintain the inactive state (20, 21). Epigenetic regulation of imprinted gene expression also differs between embryonic and extraembryonic lineages, with histone modifications maintaining imprinting in extraembryonic cells independently of DNA methylation (22, 23). Global differences in epigenetic status between embryonic and extraembryonic lineages also exist. DNA methylation levels are lower in extraembryonic cells compared to embryonic cells (24, 25). However, recent genomewide studies comparing promoter methylation in TS and ES cells have shown that the majority of differences are likely to be located in nongenic regions (26). Additionally, immunocytochemistry revealed lower levels of several histone modifications, including H3K27me3 and histone H3 lysine 9 acetylation (H3K9ac), in TE compared to EPI (11, 12). Whether these lineage-specific differences in histone marks impact transcriptional regulation and are involved in the establishment or maintenance of lineage progenitors is not known. Therefore, comparing the histone methylation status of the three lineages derived from the mouse blastocyst at a gene-specific level remains an important goal. To this end, we examined the genomewide location of active and repressive histone modifications in ES, TS, and XEN cells. Our results show that the activating modification H3K4me3 has a similar role in the three stem cell types, but in contrast, the repressive modification H3K27me3 varies in abundance and genomewide distribution. We find that very few promoters are marked by H3K27me3 in TS and XEN cells and therefore H3K4me3/H3K27me3 bivalent domains are not a common epigenetic feature of all blastocyst-derived stem cells. It is also important to determine whether these epigenetic mechanisms arise during selection for stem cell self-renewal in vitro or truly reflect the status of early lineage progenitor cells in the embryo itself. Using carrier chomatin immunoprecipitation (cChIP) we were able to show that the bivalent domains seen in ES cells exist in pluripotent cells of the early mouse embryo. However, the epigenetic status of extraembryonic progenitor cells in the embryo did not entirely reflect the extraembryonic stem cell lines. These studies indicate that histone modification mechanisms may differ between early embryo lineages and emphasize the importance of examining in vivo and in vitro progenitor cells.
Results
Bivalent Domains Are Not Common in TS and XEN Cells.
To compare the histone methylation status of embryonic and extraembryonic stem cells, we examined the genomewide location of active and repressive histone modifications in ES, TS, and XEN cells (Fig. 1A). We performed native chromatin immunoprecipitation (ChIP) to isolate DNA associated with H3K4me3 or H3K27me3 modified histones, which was sequenced using an Illumina Genome Analyzer. Unique reads were mapped to the genome (Fig. S1A). Genomic regions exceeding a threshold signal, as defined by a randomization model (27), were categorized as significantly enriched for either histone modification (details in SI Methods). Importantly, our data from ES cells were highly similar to previously published studies (Fig. S1B) (16).
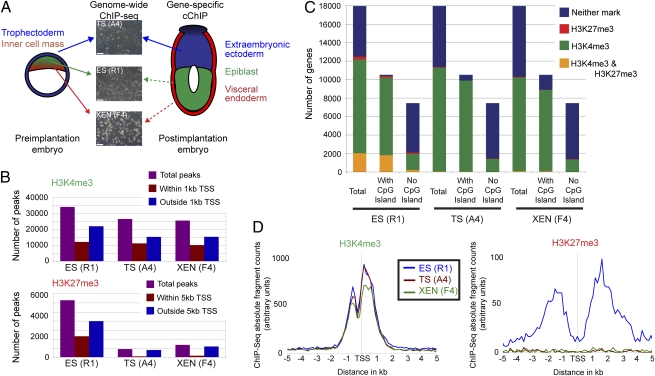
Fig. 1.
Analysis of genomewide histone states reveal differences in H3K27me3 between ES (R1), TS (A4), and XEN (F4) cells. (A) Overview of the experimental approach used to examine histone modifications of lineage progenitors from early mouse embryos. Arrows indicate source of tissue used for ES, TS, and XEN cell derivation, with dashed arrows indicating cells of the same developmental lineage. (Bar, 100 μM.) (B) Analysis of chromosome 19 reveals that a similar number of regions within 1 kb from TSS contain H3K4me3 peaks in ES, TS, and XEN cells. In contrast, the number of H3K27me3 peaks is lower in TS and XEN cells, and a smaller proportion of these peaks are located within 5 kb from TSS compared with ES cells. (C) The number of gene promoters in the genome that are modified with H3K4me3, H3K27me3, H3K4me3 and H3K27me3 or neither. Fewer promoters are marked by H3K27me3 in TS and XEN, compared to ES cells. Genes are further subdivided into those with or without a CpG island. Although the number of H3K4me3/H3K27me3 marked genes is low in TS and XEN cells, gene ontology analysis revealed that genes within this category were involved in organ development, chromatin assembly, metabolic processes, and cell cycle (Fig. S7). (D) Distribution of H3K4me3 relative to the nearest TSS reveals a highly similar profile between ES, TS, and XEN cells (ES, n = 13,131 genes; TS, n = 11,637; XEN, n = 10,660). Distribution of H3K27me3 relative to the nearest TSS shows that H3K27me3 is localized to promoter regions in ES cells only (ES, n = 2,320 genes; TS, n = 104; XEN, n = 171).
Numerous similarities in H3K4me3 location were detected among ES, TS, and XEN cells. In particular, a similar number of sequencing peaks were detected in all three cell lines, with ~40% of the peaks mapping to within 1 kb of a transcriptional start site (TSS) (Fig. 1B). Furthermore, H3K4me3 marked a similar number of genes in each cell type (Fig. 1C), was located at the same position relative to TSS (Fig. 1D), and correlated with high gene expression levels (Fig. S1 C and D). For all three cell types, genes without CpG island promoters tended to be devoid of H3K4me3 and were generally expressed at lower levels (Fig. 1C and Fig. S1D).
In contrast, H3K27me3 was different in TS and XEN cells compared to ES cells. Most strikingly, the number of sequencing peaks was ~7-fold lower in TS cells and ~5-fold lower in XEN cells, as compared to ES cells (Fig. 1B). Whereas ~40% of the peaks mapped to within 5 kb of a TSS in ES cells, this category included only ~10% of peaks in TS and XEN cells (Fig. 1B). These findings impact the total number of genes significantly marked by H3K27me3: 0.6% (104/18,025 genes) for TS cells and 0.9% (171/18,025) for XEN cells, compared to 13% (2,320/18,025) for ES cells (Fig. 1C). The overall distribution of H3K27me3 in TS and XEN cells was not localized to the TSS as for ES cells, but found at low levels throughout the genome (Fig. 1D). Furthermore, the median width of H3K27me3 modified regions was greater in ES cells (2.2 kb; 10% of all regions are >5 kb), compared to TS and XEN cells (1 kb; 1% of all regions are >5 kb) (Fig. S2A). These data demonstrate lineage-specific differences in H3K27me3.
We undertook several steps to confirm that our observation of low H3K27me3 levels in TS and XEN cells was not due to technical artifacts. The sequencing depth was similar for all samples (Fig. S1A) and the false detection algorithm incorporates the total number of sequencing reads into the threshold calculation, suggesting that low H3K27me3 levels were not an artifact of differential sequencing depth. Nor did this result from the threshold filter being too stringent, as we were able to process raw, previously published, H3K27me3 ChIP-sequencing data from embryonic fibroblasts and obtain near identical results to the published report (16). In addition, an alternative strategy of genome binning and pairwise comparison of ChIP-sequencing reads confirmed fewer H3K27me3 peaks in TS and XEN cells. This analysis also revealed that background levels and average peak height are highly similar between ES, TS and XEN cells, which would be expected only if the ChIP efficiency was the same between the three cell lines (Fig. S2 B–D). Therefore, differences in H3K27me3 levels between cell types was not due to an artifact of data processing or altered ChIP efficiency, and thus ES, TS, and XEN cells appear to differ in use of repressive epigenetic mechanisms.
The lower levels of H3K27me3 impacts the number of putative bivalent domains detected in the three stem cell lines as well. Thus, only 56 genes could be classified as bivalently modified in TS cells and 96 genes in XEN cells, compared to 2,033 in ES cells (Fig. 1C). Instead, the majority of genes in TS and XEN cells were categorized as H3K4me3 without H3K27me3; 62% (11,248/18,025) for TS cells and 56% (10,135/18,025) for XEN cells. Neither modification was detected at 37% of genes (6,673/18,025) in TS cells and 43% of genes (7,719/18,025) in XEN cells (Fig. 1C). Thus, bivalent domains involving H3K4me3 and H3K27me3 are not a common epigenetic mark in all blastocyst-derived stem cell lines.
Lower Levels of H3K27 Methylation Machinery in Extraembryonic Stem Cells.
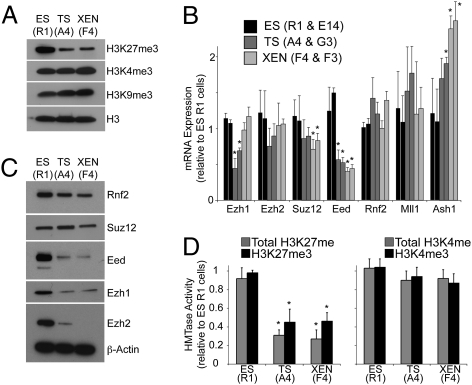
Western blot analysis confirmed that TS and XEN cells were globally low in H3K27me3, compared to ES cells (Fig. 2A). To understand why extraembryonic stem cells have lower levels of H3K27me3, we examined the transcript levels, protein abundance, and enzymatic activity of histone methylation core machinery. Methylation of H3K27 is catalyzed by the canonical Polycomb repressive complex 2 (PRC2), which contains core components Suz12, Eed, and Ezh2 (28–31). Reverse transcription–qPCR (RT-qPCR) revealed that the level of Eed mRNA was significantly less (by ~65%) in TS and XEN cells, compared to ES cells (P < 0.02, Student's t test; Fig. 2B). It has previously been shown that removal of Eed can disrupt PRC2 stability and lead to loss of Ezh2 protein (32, 33). Our Western blot analysis was consistent with this, showing a near complete absence of Eed and Ezh2 protein in TS and XEN cells, compared to ES cells (Fig. 2C). Similarly, the noncanonical PRC2 component Ezh1 was detected at lower levels in TS and XEN cells, compared to ES cells, suggesting that Ezh1 stability may be dependent on incorporation into a functional PRC2 complex (Fig. 2C). In contrast, Suz12 and the Polycomb repressive complex 1 (PRC1) component Rnf2 were detected at similar levels in all three stem cell types (Fig. 2C). Additionally, histone methyltransferase activity associated with total H3K27 methylation and H3K27me3 was significantly lower by 2.1- to 3.4-fold in TS and XEN cells, compared to ES cells (P < 0.05, Student's t test; Fig. 2D). Together, these data suggest that lower Eed levels may result in lower levels of functional PRC2 in extraembryonic stem cells. By contrast, expression levels of two different histone H3K4 methyltransferases showed either no change or were higher in TS and XEN cells compared to ES cells (Fig. 2B). In addition, no significant difference in H3K4 methyltransferase activity was detected between ES, TS, and XEN cells (Fig. 2D).
Fig. 2.
PRC2 expression and activity are low in extraembryonic stem cells. (A) Western blot analysis shows that global levels of H3K27me3 are lower in TS (A4) and XEN (F4) cells, compared to ES (R1) cells. H3K4me3 and H3K9me3 are detected at similar levels in the three stem cell types. Unmodified histone H3 was used as a loading control. (B) qRT-PCR analysis of levels of PRC2 components Ezh1, Ezh2, Suz12, and Eed; PRC1 component, Rnf2, and two H3K4 methyltransferases, Mll1 and Ash1. Data represent mean plus SD from three biological replicates. For each stem cell type, two independent lines were analyzed. Asterisks indicate statistically significant difference as compared to ES cells, P < 0.05 (Student's t test). (C) Western blot analysis shows that expression of PRC2 components Eed, Ezh1, and Ezh2 is lower in TS (A4) and XEN (F4) cells, compared to ES (R1) cells. Suz12 and Rnf2 are expressed at similar levels. β-Actin was used as a loading control. (D) Total H3K27 and H3K27me3-specific histone methyltransferase activity is significantly lower in TS (A4) and XEN (F4) cells, compared to ES (R1) cells. No significant differences in total H3K4 or H3K4me3-specific histone methyltransferase activity were detected. Data represent mean plus SD from three biological replicates. For each stem cell type, two independent cell lines were analyzed and data combined. Asterisks indicate statistically significant difference as compared to ES cells, P < 0.05 (Student's t test).
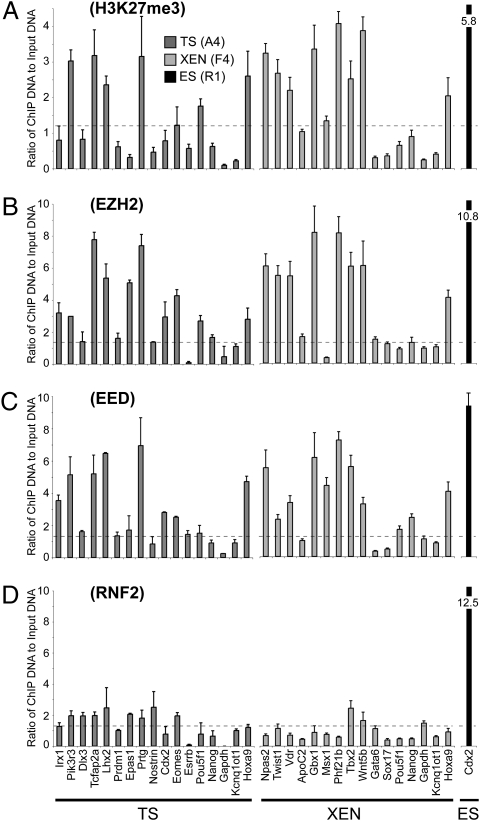
We next examined whether H3K27me3 marks in TS and XEN cells were functionally equivalent to H3K27me3 marks in ES cells, despite their reduced numbers, by assessing their ability to recruit appropriate proteins to mediate transcriptional repression. The H3K27me3 mark is deposited by PRC2 and functions at bivalent domains in ES cells by recruiting PRC1, which maintains the underlying promoter in a poised transcriptional state (34–36). Using ChIP-qPCR, we detected binding of PRC2 components Ezh2 and Eed above background at H3K27me3-marked genes in TS and XEN cells, suggesting that H3K27me3 is actively maintained in extraembryonic stem cells (Fig. 3 A–C). In contrast, binding of PRC1 component Rnf2 was low or negligible in TS and XEN cells (Fig. 3D) despite being present at similar levels to ES cells (Fig. 2C), suggesting that H3K27me3 marks in TS and XEN cells are not functionally equivalent to bivalent domains in ES cells. The domains detected in TS and XEN cells appear similar to PRC1-negative domains in ES cells, which show poor retention of H3K27me3 and correlate poorly with functional repression (34). Together, these data show that H3K27me3 is not a predominant mark in undifferentiated extraembryonic stem cells, suggesting that alternative epigenetic modifications must be involved in transcriptional repression.
Fig. 3.
Differences in PRC1 binding between TS/XEN cells and ES cells. (A–D) The ability of H3K27me3 to recruit downstream mediators in TS and XEN cells was investigated using ChIP-qPCR on a panel of candidate gene promoters. We included H3K27me3-modified and unmodified promoters that were identified by our genomewide analysis. The panel also contains bivalent promoters classified in ES cells as PRC1-positive (Irx1, Dlx3, Tcfap2a, Lhx2, Npas2, Gbx1, Msx1, Tbx2, Gata6, and Sox17) and PRC1-negative (Pik3r3, Prtg, and Nostrin) (34). Positive (Hoxa9 and Pou5f1) and negative (Gapdh and Kcnq1ot1) control promoters for H3K27me3 were included (61). Ezh2 and Eed binding was detected above background levels at H3K27me3-modified promoters in TS (A4) and XEN (F4) cells. However, binding of the downstream mediator Rnf2 was detected only at low or negligible levels, irrespective of H3K27me3, Ezh2, or Eed status. As we could not identify a promoter that was bound by Rnf2 in TS and XEN cells, we used the Cdx2 promoter in ES (R1) cells as a positive control (indicated by the black bar) (34). Data represent mean plus SD from three biological replicates. Dashed lines indicate 2-fold of mean background levels, as determined using a nonspecific control antibody (background data shown in Fig. S8A).
H3K9me3 Marks Transcriptionally Repressed Genes in TS and XEN Cells.
Given the low prevalence of H3K27me3-modified promoters in TS and XEN cells, we next undertook a candidate approach to identify possible alternative repressive modifications in extraembryonic stem cells.
We initially examined DNA methylation, but using two different experimental approaches we found no evidence that this modification is providing a compensatory mechanism to regulate gene transcription in TS cells. First, integration of a genomewide DNA methylation dataset (26) with our genomewide histone methylation data revealed that the majority of genes with high levels of DNA methylation lacked histone methylation marks and were transcriptionally repressed: 67% (746/1,108 genes) for ES cells and 66% (1,141/1,734) for TS cells (Fig. S3). Less than 7% of promoters in ES and TS cells that are modified by H3K4me3 had high levels of DNA methylation (Fig. S3; ES: n = 4,451 genes, TS: n = 6,586) and these promoters are not preferentially induced upon differentiation, as would be predicted if DNA methylation was compensating for the absence of H3K27me3 at poised regulatory genes (P > 0.2, χ2 test, 1 df; Dataset S1). Second, we used pyrosequencing to quantify DNA methylation levels at the promoters of genes that are induced or repressed as TS cells undergo differentiation. After TS cell differentiation, DNA methylation levels of gene promoters remained largely unchanged and showed no correlation with changes in gene expression levels (Fig. S3C). Together, these data suggest that DNA methylation is not providing a compensatory method of epigenetic repression in TS cells.
We next examined four alternative histone modifications associated with transcriptionally repressed chromatin, including H3K9me2, H3K9me3, H3K79me3, and H4K20me3. We assayed these marks by ChIP-qPCR in genes that are either induced or repressed as TS cells undergo differentiation. Of these, only H3K9me3 correlated inversely with changes in gene expression (Fig. 4 A and B). Eight out of nine genes that are induced showed a reduction in H3K9me3 after TS cell differentiation, and this was statistically significant for five of the genes (P < 0.05, Student's t test, P < 0.3 for the other three genes; Fig. 4B). Additionally, all five genes studied that were repressed during TS cell differentiation showed an increase in H3K9me3, which was statistically significant for three of the genes (P < 0.05, Student's t test, P < 0.08 and P < 0.3 for the other two genes; Fig. 4B). These results were confirmed with an independent TS cell line (Fig. S4A).
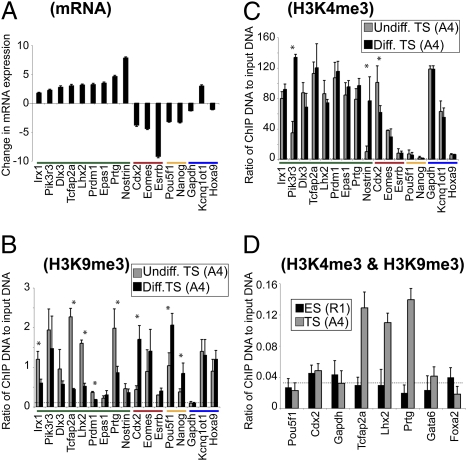
Fig. 4.
H3K9me3 correlates inversely with gene transcription during TS cell differentiation. (A) qRT-PCR analysis reveals fold change in expression of mature mRNA transcripts in differentiated TS cells, as compared to undifferentiated TS cells (log 2 scale; TS cell line A4). For reference, genes are grouped according to their change in expression status upon TS cell differentiation: induced (green), extraembryonic factors that are repressed (red), and embryonic factors that are repressed (yellow). Gapdh, Kcnq1ot1, and Hoxa9 (blue) promoters provide control regions (46, 61). Data represent mean plus SD of three biological replicates. ChIP experiments demonstrate changes in (B) H3K9me3 and (C) H3K4me3 that accompany gene transcription during TS (A4) cell differentiation. Dashed lines indicate 2-fold of mean background levels, as determined using a nonspecific control antibody (Fig. S8B). Asterisks indicate a statistically significant difference between undifferentiated and differentiated TS cells, P < 0.05 (Student's t test). Data represent mean plus SD of three biological replicates. The same experiments were performed using an additional TS cell line (G3) and gave highly similar results (Fig. S4). (D) Sequential ChIP experiments confirm the coexistence of H3K4me3 and H3K9me3 in undifferentiated TS (A4) cells. Cdx2 and Gapdh in TS cells and Pou5f1 and Gapdh in ES cells are known to be modified by H3K4me3 only and served as controls for the specificity of the H3K9me3 immunoprecipitation. Dashed lines indicate 2-fold of mean background levels, as determined using anti-H3K4me3 in the first immunoprecipitation and a nonspecific control antibody in the second immunoprecipitation (Fig. S8C). Data represent mean plus SD of three biological replicates.
Interestingly, our candidate gene ChIP analysis in TS cells indicated that certain promoters may be simultaneously marked by active H3K4me3 and repressive H3K9me3 histone modifications (Fig. 4 B and C). To test whether these marks are present on the same chromatin fragment, we performed sequential ChIP analysis, where chromatin is first immunoprecipitated with one antibody and then subjected to an additional immunoprecipitation with a second antibody. We detected enrichment ~10-fold compared to control samples, suggesting that, at the three promoters analyzed, the majority of chromatin containing H3K4me3 is also modified by H3K9me3 in TS cells (Fig. 4D and Fig. S4A). This enrichment value is similar to that obtained with sequential H3K4me3/H3K27me3 ChIP at known bivalent domains in ES cells (~12-fold; Fig. S4B). In contrast to TS cells, we did not detect enrichment of H3K4me3/H3K9me3 comodification above controls in ES cells (Fig. 4D).
Encouraged by our observations in TS cells, we assayed for the presence of H3K4me3/H3K9me3 domains on a panel of candidate promoters in XEN cells. We detected both H3K4me3 and H3K9me3 above background at three promoters of genes transcriptionally repressed in XEN cells but that are expressed in differentiated cells of the extraembryonic endoderm lineage (Fig. S4C) (37). Sequential ChIP confirmed that these modifications coexisted on the same chromatin fragment in XEN cells (~11-fold enrichment over controls; Fig. S4D). Taken together, we have found that H3K9me3 is associated with transcriptionally repressed promoters in TS and XEN cells and using sequential ChIP have shown that certain promoters are modified by H3K4me3 and H3K9me3 in the same chromatin region.
Epigenetic States During Lineage Development.
To begin to understand whether the epigenetic status of ES, TS, and XEN cells arise during selection for stem cell self-renewal in vitro or truly reflect the status of lineage progenitor cells in vivo, we examined histone methylation levels at candidate promoter regions in tissues microdissected from early mouse embryos. We focused on early postimplantation embryos because these tissues represent the best stages at which to obtain homogeneous populations of undifferentiated lineage progenitors that arise from the early EPI, TE, and PE (1, 38) in sufficient numbers to allow examination of gene-specific histone modifications. Due to the technical difficulty in isolating lineage progenitors from these early postimplantation stages, in combination with the small number of cells within each embryo, these tissues have not been examined previously for gene-specific histone modifications.
We chose to use cChIP for this analysis, which is designed to analyze histone modifications in small cell samples (39). We microdissected embryos at embryonic day E5.5 to isolate EPI, extraembryonic ectoderm (ExE), and visceral endoderm (VE) (Fig. S5A) and subjected the tissues to RT-qPCR analysis of gene expression and to cChIP-qPCR for analysis of gene-specific H3K4me3 and H3K27me3. In parallel experiments, we subjected an equivalent number of ES, TS, or XEN cells to the same analyses to provide a positive control for the small-scale studies.
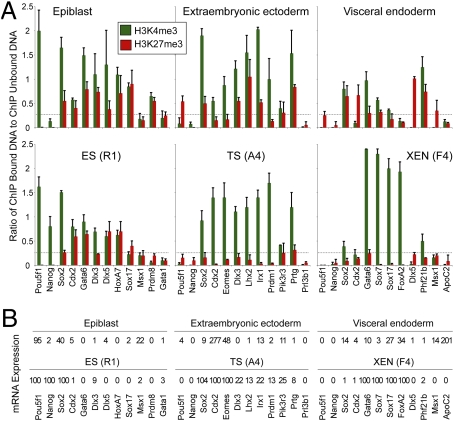
cChIP analysis of pluripotent EPI showed that promoters of lineage-specific transcriptionally active genes, such as Pou5f1 (also known as Oct4), were modified with H3K4me3 and not H3K27me3 (Fig. 5). Genes encoding developmental regulators such as Cdx2, Gata6, Hoxa7, and Sox17, which are bivalently modified in ES cells, appeared to be bivalent in EPI, with high H3K4me3 and H3K27me3 (Fig. 5A). In addition, we detected low or negligible mRNA transcripts from these genes (Fig. 5B). Overall, the histone status of most genes examined was very similar between EPI and ES cells. One difference was H3K4me3 marking of Nanog, which was high in ES cells and low in EPI, consistent with the higher expression level of Nanog in ES cells as compared to EPI (Fig. 5).
Fig. 5.
Analysis of histone modifications in lineage progenitor cells of early mouse embryos. (A) cChIP experiments examine EPI and ES (R1) cells (Left); ExE and TS (A4) cells (Middle); VE and XEN (F4) cells (Right). Gene names are shown at the base of each column and include known lineage-specific transcription factors (Pou5f1, Nanog, Cdx2, Eomes, Gata6, and Sox7), genes known to be bivalent in ES cells (Cdx2, Gata6, Hoxa7, and Sox17), and genes that are characteristic of fully differentiated cell types (Gata1, Prl3b1, and ApoC2). Antibodies used were against H3K4me3 (green) and H3K27me3 (red). Dashed lines indicate 2-fold of mean background levels, as determined using a nonspecific control antibody (Fig. S8D). Mean plus SD are shown from two biological replicates. cChIP analyses of alternative ES, TS, and XEN cell lines show similar results (Fig. S5C). (B) Corresponding mRNA expression levels for each cell type. Values represent percentages relative to reference tissues (specified in SI Methods). Each value represents the mean from two biological replicates.
In ExE cells, which derive from the TE, Pou5f1 and Nanog are transcriptionally silent. cChIP revealed that the Pou5f1 promoter was modified by H3K27me3 and the Nanog promoter had little enrichment of H3K4me3 or H3K27me3 (Fig. 5). Transcriptionally active genes Cdx2 and Eomes showed high H3K4me3 and low H3K27me3 (Fig. 5). Intriguingly, genes that were silent in ExE but expressed by specialized placental cells in later development (Dlx3, Lhx2, and Irx1) appear to be marked bivalently in ExE, with high H3K4me3 and H3K27me3 (Fig. 5). This contrasts with the same gene promoters in TS cells, which had high H3K4me3 and low H3K27me3 (Fig. 5), as predicted from our genomewide ChIP analysis, and confirmed using an alternative TS cell line (Fig. S5C).
In VE cells, derived from PE, Pou5f1 and Cdx2 promoters were marked by H3K27me3, which is consistent with their low expression levels (Fig. 5). Known endoderm genes, Gata6, Sox7, and Sox17 were enriched for H3K4me3, although levels of this mark were low when compared to the same genes in two XEN cell lines (Fig. 5 and Fig. S5C). These endoderm genes were expressed at a much higher level in XEN cells as compared to VE, which is consistent with their histone state (Fig. 5).
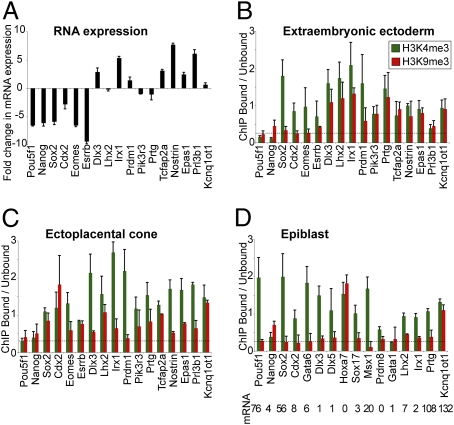
Given the changes in H3K9me3 levels during TS cell differentiation, we next used cChIP to investigate gene-specific levels of H3K9me3 during trophoblast differentiation in vivo. Embryos at E6.5 provide the ideal material for this analysis, as extraembryonic tissue at this development stage contains undifferentiated trophoblast cells in the ExE and differentiated trophoblast cells in the ectoplacental cone (EPC). Importantly, these tissues show reproducible changes in gene expression levels that are analogous to our TS cell differentiation system (Fig. 6A and Fig. S5B).
Fig. 6.
Analysis of H3K9me3 during trophoblast differentiation in early mouse embryos. (A) qRT-PCR analysis reveals fold change in expression of mature mRNA transcripts in EPC (differentiated trophoblast), as compared to ExE (undifferentiated trophoblast) (log 2 scale). (B and C) cChIP experiments reveal the gene-specific differences in H3K9me3 and H3K4me3 between (B) ExE and (C) EPC. In general, promoters that are induced upon trophoblast differentiation (Dlx3, Irx1, Tcfap2a, Nostrin, and Epas1) show an increase in H3K4me3 and a decrease in H3K9me3 when comparing EPC with ExE. Kcnq1ot1 is used as a positive control (46). Mean and SD are from two biological replicates. (D) cChIP experiments show that genes known to be bivalent in ES cells (Cdx2, Gata6, and Sox17) or marked by H3K4me3/H3K9me3 in TS cells (Dlx3, Lhx2, Irx1, and Prtg) are not modified by H3K9me3 in EPI above background levels (indicated by the dashed line). The imprinted gene Kcnq1ot1 is used as a positive control (22, 23). Mean and SD are from two biological replicates. Shown underneath are the corresponding mRNA expression levels, relative to the same reference tissues used in Fig. 5B.
cChIP analysis revealed H3K9me3 levels above background at numerous promoters in trophoblast tissues. Increased H3K9me3 levels were detected at Sox2, Cdx2, Eomes, and Esrrb promoter regions in EPC compared to ExE, which is consistent with their lower expression levels in differentiated trophoblast (Fig. 6). We also examined genes whose expression levels are higher in EPC compared to ExE. In general, their promoters were modified by H3K4me3 and H3K9me3 in ExE (Fig. 6B), whereas the same promoters in EPC tended to show higher H3K4me3 and lower H3K9me3 levels (Fig. 6C). These results recapitulate the changes in histone methylation levels during TS cell differentiation, suggesting that this configuration of histone marks may be conserved between trophoblast progenitors in vivo and in vitro.
We next investigated H3K9me3 levels in embryonic tissues at E6.5. cChIP analysis of EPI showed that genes known to be bivalent in ES cells and epiblast, and also genes shown in this study to be marked by H3K9me3 in TS cells, do not have H3K9me3-modified histones above background levels (Fig. 6D). An exception was Hoxa7, which was marked by H3K4me3 and H3K9me3. As Hoxa7 mRNA was undetectable in EPI (Fig. 6D), these epigenetic modifications are likely to reflect the unusual epigenetic marking of Hox genes in pluripotent cells (15, 40). Although we did not detect H3K4me3 and H3K9me3 at the majority of promoters examined by cChIP in EPI, recent studies have suggested that these two modifications may overlap at a subset of H3K4me3/H3K27me3 bivalent domains in ES cells (41).
Bivalent H3K4me3/H3K27me3 marks were detected at specific genes in EPI as in ES cells, demonstrating the existence of bivalent domains in early mouse development. There appears to be an epigenetic continuum between ES cells and the embryonic lineages of pre- and postimplantation stage embryos, whereby in all of these cells both H3K4me3 and H3K27me3 are highly abundant at global and gene-specific levels. In addition, we showed that high gene-specific levels of H3K27me3 were detected in extraembryonic lineages of the postimplantation embryo, but not at the same gene promoters in TS and XEN cells. As global H3K27me3 levels are low in TE and PE of preimplantation stage embryos (11) (Fig. S6A) these data suggest that the blastocyst status of histone methylation may be preserved in TS and XEN cells but reset in the postimplantation derivatives of these lineages. Overall, our results suggest that distinct histone methylation mechanisms are established in embryonic and extraembryonic progenitor cells, thus identifying an important difference in epigenetic status between early embryo lineages.
Discussion
We present here a detailed epigenetic examination of in vivo progenitor cells and in vitro stem cells that represent the first three lineages formed during mouse embryogenesis.
First, we demonstrated that H3K4me3, H3K27me3, and H3K9me3 marks generally correlated with gene expression status in the three lineages both in vivo and in vitro, consistent with a previous report that examined other modifications in the inner cell mass and TE from cultured blastocysts (39).
Second, we found that a specific set of genes encoding developmental regulators were bivalently marked with H3K4me3 and H3K27me3 in pluripotent cells in vivo as well as in vitro in ES cells. Importantly, mature mRNA transcripts of these genes were not detected in EPI, demonstrating that, similar to ES cells, the bivalent mark is repressive (14, 15). Retention of bivalent domains by both pluripotent stem cell types derived from EPI (ES cells and epiblast stem cells) suggests that this epigenetic configuration may be a conserved feature of embryonic progenitor cells (14–16, 42, 43). However, our work shows that TS and XEN cells are different because bivalent H3K4me3/H3K27me3 domains do not appear to be involved in regulation of either their undifferentiated state or their differentiation. A previous report suggested that H3K4me3/H3K27me3 status at key regulatory genes may suffice to describe developmental commitment and potential (16). Whereas this may be true for some progenitor cells, our study now shows that alternative epigenetic modifications may play a role in other early developmental progenitors.
Third, extraembryonic lineages of the blastocyst are globally low in H3K27me3 (Fig. S6A) (11), which is reflected in TS and XEN cells. This contrasts, however, with postimplantation stage extraembryonic tissues, where we detected high gene-specific levels of H3K27me3. We offer two explanations for these observations. There could be a wave of H3K27me3 deposition in extraembryonic lineages shortly after implantation, and TS and XEN cells represent a cell type before this event. Consistent with this, we have examined global H3K27me3 localization in postimplantation embryos and detected H3K27me3 in embryonic and extraembryonic cells (Fig. S6) (44). This methylation event, which coincides with the onset of Eed transcription (44), could aid lineage commitment or establish a new pattern of epigenetic marks. This would be analogous to de novo DNA methylation that occurs during preimplantation development (45). In support of this hypothesis, placentally imprinted genes acquire their differential histone marks in trophoblast between E4.5 and E7.5, and TS cells display a set of epigenetic marks before this event (46). An alternative explanation is that although global H3K27me3 is low in the extraembryonic lineages before implantation, there may still be genes marked by H3K27me3 in these tissues and these marks persist after implantation. However, these marks may be lost during TS and XEN cell derivation and propagation. This alteration would be similar to the relaxation of some repressive histone marks that occurs during ES cell derivation (39) and also consistent with proposed epigenetic differences between trophoblast progenitors and TS cells (22). Perhaps loss of epigenetic repression is necessary to convert transient lineage progenitors in the embryo into stable, self-renewing stem cell lines.
Fourth, we identified H3K9me3 as a repressive histone mark in early trophoblast development. Together with our H3K27me3 data, these findings provide support for the existence of lineage-specific histone modifications during development. How these differences could be established is unclear, although known antagonism between H3K9me3 and Polycomb protein binding suggest that histone states could depend on the relative levels of H3K9me3 and H3K27me3 (47, 48).
Loss-of-function studies in mice have revealed a requirement for numerous epigenetic modifiers in extraembryonic tissue formation and function (49). Absence of H3K27me3 in TS cells does not appear to affect their undifferentiated status, yet trophoblast with severely reduced Eed exhibits a defect in secondary giant cell formation (50, 51). Similarly, mutation of other H3K27me3 pathway components leads to defects in placental development, notably Ezh2, Suz12, and Rnf2 mutants all display abnormal amnion and chorion formation (52–54). Therefore, H3K27me3-mediated epigenetic repression may be critical to establish a specialized transcriptional program, rather than to maintain an undifferentiated trophoblast state. This would be consistent with the low prevalence of H3K27me3-marked genes that we detected in undifferentiated TS cells. The functional requirement for H3K9 methylation in extraembryonic stem cells has not been reported, although targeted deletion of the H3K9 histone methyltransferase Ehmt2 (G9a) leads to incomplete chorioallantoic fusion and midgestation lethality (55). In addition, Suv39h-null fibroblasts spontaneously become polyploid (56), raising the intriguing possibility that Suv39h-mediated H3K9me3 could be involved in the formation of polyploid trophoblast giant cells from diploid trophoblast precursors. Further detailed characterization of extraembryonic stem cell types devoid of H3K27 and H3K9 modifiers should help to address the role of epigenetic mechanisms in maintaining the undifferentiated state and in the transition to a specialized cell type.
In conclusion, our studies confirm the prediction that transcriptional memory of each early lineage is likely to be maintained by epigenetic modifications to key regulators, but suggest that the specificity of the modifications may be lineage specific.
Methods
Tissue Samples.
Mice used were ICR and B5/EGFP (57). Embryos were collected at appropriate time points from timed natural matings. Isolation of tissues was carried out as described (42). ES, TS, and XEN cell lines used in this study were male, low passage (<15), with a modal number of 40 chromosomes per nucleus and cultured in the absence of feeder cells as described in SI Methods. All experiments were carried out on ES cell line R1, TS cell line A4 and XEN cell line F4, with additional confirmation provided by alternative cell lines (ES cell line E14TG2a, TS cell line G3, and XEN cell line F3) as indicated in the text.
ChIP.
Native (unfixed) ChIP to analyze histone modifications was performed as described (58). Formaldehyde-crosslinked chromatin was analyzed using the ChIP assay kit (Millipore). Sequential ChIP was performed as described (59), except the first elution buffer contained 20 mM DTT and eluted chromatin was diluted 50-fold in RIPA buffer and used for the second immunoprecipitation. cChIP was performed as described (39), except DNA from bound and unbound samples was extracted using the DNA purification kit (Qiagen) and subjected to qPCR analysis. Detailed protocols are described in SI Methods. Illumima sequencing was performed by the BC Cancer Agency Genome Sciences Centre, Vancouver, Canada. For each stem cell type, one ChIP DNA sample was sequenced for each antibody. Sample processing and initial raw data processing was carried out as described (27).
Gene Expression Analysis.
RNA from cell lines was processed using the RNeasy mini kit (Qiagen). RNA was extracted from embryo tissues using TRIzol (Invitrogen) following the protocol for isolation of RNA from small quantities of tissue with 0.2 μg RNase-free LPA (Sigma) per sample. RNA was reverse transcribed using the SuperScript II (Invitrogen) and cDNA subjected to qPCR. Data were normalized to Hmbs. Primers targeted multiple exons in the cDNA sequence (Table S1). For microarray analysis, 100 ng of total RNA was amplified and hybridized to the Mouse Gene 1.0 ST array (Affymetrix). This service was performed by The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada.
Analysis and Integration of Histone and DNA Methylation Datasets.
ChIP-sequencing data were processed using FindPeaks (60) with a false discovery rate of 0.001 and an average fragment size of 174 bp. Gene annotations were obtained from the University of California Santa Cruz genome browser (build mm8). Full dataset is shown in Dataset S1. DNA methylation raw data for ES and TS cells (26) were processed using ChipMonk (http://www.bioinformatics.bbsrc.ac.uk/projects/chipmonk) with parameters detailed in SI Methods.
Statistical Methods.
Statistical comparison of histone methylation, DNA methylation, and microarray expression array datasets was performed in R (http://www.r-project.org) using the RKWard interface. We reported the Pearson correlation, the significance of which was calculated by a Fisher exact test and scored into bins of P values.
Supplementary Material
Acknowledgments
We thank Thomas Jenuwein (Max-Planck Institute of Immunobiology) and Prim Singh (Research Center Borstel) for providing H3K27me3 and H3K9me3 antibodies and William Stanford (University of Toronto) for sharing Polycomb antibodies. We are grateful to Yojiro Yamanaka and Roger Pedersen for advice on embryo microdissection, Adam Smith and Chunhua Zhao for assistance with pyrosequencing, Annie Lewis and Laura O'Neill for cChIP protocols and advice, and Philippe Arnaud for advice on sequential ChIP. We are indebted to Jorge Cabezas, Jodi Garner, Alison Hirukawa, Malgosia Kownacka, Andres Nieto, Nina Nouraeyan, and Valerie Prideaux for technical support. This study was funded by Grant MOP77803 (to J.R.) from the Canadian Institutes of Health Research (CIHR) and by a CIHR Bisby Fellowship (to P.J.R.-G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: All histone methylation and microarray expression datasets have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE15519).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914507107/-/DCSupplemental.
References
- 1.Rossant J, Chazaud C, Yamanaka Y. Lineage allocation and asymmetries in the early mouse embryo. Philos Trans R Soc Lond B Biol Sci. 2003;358:1341–1348. doi: 10.1098/rstb.2003.1329. discussion 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 3.Ng RK, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Yuan P, et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 7.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 9.Kunath T, et al. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 10.Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- 11.Erhardt S, et al. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- 12.Sarmento OF, et al. Dynamic alterations of specific histone modifications during early murine development. J Cell Sci. 2004;117:4449–4459. doi: 10.1242/jcs.01328. [DOI] [PubMed] [Google Scholar]
- 13.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, et al. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 22.Lewis A, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 23.Umlauf D, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 24.Chapman V, Forrester L, Sanford J, Hastie N, Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984;307:284–286. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- 25.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 26.Farthing CR, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 28.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 29.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery ND, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 33.Tie F, Stratton CA, Kurzhals RL, Harte PJ. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol. 2007;27:2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 36.van der Stoop P, et al. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS ONE. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherwood RI, et al. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev Biol. 2007;304:541–555. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 39.O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 40.Atkinson SP, et al. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells. 2008;26:1174–1185. doi: 10.1634/stemcells.2007-0497. [DOI] [PubMed] [Google Scholar]
- 41.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 43.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 44.Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2:e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 46.Lewis A, et al. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- 47.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 48.Puschendorf M, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 49.Hemberger M. Epigenetic landscape required for placental development. Cell Mol Life Sci. 2007;64:2422–2436. doi: 10.1007/s00018-007-7113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalantry S, et al. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Mager J, Schnedier E, Magnuson T. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome. 2002;13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- 52.O'Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voncken JW, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 57.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 58.Umlauf D, Goto Y, Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- 59.Noer A, Lindeman LC, Collas P. Histone H3 modifications associated with differentiation and long-term culture of mesenchymal adipose stem cells. Stem Cells Dev. 2009;18:725–736. doi: 10.1089/scd.2008.0189. [DOI] [PubMed] [Google Scholar]
- 60.Fejes AP, et al. FindPeaks 3.1: A tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terranova R, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.