Abstract
The discovery of a nonphotosynthetic plastid in malaria and other apicomplexan parasites has sparked a contentious debate about its evolutionary origin. Molecular data have led to conflicting conclusions supporting either its green algal origin or red algal origin, perhaps in common with the plastid of related dinoflagellates. This distinction is critical to our understanding of apicomplexan evolution and the evolutionary history of endosymbiosis and photosynthesis; however, the two plastids are nearly impossible to compare due to their nonoverlapping information content. Here we describe the complete plastid genome sequences and plastid-associated data from two independent photosynthetic lineages represented by Chromera velia and an undescribed alga CCMP3155 that we show are closely related to apicomplexans. These plastids contain a suite of features retained in either apicomplexan (four plastid membranes, the ribosomal superoperon, conserved gene order) or dinoflagellate plastids (form II Rubisco acquired by horizontal transfer, transcript polyuridylylation, thylakoids stacked in triplets) and encode a full collective complement of their reduced gene sets. Together with whole plastid genome phylogenies, these characteristics provide multiple lines of evidence that the extant plastids of apicomplexans and dinoflagellates were inherited by linear descent from a common red algal endosymbiont. Our phylogenetic analyses also support their close relationship to plastids of heterokont algae, indicating they all derive from the same endosymbiosis. Altogether, these findings support a relatively simple path of linear descent for the evolution of photosynthesis in a large proportion of algae and emphasize plastid loss in several lineages (e.g., ciliates, Cryptosporidium, and Phytophthora).
Keywords: Apicomplexa, Chromera velia, CCMP3155, plastid evolution, chloroplast genome
The primary obstacle in determining the evolutionary origin of the cryptic plastid in apicomplexan parasites, the apicoplast (1), has been its divergent nature. The apicoplast genome is both highly-derived (it is compact, with fast-evolving genes and a very high AT-bias), and reduced (it has lost all genes related to photosynthesis), making clear comparisons with other plastids difficult (2). Moreover, the closest algal relatives to the apicomplexans are dinoflagellates, and dinoflagellate plastids are equally derived but in different ways. Characterized dinoflagellate plastid genomes encode only 12–14 genes, which are extremely fast-evolving and are localized on minicircles with one or a few genes (3). Most importantly, however, nearly all of the genes retained in the dinoflagellate plastid encode photosystem proteins, so the two genomes are virtually incomparable in their gene content. For these reasons, hypotheses for the apicoplast origin rest on analyses of its divergent genes in the absence of dinoflagellate homologs (4–6), plastid-derived genes encoded in the nucleus (7), or genes with no connection to the plastid whatsoever (8, 9). Not surprisingly, these data have led to completely inconsistent conclusions either for a green or red algal origin of the apicoplast. The hypothesis that the apicoplast is derived from a red alga is also tied to the broader “chromalveolate” hypothesis, which posits that the endosymbiosis that gave rise to the apicoplast is much more ancient and also gave rise to plastids in dinoflagellates, heterokonts, and hacrobians (cryptomonads and haptophytes) (10). Although this notion minimizes the number of endosymbiotic events required to explain plastid diversity, it also leads to complexity in other ways because each of the chromalveolate lineages contains early-branching members or sister groups where no plastid is known. Minimizing endosymbiotic events therefore increases the number of times photosynthesis or plastids must have been lost. Alternatively, each of these lineages could have obtained its plastid from an independent red algal endosymbiosis (11) or from another eukaryote already containing a red algal plastid through serial tertiary endosymbioses (12, 13). The apicomplexans and dinoflagellates illustrate this discrepancy well, because recognizable plastids appear to be absent in basal subgroups of both lineages and the presence of photosynthesis in their common ancestor would require between five and nine independent losses of photosynthesis (and in some cases plastids) just among the early-branching lineages, and probably another dozen losses within dinoflagellates as a whole. Distinguishing between the early vs. late origin of these plastids has proven extraordinarily difficult and has led to a passionate debate over likelihood of plastid gain versus loss and how to interpret genomic data from various nonphotosynthetic groups related to red plastid-containing lineages like ciliates, oomycetes, and rhizarians (13, 14). Comparisons between complete plastid genomes would constitute the most direct way to test these hypotheses, but this has historically precluded both apicomplexans and dinoflagellates because their genomes are so reduced and divergent that their relationship to one another and other plastid lineages remains obscure. A potential breakthrough to this stalemate was the recent description of a photosynthetic relative of the apicomplexans, Chromera velia (15). Although related to apicomplexans, C. velia is photosynthetic, so if the C. velia plastid genome retains characteristics ancestral to both apicomplexan and dinoflagellate plastids, it has the potential to settle many debates conclusively. Unfortunately, to date only three of its genes have been characterized (15, 16), and nothing is known of its gene content, organization, or structure. Here, we describe the complete plastid genome sequences and other plastid-associated data from C. velia, and also from a second independent lineage of photosynthetic alveolate, represented by the undescribed species CCMP3155. These data provide several lines of evidence (e.g., shared gene content, genome structure, processing pathways, lateral gene transfers, as well as gene phylogenies) that the common ancestor of apicomplexans and dinoflagellates contained a plastid and that the extant plastids in these lineages descended from that ancestral organelle. Phylogenetic reconstruction also provides direct evidence supporting the common origin of this plastid with those of heterokont algae.
Results and Discussion
Plastid Genomes from Two Photosynthetic Relatives of Apicomplexa.
C. velia has been shown previously to be a photosynthetic alga that is found associated with corals and is related to apicomplexans (15). CCMP3155 is another photosynthetic alveolate originally isolated from bodies of reef corals, and we have investigated it as a possible second such lineage. CCMP3155 oscillates between a coccoid stage and a flagellate stage closely reminiscent of colpodellids. The coccoid cell contains a single plastid surrounded by four membranes, like the plastid of C. velia and apicomplexans, and has thylakoids stacked in triplets, similar to C. velia and dinoflagellates (Fig. S1) (15, 17).
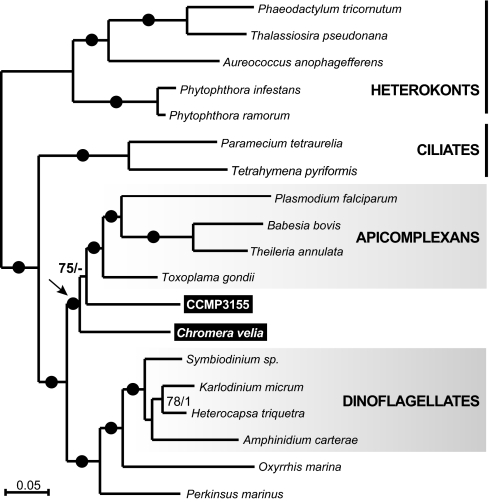
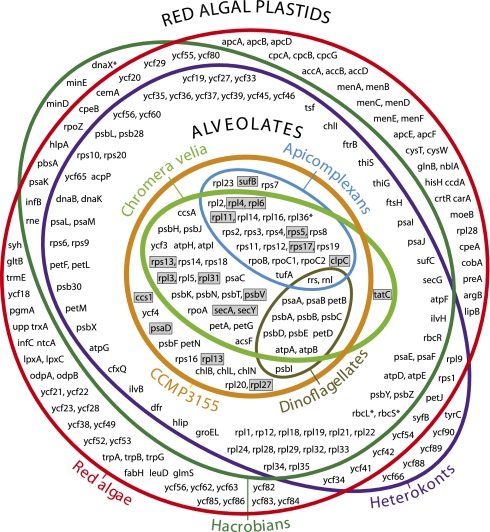
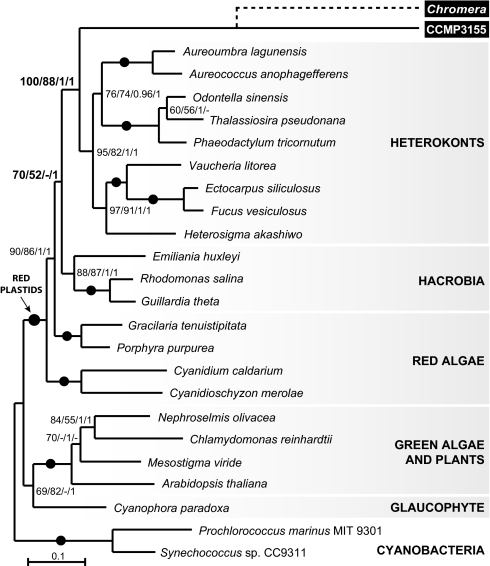
The relationship of CCMP3155 to other alveolates is unknown and the position of C. velia has been inferred only from single-gene analyses (15, 16), so we first sought to clarify the phylogeny. Their position relative to apicomplexans and dinoflagellates is critical to any interpretation of plastid characters—especially if either proved to be specifically related to photosynthetic dinoflagellates rather than apicomplexans. Total genomic DNA from both organisms was sequenced by 454-pyrosequencing and selected nuclear genes extended by PCR, RT-PCR, and 3′RACE into a concatenated dataset of eight nuclear genes consisting of 7,137 characters. All phylogenetic analyses consistently showed with strong support that C. velia and CCMP3155 are closely related to apicomplexans and, strikingly, form two distinct lineages with CCMP3155 more closely related to apicomplexans (Fig. 1). Analyses of plastid genes (see Plastid Phylogeny below) support their separation, but the positions of C. velia and CCMP3155 are interchanged, so all evidence indicates that they represent two distinct photosynthetic lineages that are closely related to apicomplexans. The plastid genomes of both organisms were assembled into single contigs from 454 sequence data. The CCMP3155 plastid DNA maps as a circle (Fig. S2), whereas in C. velia a single gap between the two copies of psbA could not be filled, but the sequenced genome size (119.8 kb) corresponds closely to the size estimated from pulse-field gel electrophoresis and Southern hybridization (121.2 kb) (Fig. S3), suggesting a small gap with few or no genes. It is also possible that the majority of C. velia plastid genome exists as a linear molecule. The C. velia plastid genome is larger than that of CCMP3155 (121.2 kb vs. 85.5 kb), with a lower gene density and stronger strand polarity (Fig. S2). The C. velia plastid uses a noncanonical genetic code (UGA encodes tryptophan) (15), whereas CCMP3155 uses the universal genetic code. At 47.74% GC, the CCMP3155 plastid is one of the least AT-biased plastid genomes known, contrasting to the extremely AT-rich apicoplast (86.86% AT in Plasmodium falciparum). The ribosomal RNA operon of CCMP3155 is also of interest as it is interrupted by a gene for phosphonopyruvate decarboxylase, which appears to be a rare case of lateral gene transfer to the plastid. Comparing gene content among alveolate plastids (Fig. 2, center rings), reveals the nearly mutually-exclusive gene sets of apicomplexans and dinoflagellates (they only share rRNAs and a handful of tRNAs). Plastid genomes of C. velia and CCMP3155 have a relatively modest gene complement, but nevertheless both contain all genes found in either apicomplexans or dinoflagellates, plus numerous other genes. This is consistent with alveolate plastid genomes originating by reduction from a common ancestor. Similarly, the complete set of alveolate plastid genes is also retained in heterokont, hacrobian, and red algal plastids (Fig. 2, outer rings). Significantly, 18 genes found in the alveolate collective gene set are never found in green algal or plant plastids (Fig. 2, boxes), but all are present in plastids of heterokonts, hacrobians, and red algae, consistent with the red algal origin of all alveolate plastids. Another characteristic uniting red algal plastids and their descendants is the ribosomal superoperon, which originated by the fusion of the str and S10+spc+alpha operon clusters (18, 19). The ribosomal superoperon is also present in apicomplexans and CCMP3155 (with several internal rearrangements and gene losses) (Fig. S4), consistent with a red algal origin of these plastids (in C. velia the superoperon has broken up, but the region surrounding the original fusion has been retained in rearranged form on one fragment) (Fig S4). The alveolate superoperons are not as well conserved as those of other algae, but they do share several gene losses in common, including three genes (rpl22, rps9, rps10) that are present in all other red algal plastids. These may have been lost independently, but, given their apparent relationship, a common ancestral loss is perhaps more likely. In addition, the unusual transposition of rpl31 within the superoperon is unique to CCMP3155 and C. velia plastids, suggesting it may also be ancestral to apicoplasts, which have lost the gene (Fig S4). A detailed analysis of gene order throughout alveolate plastids revealed little conservation in C. velia but significant colinearity between CCMP3155 and some apicomplexans. Comparison using a tool for analyzing rearrangements between pairs of genomes (GRIMM, SI Materials and Methods) resulted in a minimum of four inversions (three in the large single copy region and one in the rRNA operon) to explain differences between CCMP3155 and Plasmodium protein- and rRNA-encoding genes. The majority of tRNA genes was also found in orthologous gene blocks in both species, although they are generally more prone to rearrangements as is the case in other plastid genomes (Fig S5). In six cases, the order of genes in blocks ranging from four to two genes are unique to CCMP3155 and apicoplasts among all known plastid genomes (Fig S5). This level of conservation lends further support to the common origin of plastids in apicomplexans and CCMP3315, but more importantly shed some light on how the apicoplast genome reduced to its current state. This is because the blocks of conserved gene order are generally not identical due to the presence of many genes in CCMP3155 that are absent in the apicoplast, nearly all of which are related to photosynthesis (Fig S5). This suggests that the transition from a CCMP3155-like genome to an apicoplast involved surprisingly little reorganization and primarily involved gene loss. Indeed, except for genes associated with photosynthesis, the C. velia and CCMP3155 plastids respectively contain only 4 and 11 additional genes that are absent from the collective apicoplast gene set, providing an interesting glimpse into how the apicoplast reduced.
Fig. 1.
The nuclear phylogeny of C. velia and CCMP3155 show they are two independent photosynthetic lineages closely related to apicomplexan parasites (arrow). The RAxML tree is derived from concatenation of eight nuclear encoded genes (7,137 characters). RAxML/ MrBayes supports are shown above branches; solid circles indicate 100/1 supports.
Fig. 2.
Venn diagram of plastid genome contents in various red plastid lineages. Overlap between the four lineages of alveolates represented by the center rings reveals that plastids of C. velia and CCMP3155 collectively encode all genes found in both apicoplasts and dinoflagellate plastids. Gray boxes highlight 18 genes that are absent in plastids of plants and green algae but all found in alveolate and red algal derived plastids. Genes that originated through horizontal gene transfer are marked with an asterisk. The diagram does not include genes for tRNAs, other small RNAs (5S rRNA, ffs, tmRNAs, rnpB) and the ppd gene horizontally transferred to the CCMP3155 plastid genome.
Ancient Horizontal Transfer of Form II Rubisco to Alveolates.
Neither C. velia nor CCMP3155 plastid genomes encode a form I Rubisco, at least one subunit of which is present in all photosynthetic plastid genomes with the single exception of dinoflagellates. In dinoflagellates, the form I Rubisco has been replaced by a nucleus-encoded, single subunit form II Rubisco acquired by horizontal gene transfer from a proteobacterium (20, 21). We identified a form II Rubisco gene in the nonplastid 454 data from both C. velia and CCMP3155. To confirm that the C. velia Rubisco gene is nucleus-encoded, we used 3′ and 5′ RACE to show the transcript is polyadenylated and encodes an N-terminal extension with characteristics required for plastid-targeting. Both features were confirmed, and the N-terminal extension was found to encode a readily identifiable signal peptide (P = 0.998 in SignalP-HMM) followed by a positively charged region, features consistent with the bipartite leader required for plastid-targeting in apicomplexans and dinoflagellates (22, 23).
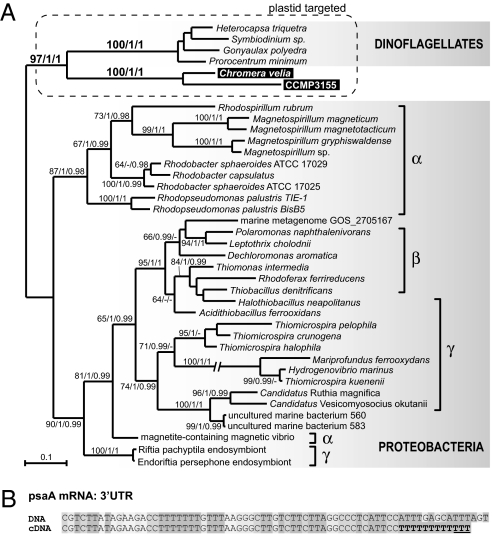
Unrooted phylogenetic analyses demonstrate both C. velia and CCMP3155 Rubisco genes are closely related to homologs from dinoflagellates (Fig. 3A). Rooting this tree using distantly related nonform II Rubisco genes (24) did not affect this: the root fell in various positions among proteobacteria but never within alveolates. Altogether, these data show that the horizontal gene transfer that gave rise to the nuclear-encoded form II Rubisco in dinoflagellates actually took place in the common ancestor of dinoflagellates, apicomplexans, C. velia, and CCMP3155, once again supporting the common origin of their plastids.
Fig. 3.
Form II Rubisco and polyU tails in plastid mRNA link the plastids of C. velia, CCMP3155, and dinoflagellates. (A) RaxML phylogenetic tree of form II Rubisco shows the C. velia and CCMP3155 form II Rubisco genes are closely related to homologs in dinoflagellates. (B) 3′ tails of C. velia plastid mRNAs, represented here by the psaA transcript, contain short polyU tails absent from plastid DNA, a feature previously considered unique to dinoflagellate plastids. Transcripts from other genes are shown in Fig. S6. Underlined thymidines may correspond to the 5′UTRs of the circularized transcripts (Materials and Methods).
Ancient Origin of mRNA Polyuridylylation.
Another feature thought to be unique to dinoflagellate plastids is the 3′ polyuridylylation of transcripts (25). To see if this too may be ancestral to dinoflagellates and apicomplexans, we carried out RT-PCR on circularized mRNAs from three C. velia photosystem genes (psbB, psbC, and psaA). Multiple mRNAs from all three genes were found to be polyuridylylated (Fig. 3B and Fig. S6). This result was confirmed and extended using 3′RACE with a polyU-complementary primer on transcripts from eight other functionally diverse plastid genes. For all eight genes, polyU-specific products were characterized, suggesting polyuridylylation is common to all plastid transcripts in C. velia. This form of processing is otherwise known only in dinoflagellates (25), so once again the presence of this feature in C. velia suggests it was present in a common ancestor of apicomplexan and dinoflagellate plastids (Fig. 3). This would in turn suggest that the character is also either present in apicomplexans and CCMP3315, or that it was lost in one or both of those lineages, but to date we are aware of no data from either to distinguish.
Plastid Phylogeny Supports a Common Origin of Alveolate and Heterokont Plastids.
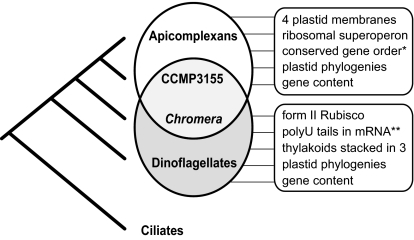
Reduced gene content severely restricts any direct comparisons between apicomplexan and dinoflagellate plastids, and plastids in other algal lineages. The C. velia and CCMP3155 plastid genes are also divergent and phylogenies based on them need to be interpreted carefully, however, they nevertheless provide the means to test alveolate relationships in another way. The relationship between apicomplexan and dinoflagellate plastids abundantly supported by the gene content, gene order, and rare genomic characters described above was tested by evaluating the relationship of each group individually to C. velia and CCMP3155 using the gene set common to each. In both cases the monophyly of alveolate plastids and their relationship to red algae are supported under all analytic models (Fig. S7), reinforcing the conclusion that the ancestor of apicomplexans and dinoflagellates possessed a red algal plastid, that their extant plastids are direct descendents of that organelle, and that each retains different subsets of its ancestral characteristics (Fig. 4). The plastid genomes of C. velia and CCMP3155 also provide an opportunity to examine the deeper history of this endosymbiosis. The reduction of apicomplexan and dinoflagellate plastids not only made their direct comparison difficult but also challenged any comparisons with other plastid lineages. In contrast, the C. velia and particularly CCMP3155 genomes are the most slowly evolving, gene-rich alveolate plastid genome known and are therefore more readily comparable to other plastid genomes. In phylogenetic analyses of whole-plastid genomes, CCMP3155 consistently groups as a sister lineage to heterokonts with strong support (Fig. 5 and Figs. S7 A and B and S8). Alveolates and stramenopiles are also related in nuclear gene trees (26, 27), so their affiliation in whole-plastid phylogenies provides evidence that their plastids are also ancestral. The common ancestry of hacrobian plastids (cryptophytes and haptophytes) also received strong support in all analyses (Fig. 5 and Figs. S7A and S8) and is consistent with the horizontal replacement of rpl36 in their plastid genomes (28) and analyses of nuclear genes (29, 30). Many analyses recovered a monophyletic lineage including all red algal derived plastids (the chromalveolates), but this is not as strongly supported as the alveolate/heterokont or hacrobian groupings. Trees including all plastid genes recovered chromalveolates with weak support (Fig. S8), whereas trees restricted to the slowest evolving 34 and 11 genes recovered chromalveolates with modest and strong support, respectively (Fig. 5 and Fig. S7A). These genes are mostly photosystems, which have been shown to be less likely to lead to spurious results than the housekeeping genes (31, 32). Overall we conclude the plastid genomes support the monophyly of two major groups, the alveolate/heterokont group and the hacrobian group—whether they form a single chromalveolate group is not yet certain.
Fig. 4.
Summary of plastid evolution in alveolates. The plastid genomes of C. velia and CCMP3155 provide a direct link between the plastids of apicomplexans and dinoflagellates because they retain ancestral features that were previously thought to be exclusive to one or the other of these lineages (boxed at the right). Relationships between the lineages based on nuclear data are shown at the left. An asterisk indicates that several regions of conserved gene order are found between the plastid genomes of apicomplexans and CCMP3155, and CCMP3155 and C. velia. Double asterisk indicates that the presence of polyUs in CCMP3155 plastid transcripts has not yet been determined.
Fig. 5.
Plastid phylogenies of 34 conserved plastid genes confirm that CCMP3155 contains a red algal plastid and strongly support its close relationship to plastids of heterokonts. The dotted branch indicates the placement of C. velia sequence, which received complete support in all analyses, but was excluded because of its high rate of substitution. The RAxML tree shows RAxML/PhyML-CAT/MrBayes/PhyloBayes supports over branches; solid circles indicate 100/100/1/1 support. Only ≥60/≥50/≥0.98/≥0.98 branch supports are shown as significant.
Simple Hypothesis for Plastid Evolution.
The plastid genomes of C. velia and CCMP3155 provide multiple lines of evidence for a common origin of red algal plastids in apicomplexans, dinoflagellates, and heterokonts. This, together with parallel evidence for a relationship between the host lineages (26, 27), supports a rather simple picture of plastid evolution by direct descent in these lineages. Recently, a number of more complex theories involving serial tertiary endosymbiosis have been proposed and expanded, in particular, some that suggest either dinoflagellate and apicomplexan plastids were acquired recently from different sources (13, 33). Our data are explicitly inconsistent with this notion, because extant plastids of dinoflagellates and apicomplexans can be linked through C. velia and CCMP3155.
Although serial transfers of plastids could formally explain the relationship of alveolate and hetereokont plastids, congruent relationships inferred from plastid (this study) and nuclear genes (26, 27) suggest simple descent from a common ancestor a more likely explanation. Indeed, serial acquisition of eukaryotes with complex (four membrane) plastids remain a theoretical model that has never been observed outside a few lineages of dinoflagellates, and we show here that all other dinoflagellate plastids were inherited by descent from a common ancestor with apicomplexans. A similar situation is found in hacrobians. Plastid and nuclear gene phylogenies combined with the shared presence of horizontally transferred plastid rpl36 argue strongly for a common plastid acquisition. Overall, available data provide support for at most two and perhaps only one secondary endosymbiosis of a red alga (one in hacrobians and the other in the ancestor of heterokonts and alveolates), and there is as yet no direct evidence for any major algal group acquiring a plastid by tertiary endosymbiosis. Another potential twist in plastid evolution is the notion modern plastids might have supplanted older plastids in some algal lineages, and the history of this original organelle is now only recognizable through the presences of relict genes. Such a case was recently made for chromalveolates using molecular data from some members of the heterokonts (34). The present results neither confirm nor undermine these hypotheses because our conclusions derive from the plastid genome itself and relate specifically to extant plastids.
Role of Loss in Plastid Evolution.
A common origin of alveolate plastids impacts how we view the importance of photosynthesis and plastid loss in evolution. Many alveolate lineages are nonphotosynthetic, but if the ancestors of alveolates and heterokonts had a plastid, then all nonphotosynthetic members of these groups had photosynthetic ancestors. Whether they lost plastids or just photosynthesis remains unknown in most cases: the recent discovery of a cryptic plastid in Perkinsus marinus (35), and several plastid-targeted genes in Oxyrrhis marina (36) (both deep-branching members of the dinoflagellate lineage), highlights this distinction and the need for direct evidence of plastid ancestry in such lineages. In other lineages the abundance of data allows more solid conclusions. In particular, the data now supporting the photosynthetic ancestor of apicomplexans and dinoflagellates lead us to infer that the ancestor of the apicomplexan Cryptosporidium had a plastid despite the absence of plastid ultrastructure or genes for plastid-targeted proteins. A similar case can be made for ciliates and various nonphotosynthetic heterokonts (in particular oomycetes) where whole genomes again confirm the absence of a plastid (37, 38), although claims of relict plastid endosymbiont genes have been made (38, 39). Overall, the apicomplexans, dinoflagellates, and their close relatives are a hotspot for loss of plastids and photosynthesis and further research on this group will likely give us important clues about plastid and photosynthesis loss in other algal lineages.
Implications for Plastid Genome Evolution in Apicomplexans and Dinoflagellates.
Apicomplexan plastid genome reduction is commonly linked to the loss of photosynthesis, although in dinoflagellates the transformation of the plastid genome into single gene minicircles (3) could be interpreted as having allowed the massive transfer of genes to the nucleus (40, 41). However, comparing gene content across all red plastids (Fig. 2) reveals that many genes missing from the plastids of apicomplexans and/or dinoflagellates are also absent from those of C. velia and CCMP3155, and probably were already missing in their common ancestor. This indicates a massive loss or migration to the nucleus of at least 81 genes took place before either apicomplexans or dinoflagellates evolved. Searching the C. velia and CCMP3155 nonplastid sequence revealed fragments of several of these “missing” genes, and 3′RACE showed that transcripts of at least three such genes (chlI, rps9, and rpl21) are polyadenylated, confirming their nuclear localization. Therefore, a major plastid genome reduction by migration to the nucleus took place early in alveolate evolution, and although it continued in both dinoflagellates and apicomplexans (likely for different reasons), the process was not necessarily triggered by specific changes in these lineages. Indeed, it is even possible that this ancient wave of transfers might have precipitated some of the lineage-specific transformations that we observe in their plastids today. These questions and others may be difficult to answer now, but this might change if additional photosynthetic lineages are found to fall in this region of the tree—there are suspiciously few deep-branching photosynthetic alveolates, and the genomes of any additional lineages found might led to further revisions of plastid history.
Materials and Methods
C. velia (CCMP2878) and CCMP3155 were cultivated in L1 seawater medium at 22 °C in 16/8 light/dark cycle. Transmission electron microscopy of CCMP3155 was performed as described previously (15). Genomic DNA was extracted as described previously (15), and sequenced by 454-pyrosequencing. The assembly of plastid genomes resulted in 13.46× and 92.65× coverage in C. velia and CCMP3155, respectively. The assembly in C. velia was verified by direct pair-end sequencing of specific PCR products spanning the entire genome, either connecting conserved plastid genes or directly overlapping. The nuclear dataset was constructed by cloning and sequencing PCR, RT-PCR, and 3′RACE products with specific primers designed to extend fragments of nuclear genes found in the 454 data (see the list of genes below). For the size estimate of C. velia plastid genome, cells were embedded in low-melting agarose plugs, treated with 2% N-laurylsarcosine detergent and 2 mg/mL proteinase K for 30 h at 56 °C and run on pulse-field electrophoresis at U = 6 V/cm (pulses 0.5–25 s) for 20 h in 0.5× TBE. Separated DNA was then blotted onto a membrane, which was hybridized with a radioactively marked psbA probe and the final size of the plastid genome was calculated as an average of five lane measurements from two independent pulse field gel electrophoresis (PFGE) runs. Total RNA was isolated by using TRIzol, treated with DNase I for 10 min, and purified using RNeasy MinElute Cleanup Kit (Qiagen). 3′RACE and 5′RACE were performed using FirstChoice RLM-RACE kit (Ambion). 3′ regions of chlI, rps9, and rpl21 transcripts were cloned using a standard protocol and three clones from each sequenced and assembled into single contigs. For determining polyU tails in plastid transcripts, purified total RNA was treated with RNA ligase, reversely transcribed into first-strand cDNA and PCR products amplified using outwards-facing, plastid gene specific primers. All PCR products amplified from cDNA were cloned and one to three clones sequenced. Phylogenetic methods are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank H. Khan and J. M. Archibald (Dalhousie University, Halifax, Canada) for their dataset of plastid genes. This work was supported by a grant to P.J.K. from the Canadian Institutes for Health Research (MOP 84265). J.J. was supported by a University Graduate Fellowship from the University of British Columbia. A.H. was supported by a grant to the Centre for Microbial Diversity and Evolution from the Tula Foundation. M.O. was supported by the Grant Agency of the Academy of Sciences of the Czech Republic (IAA601410907). J.L. was supported by the Czech Ministry of Education (6007665801) and the Praemium Academiae Award. P.J.K. is a Fellow of the Canadian Institute for Advanced Research and Senior Scholar of the Michael Smith Foundation for Health Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HM222967-68 and HM245036-60).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003335107/-/DCSupplemental
References
- 1.McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- 2.Wilson RJ, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 4.Köhler S, et al. A plastid of probable green algal origin in Apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, Fuller AL, McDougald LR, Zhu G. Apicoplast genome of the coccidian Eimeria tenella. Gene. 2003;321:39–46. doi: 10.1016/j.gene.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Lau AO, McElwain TF, Brayton KA, Knowles DP, Roalson EH. Babesia bovis: A comprehensive phylogenetic analysis of plastid-encoded genes supports green algal origin of apicoplasts. Exp Parasitol. 2009;123:236–243. doi: 10.1016/j.exppara.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Fast NM, Kissinger JC, Roos DS, Keeling PJ. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 8.Funes S, et al. A green algal apicoplast ancestor. Science. 2002;298:2155. doi: 10.1126/science.1076003. [DOI] [PubMed] [Google Scholar]
- 9.Waller RF, Keeling PJ, van Dooren GG, McFadden GI. Comment on “A green algal apicoplast ancestor.”. Science. 2003;301:49. doi: 10.1126/science.1084684. and author reply (2003) 301:49. [DOI] [PubMed] [Google Scholar]
- 10.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 11.Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Puerta MV, Delwiche CF. A hypothesis for plastid evolution in chromalveolates. J Phycol. 2008;44:1097–1107. doi: 10.1111/j.1529-8817.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- 13.Bodył A, Stiller JW, Mackiewicz P. Chromalveolate plastids: Direct descent or multiple endosymbioses? Trends Ecol Evol. 2009;24:119–121. doi: 10.1016/j.tree.2008.11.003. author reply 121–122. [DOI] [PubMed] [Google Scholar]
- 14.Lane CE, Archibald JM. The eukaryotic tree of life: Endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 16.Oborník M, Janouskovec J, Chrudimský T, Lukes J. Evolution of the apicoplast and its hosts: From heterotrophy to autotrophy and back again. Int J Parasitol. 2009;39:1–12. doi: 10.1016/j.ijpara.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Obornik M, et al. Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of apicomplexa, Chromera velia. Protist. doi: 10.1016/j.protis.2010.02.004. in press. [DOI] [PubMed] [Google Scholar]
- 18.Blanchard JL, Hicks JS. The non-photosynthetic plastid in malarial parasites and other apicomplexans is derived from outside the green plastid lineage. J Eukaryot Microbiol. 1999;46:367–375. doi: 10.1111/j.1550-7408.1999.tb04615.x. [DOI] [PubMed] [Google Scholar]
- 19.McFadden GI, Waller RF. Plastids in parasites of humans. Bioessays. 1997;19:1033–1040. doi: 10.1002/bies.950191114. [DOI] [PubMed] [Google Scholar]
- 20.Whitney SM, Shaw DC, Yellowlees D. Evidence that some dinoflagellates contain a ribulose-1,5-bisphosphate carboxylase/oxygenase related to that of the alpha-proteobacteria. Proc Biol Sci. 1995;259:271–275. doi: 10.1098/rspb.1995.0040. [DOI] [PubMed] [Google Scholar]
- 21.Morse D, Salois P, Markovic P, Hastings JW. A nuclear-encoded form II RuBisCO in dinoflagellates. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 22.Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- 24.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc Lond B Biol Sci. 2008;363:2629–2640. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Morse D. The plastid-encoded psbA gene in the dinoflagellate Gonyaulax is not encoded on a minicircle. Gene. 2006;371:206–210. doi: 10.1016/j.gene.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4:366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burki F, et al. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, telonemia and centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;2009:231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: Gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17:887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 30.Hackett JD, et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 31.Hagopian JC, Reis M, Kitajima JP, Bhattacharya D, de Oliveira MC. Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastids. J Mol Evol. 2004;59:464–477. doi: 10.1007/s00239-004-2638-3. [DOI] [PubMed] [Google Scholar]
- 32.Khan H, et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: Lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24:1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- 33.Bodyl A. Do plastid-related characters support the Chromalveolate hypothesis? J Phycol. 2005;41:712–719. [Google Scholar]
- 34.Moustafa A, et al. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- 35.Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A cryptic algal group unveiled: A plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol Biol Evol. 2008;25:1167–1179. doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- 36.Slamovits CH, Keeling PJ. Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol Biol Evol. 2008;25:1297–1306. doi: 10.1093/molbev/msn075. [DOI] [PubMed] [Google Scholar]
- 37.Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 39.Reyes-Prieto A, Moustafa A, Bhattacharya D. Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr Biol. 2008;18:956–962. doi: 10.1016/j.cub.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackett JD, et al. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr Biol. 2004;14:213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 41.Bachvaroff TR, Concepcion GT, Rogers CR, Herman EM, Delwiche CF. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist. 2004;155:65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.