Abstract
Technological advances hold the promise of rapidly catalyzing the discovery of pathogenic variants for genetic disease. However, this possibility is tempered by limitations in interpreting the functional consequences of genetic variation at candidate loci. Here, we present a systematic approach, grounded on physiologically relevant assays, to evaluate the mutational content (125 alleles) of the 14 genes associated with Bardet–Biedl syndrome (BBS). A combination of in vivo assays with subsequent in vitro validation suggests that a significant fraction of BBS-associated mutations have a dominant-negative mode of action. Moreover, we find that a subset of common alleles, previously considered to be benign, are, in fact, detrimental to protein function and can interact with strong rare alleles to modulate disease presentation. These data represent a comprehensive evaluation of genetic load in a multilocus disease. Importantly, superimposition of these results to human genetics data suggests a previously underappreciated complexity in disease architecture that might be shared among diverse clinical phenotypes.
Keywords: epistasis, ciliopathy, zebrafish, in vivo assays
Exome and whole-genome resequencing is likely to catalyze a paradigm shift in the identification of genetic lesions in patients (1). At the same time, even within the confines of the coding genome, such technologies pose an interpretive problem, in that the pathogenic candidacy of mutations can only be derived by narrow genetic models and limited computational predictive tools, both of which are likely to under- and misinterpret the effect of some mutations. Moreover, inter- and intrafamilial variability, a phenomenon prevalent in most genetic traits, remains a major confounding factor because both allelic variation at a single locus and second-site trans modifiers can exert a significant influence on penetrance and expressivity through additive and epistatic effects (2).
Bardet–Biedl Syndrome (BBS) is a useful model for dissecting epistasis because most of the 14 BBS genes can also contribute epistatic alleles (3–17). BBS is also a representative of the ciliopathy disease spectrum, a group of disorders characterized by defects in ciliary structure and/or ciliary signal output (18). Hallmarks of BBS include retinal degeneration, obesity, hypogonadism, polydactyly, renal dysfunction, and mental retardation (19).
We and others have shown that the zebrafish provides experimentally tractable and physiologically relevant models of important aspects of ciliary dysfunction (2, 15, 17, 20–23). Moreover, human mRNA for ciliopathy genes can rescue both morphant and mutant zebrafish phenotypes efficiently, providing a robust platform for interpretation of the pathological relevance of identified missense alleles, whose causal relation to the disorder cannot be proven definitively with genetic arguments alone (15, 17, 22, 23).
Here, we have integrated multiple independent in vivo assays, followed by in vitro validations, to interrogate aspects of the functionality of each of 125 alleles and to determine their contribution to genetic disease as a function of total mutational load across all the known BBS genes.
Results
Modeling BBS Developmental Defects in Zebrafish.
Injection of morpholino against each BBS gene results in gastrulation defects.
We have shown previously that suppression of some BBS proteins in zebrafish causes gastrulation defects that include shortened body axes, longer somites, and broad and kinked notochords (14, 16, 17, 20, 21). These phenotypes are consistent with abnormal planar cell polarity (PCP) signaling (21, 22, 24, 25), which likely underlies several clinical phenotypes in patients who have BBS, including hearing defects (20), neural tube closure abnormalities (26), renal cyst formation (27), and possibly obesity and cognitive impairment (28). The fact that these observations are likely relevant to the etiopathology of BBS (20, 21, 23–25), and true for all seven bbs orthologues tested (bbs1, bbs4, bbs6, bbs10, bbs12, mks1, and cep290) as well for as the three BBS modifiers mgc1203, mks3, and rpgrip1l, suggested that they might represent useful assays for all BBS genes. We therefore designed translation-blocking morpholinos (MOs) against bbs1–12 ( and SI Appendix, Table S1) and injected each into WT embryos at varying concentrations to establish a survival curve (SI Appendix, Table S3) from which to derive the optimal working MO concentration (minimal cytotoxicity, maximal phenotype). Scoring of 8 to 10 somite stage embryos revealed that knockdown of each bbs gene results in defects similar to those described previously (Fig. 1A). These phenotypes were unlikely to be caused by nonspecific toxicity because they were fully reproduced by injection of splice-blocking MOs, for which efficient knockdown could be demonstrated by RT-PCR (SI Appendix, Fig. S1 A–C and Table S3). These effects were corroborated by quantification of the body axis shortening defect by in situ hybridization (SI Appendix, Fig. S2) and by quantification of defects in gastrulation movements (SI Appendix, Fig. S3).
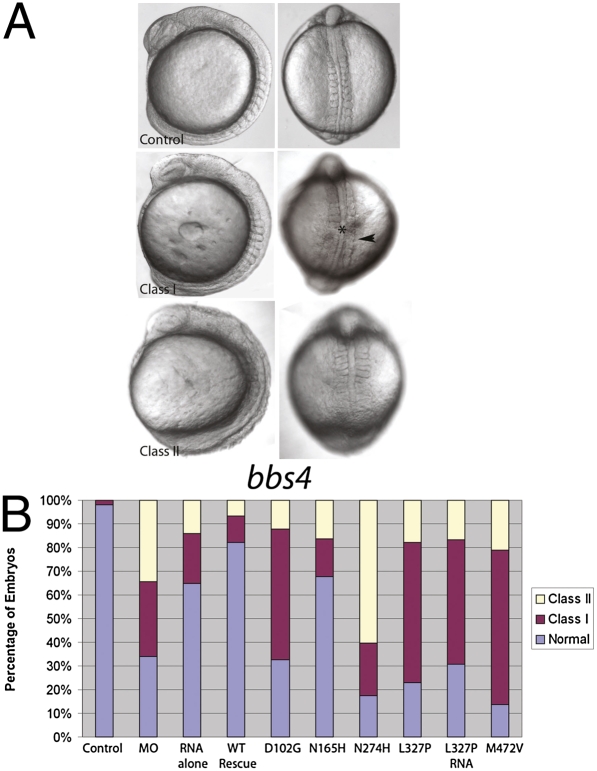
Fig. 1.
Suppression of BBS4 produces specific defects. (A) Injection of MOs against individual bbs genes (bbs4 shown here) into zebrafish embryos produces gastrulation defects, including a short body axis, a widened and kinked notochord (*), and broadened somites (arrowhead), which can be rescued by WT human mRNA. Coinjections of mutant mRNAs produce a spectrum of defects. (B) Whereas WT RNA rescues the MO phenotypes (WT RNA), coinjection of hypomorphic mutations (N165H) partially rescues the phenotype; however, null mutations (D102G, N274H, and M472V in BBS4) do not rescue, and dominant-negative mutations (L327P) exacerbate the phenotype and produce defects by injection of RNA alone.
Human BBS mRNA rescues morphant phenotypes.
Given the high conservation between human and zebrafish bbs orthologues (SI Appendix, Table S2) and the previously reported rescue of bbs11, mks1/bbs13, and cep290/bbs14 with human mRNAs (15, 17), we reasoned that expression of other human BBS proteins should also be able to rescue the morphant phenotypes. We therefore performed rescue experiments for each bbs MO (bbs1–12) with full-length human mRNA.
On scoring of nine (±1) somite stage embryos, we found efficient rescue in each case [SI Appendix, Figs. S1–S4 and Table S4, and an online database (http://misc.ciliaproteome.org/bbs_mutations;)], confirmed by quantification by in situ hybridization as well as scoring of epiboly movements, each of which was similar to uninjected controls (Figs. 2 and 3 and SI Appendix). To verify further the specificity of each BBS mRNA to rescue the corresponding bbs MO, we coinjected each bbs MO with a different BBS mRNA, and vice versa, and did not see any rescue (SI Appendix, Fig. S1D). Importantly, in comparison of blindly scored embryos, phenotypes among rescued embryo cohorts were consistently statistically improved from the MO cohort (SI Appendix, Table S4).
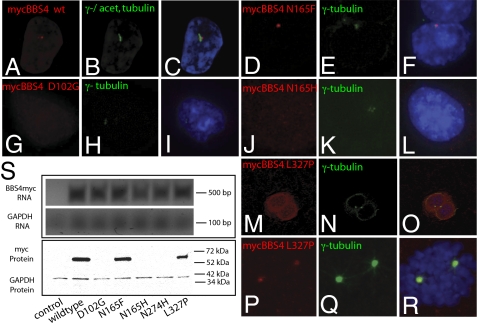
Fig. 2.
In vitro validation of mutation effects. (A–C) Localization of myc-tagged WT BBS4 protein (red) in IMCD3 cells shows presence at the basal body, as indicated by colocalization with γ-tubulin and acetylated α-tubulin (green). (D–F) Introduction of an artificial N165F mutation in BBS4 has no observable effect on localization. Expression of the D102G mutant, characterized as null by in vivo scoring, is undetectable 2 d after transfection (G–I), whereas the hypomorphic N165H variant is expressed but mislocalized (J–L). The dominant-negative mutation L327P results in mislocalization in postmitotic cells (M–O) but not in dividing cells (P–R). (S) Expression of MYC-tagged constructs 4 d after transfection is undetectable for null mutations (D102G, N274H, and N165H), although RT-PCR showed message in each case.
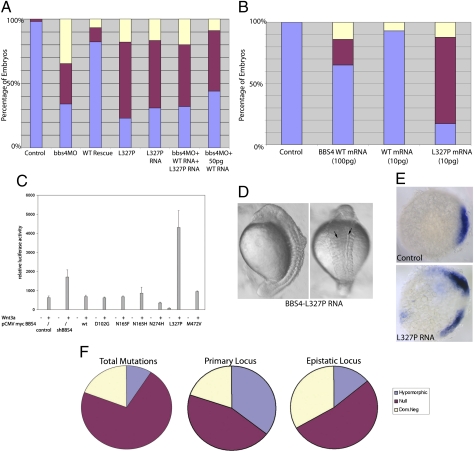
Fig. 3.
The dominant-negative effect of BBS4 L327P. (A) Coinjection of 3 ng of bbs4 MO and 100 pg of WT human mRNA rescues the morphant phenotype, and coinjection of 100 pg of L237P bearing BBS4 mRNA produces defects similar to, or more severe than, MO alone. Introduction of equimolar amounts (50 pg each), however, of both WT and mutant mRNA attenuates the ability of 50 pg of WT mRNA alone to rescue partially the morphant phenotype. (B) Injection of a suboptimal dose (10 pg) of L327P mRNA produces significant phenotypes. (C) Overexpression of the dominant-negative BBS4 mutation L327P but not of the loss-of-function alleles activated canonical Wnt signaling by the Super TOP-Flash luciferase assay in a manner similar to BBS4 suppression. Overexpression of L327P mRNA at a high concentration (300 pg) results in dorsalized embryos and double axes (arrows) (D) and ectopic chordin expression (E). (F) In the context of oligogenic inheritance, the primary (recessive) locus has a significantly increased likelihood of being hypomorphic (P < 0.0001), whereas the epistatic locus is more likely to be severe, with significant enrichment of dominant-negative lesions (P = 0.0004).
Rescue of gastrulation phenotypes yields a spectrum of severity.
The rescue of bbs morphants with human mRNA for all BBS genes offered us the opportunity to test the effect of all nonsynonymous amino acid changes found in patients in the context of the human protein sequence. First, we engineered mutant constructs by introducing single nucleotide changes in each human gene (n = 109). We then assayed the efficiency of rescue of each construct by injecting WT embryos with either (i) a negative control, (ii) MO alone, (iii) MO plus a predetermined molar amount of WT human RNA, (iv) MO plus the same molar amount of mutant mRNA, (v) WT mRNAs alone, or (vi) mutant mRNAs alone. Each coinjection was scored as a comparison with the MO alone or with the WT rescue (P values in SI Appendix, Table S4).
Fourteen alleles showed no appreciable differences in rescue efficiency (SI Appendix, Fig. S4 and Table S5), suggesting that they were benign with respect to the function(s) assayed. For example, BBS6 R518H mRNA fully rescued the morphant phenotype (SI Appendix, Figs. S2–S4). By contrast, 15 alleles yielded phenotypes significantly improved from MO-injected embryos but worse than embryos coinjected with WT mRNA, suggesting that these alleles are functional hypomorphs, as exemplified by BBS4 N165H (Fig. 1). A third set of 45 alleles yielded no rescue, and they were therefore scored as nulls (e.g., BBS4 D102G, N274H; Fig. 1). Finally, 35 alleles produced phenotypes significantly worse than MO alone and mutant mRNA produced phenotypes similar to or worse than MO alone, suggesting that these might be dominant-negative alleles (e.g., BBS4 L327P; Fig. 1). Importantly, we randomly assayed 26 mutations by epiboly tracking quantification and somite trunk length measurements and found full concordance across all assays, suggesting that scoring bias is unlikely to introduce false-positive results.
This systematic evaluation, in addition to our previously reported data for BBS13-14, MKS3, and RPGRIP1L (17, 23), enabled us to generate a complete allelic series for 125 nonsynonymous coding variants across all BBS-associated genes (Fig. 1 and SI Appendix, Fig. S4 and Table S5).
Sensitivity and specificity: Genetic validations.
Segregation and population-based analysis or in vitro functional data for a subset of mutations found in patients who have BBS have provided strong evidence for pathogenicity, affording us the opportunity to test the sensitivity of our in vivo scoring. We therefore selected 49 alleles of known pathogenicity in humans and asked how they were scored in vivo (SI Appendix, Table S5). With the exception of the S329L allele in BBS10 (scored benign), 48 of 49 alleles were predicted correctly as pathogenic, suggesting that the in vivo assays have a sensitivity of 98%.
A similar approach can be implemented to test specificity. We selected 12 changes (BBS2 123IV; BBS6 C517R; BBS9 A455T; BBS10 D142N; BBS12 I170V, R235M, Q386R, and S429T; MKS1 L491I; MKS3 D261N; and L437V and RPGRIP1L G1025S) common in human populations (minor allele frequency >15%) and thereby found frequently in unaffected parents and siblings of patients, sometimes in homozygosity. We also tested a 13th allele, BBS4 N165F, an induced change that does not alter the properties of the protein (see below), and four BBS6 alleles (G52D, I339V, S511A, and R518H) that behaved indistinguishably from WT in cultured cells (29); 14 of 17 changes were benign in all three in vivo assays, suggesting a specificity of >82%.
In vitro assays.
Phenotypes related to some of the BBS proteins have been observed in cultured cells (8–10, 29–34). Therefore, we asked whether some of the aberrant activity in zebrafish embryos could be explained by defective behavior in mammalian cells.
We focused initially on BBS4 because, in contrast to other BBS proteins, its localization near the two centrioles can be achieved reproducibly by overexpression of tagged protein in various transformed cell lines (31, 32). We engineered six BBS4 missense mutations into an N-terminal MYC-tagged mammalian expression vector. Three were predicted to be null (D102G, N274H, and M472V), one was a hypomorph (N165H), one was dominant-negative (L327P), and one was the benign variant N165F. Subsequent to transfection into IMCD3 cells, we found our observations to be concordant with the in vivo assessments. Although the benign construct N165F showed a pattern of localization indistinguishable from that of WT (Fig. 2 A–F), each of the seven nonneutral mutations showed reproducible phenotypes present in >70% of transfected cells. The null D102G and N274H mutations were undetectable 48 h after transfection (Fig. 2 G–I and S), whereas the hypomorphic allele N165H was also degraded, albeit more slowly (96 h after transfection; Fig. 2 J–L and S). Finally, the dominant-negative allele L327P showed stability indistinguishable from that of WT (Fig. 2S) yet failed to colocalize with γ-tubulin in postmitotic cells (Fig. 2 M–O). Interestingly, we observed centriolar localization of L327P in the mitotic spindle of dividing cells, suggesting that this mutation exerts a specific postmitotic effect (Fig. 2 P–R). Expression of each construct was verified by RT-PCR and with a GFP cotransfection marker (SI Appendix, Figs. S2 and S6).
These observations were not unique to BBS4; we engineered mammalian expression constructs for a random set of 17 BBS mutations (14%). Even though most overexpressed WT proteins show a consistently diffuse cytoplasmic distribution, similar to previous reports (34), we were still able to evaluate differences between WT and mutant constructs. Several alleles predicted to be detrimental to protein function showed a localization pattern distinct from WT (SI Appendix, Fig. S6). Notably, we observed several examples in which the mutation drove the localization of the protein to subcellular regions never seen with WT constructs. For example, WT BBS7 is detected throughout the cytoplasm but is largely excluded from the nucleus (SI Appendix, Fig. S6L). By contrast, we found mutants T211I and H323R in the nucleus of a significant proportion of cells (SI Appendix, Fig. S6 M and N).
We did find one discrepant result: the BBS6 T112A allele was benign in zebrafish, yet a 112A-expressing construct showed aberrant localization in the nucleus in mammalian cells (SI Appendix, Fig. S6). Although it is not clear which of the two assays is more informative, we concluded that, conservatively, our in vivo strategy has an accuracy rate when compared with in vitro mammalian cell data of 94% (comparable to the level of sensitivity observed across the three in vivo assays). Importantly, our observations from IMCD3 cells were fully corroborated by localization studies in ARP19 and hTERT human ciliated cells (online database).
Dominant-negative mutations.
Our in vivo studies revealed 35 candidate dominant-negative alleles (28%). To examine this possibility, we performed two independent studies. We first revisited the in vivo zebrafish assay and reasoned that if these alleles were dominant-negative, the following two statements should be true: Injection of mRNA encoding dominant-negative alleles alone should phenocopy the morphant phenotype, and equimolar injection of dominant-negative and WT mRNA should attenuate the rescue effect of the WT message.
We found the first statement to be true for all dominant-negative alleles; injection of dominant-negative mRNA alone produced phenotypes similar to MO alone for each of the 35 mutations (Fig. 3A, SI Appendix, Fig. S7 A–E, and online database). This is true even at suboptimal concentrations, at which WT mRNA produces minimal defects (Fig. 3B and SI Appendix, Fig. S7 F and G). Moreover, further testing suggested the second statement also to be true. For example, for bbs4, coinjection of MO and 100 pg of WT mRNA efficiently rescued the morphant phenotype. Coinjection of 100 pg of 327P mRNA with MO, however, gave rise to a significantly worse embryo phenotype than when MO alone was injected, as did injection of 100 pg of mutant mRNA alone. Further, whereas coinjection of bbs4 MO and 50 pg of WT mRNA partially rescued the morphant phenotype, addition of 50 pg of 327P coding mRNA (for a total of 100 pg of mRNA) attenuated the ability of the WT mRNA to rescue the phenotype (Fig. 3A). We observed similar scenarios for all the alleles tested (SI Appendix, Fig. S7 A–E).
Seeking additional evidence, we turned to a mammalian cell-based biochemical assay. We have shown previously that suppression of BBS4 leads to elevated canonical Wnt signaling activity, as monitored by the Super TOP-Flash luciferase assay, whereas overexpression of WT BBS4 has no effect (21). If we have interpreted the zebrafish data correctly, the sole BBS4 allele predicted to be dominant-negative, L327P, should transactivate the β-catenin reporter in a manner similar to the BBS4shRNA. We therefore expressed L327P as well as WT, four loss-of-function BBS4 mutations (D102G, N165H, N274H, and M472V), and the artificial neutral variant N165F. Blinded analysis of the data correctly predicted L327P to be dominant-negative in three separate experiments (Fig. 3C). To determine if this effect can be replicated in vivo, we overexpressed L327P mRNA at a high concentration (300 pg) and observed dorsalization, including double axis formation in a small proportion of embryos (Fig. 3D), a hallmark of up-regulation of canonical Wnt signaling; consistently, these embryos exhibit ectopic expression of the canonical Wnt target gene, chordin (21), at midepiboly stages (Fig. 3E).
The discovery of a significant number of dominant-negative mutations is inconsistent with the conceptual notion of loss-of-function effects in recessive disease; that the observations are artifacts is unlikely because of their reproducibility across multiple repetitions and independent assays. We therefore asked whether genetic insights could be gleaned by examining the distribution of candidate dominant-negative alleles in the 218 patients and families in our cohort. We found that dominant-negative alleles account for a minority (18%) of all BBS alleles. However, we and others have reported numerous families in which a third trans mutation interacts genetically with recessive alleles at a primary locus to modulate disease penetrance and expressivity (34–37). When mapping the position of dominant-negative alleles (primary locus vs. epistatic locus), we found a significant increase of dominant-negative mutations in epistatic loci (33% in modifiers vs. 18% contribution to total mutations; P = 0.0004; Fig. 3F), suggesting that a majority of modifier phenomena are inherited dominantly.
Common variants in BBS.
Our functional assays were not restricted to rare or private mutations but encompassed all known allelism in BBS genes. Both in vivo tests and cell localization studies indicated that 7 of 14 alleles formally classified as polymorphisms (>1% population frequency) are also detrimental to protein function, and might therefore participate in the disease process (SI Appendix, Fig. S5 and Table S6). Given that most of these variants can be found in homozygosity in control individuals suggests that they are insufficient to cause disease. However, we wondered whether they might interact with strong mutations to potentiate BBS, as shown recently for one of them, the A229T allele in RPGRIP1L (23).
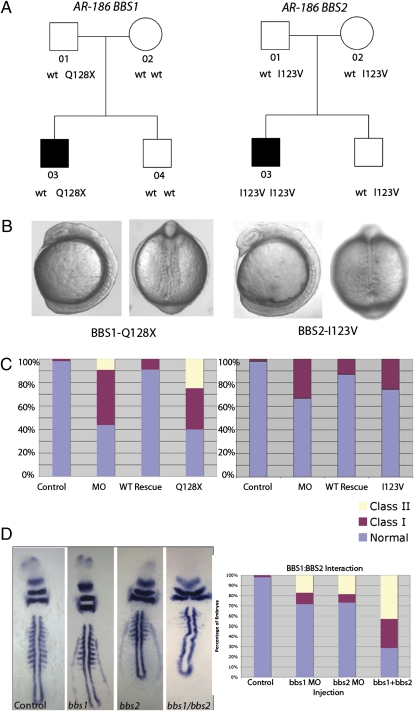
These data suggested that population frequency of alleles is not necessarily an indicator of functionality, which led us to query the segregation of all nonsynonymous polymorphisms (n = 18) in our families. Not surprisingly, the seven benign SNPs segregated in patterns unrelated to the disease (SI Appendix, Fig. S5 and Table S6). For other alleles, however, the combination of in vivo data and segregation was potentially able to account for the transmission of the disorder in families. We and others have reported numerous pedigrees in which only one heterozygous pathogenic mutation has been found in any given BBS gene (4, 5, 7, 14, 16, 17, 38). The assumption has been that the second mutation might lie in a regulatory element. However, our data offer an alternative hypothesis, by which, for some families, the disease may be explained by nonallelic noncomplementation among rare and common alleles. Although most of these alleles were too infrequent in our cohort to perform formal genetic tests for this hypothesis, this was not the case for 123IV in BBS2 [13.3% in our controls (n = 240), 13% in Utah residents with ancestry from northern and western Europe (CEU) HapMap]. In Northern European family AR186, the patient was haploinsufficient for BBS1 (with a bona fide loss-of-function Q128X mutation, as determined by the in vivo assay (Fig. 4 B and C), did not have an obvious deletion in the other allele (as judged by SNP heterozygosity at the ~1-kb level of resolution across the locus), and did not have any other candidate pathogenic alleles in BBS1. However, this patient was also homozygous for the 123IV SNP in BBS2, a functional hypomorph (Fig. 4 A–C). Importantly, although the 123V allele might appear to be sufficient for pathogenesis in this family, the presence of 123V homozygotes in the general population (3% in our controls, 3.3% in CEU HapMap) argues that its combination with the BBS1 nonsense allele might potentiate the disease state.
Fig. 4.
Reevaluation of genetic data using in vivo scoring. Effects of mutations based on in vivo analyses suggest that the severity of BBS mutations is consistent with segregation of disease in families with BBS. (A) Although the heterozygous nonsense BBS1 mutation Q128X present in the individual who has BBS in family AR186 is functionally null (B and C), its presence does not fully account for the disease. (B and C) Affected individual, however, is also homozygous for the BBS2 polymorphism, I123V, predicted to be a mild hypomorph. (D) Injection of either bbs1 or bbs2 MO alone at suboptimal concentrations results in mildly affected embryos. Coinjection of both MOs together produces severely affected embryos with phenotypes not seen by injection of either MO alone, including near-complete loss of somite definition (labeled by myoD) and loss of eye field (labeled by pax2).
Because the 123IV change is sufficiently frequent in Europeans, we were able to test whether the in vivo prediction of nonneutrality was true by asking whether the 123V allele of BBS2 was enriched and overtransmitted in patients who have BBS. We observed both to be true: We found 123V in 55 (19.3%) of 284 chromosomes from Northern European patients who had BBS compared with 32 (13.3%) of 240 chromosomes from ethnically matched controls, which is a significant difference (χ2 = 8.78, P = 0.003), although transmission disequilibrium testing showed that of 25 informative trios analyzed (no parent homozygous for the minor allele and excluding the “discovery” family AR186), 123V was transmitted to the affected individual in 22 instances (88%; P = 0.02).
These data, although of modest numbers, are both consistent with the in vivo analyses and suggestive of an interaction between BBS1 and BBS2. We therefore tested whether subeffective loss of BBS2 can modulate the phenotype of BBS1 loss of function. We found strong evidence that this is true: Injection of both bbs1 and bbs2 MOs produced phenotypes significantly more severe than injection of either MO alone, some of which, such as the near-complete loss of somite definition and loss of the eye field, were not observed in either single MO injection, even at higher concentrations (Fig. 4D).
Discussion
We used a broad and comprehensive in vivo analytic strategy validated by multiple independent in vitro and genetic tools to assess the functional consequences of nonsynonymous variants in BBS genes. The application of such comprehensive in vivo assays is not unique. For example, misexpression of human genes in Drosophila has been used to assess the pathogenic potential of the genes/alleles associated with cardiomyopathy (39–41). Advantages of our system include the use of a vertebrate that shares many developmental processes and anatomical features with humans and the assaying of a physiologically relevant phenotype of known significance to the syndrome in humans. We note that we are assaying only this aspect of the BBS phenotype and other components of the disorder that are PCP-independent might not be captured. Notably, however, our data were also consistent with all published observations of allele pathogenicity, suggesting that these assays are reasonable surrogates. We found 48 of 49 mutations predicted to be pathogenic from human genetic analysis to be such, whereas our interpretation of the BBS1 M390R allele as a hypomorph is consistent with the M390R knock-in mouse model (42) and our prediction of BBS11 P130S to be pathogenic is consistent with a previous report of failure of this allele to rescue melanosome transport (15). Further, our in vivo data for BBS3 and BBS6 are in agreement with two recent biochemical studies (29, 30).
We were initially surprised to find a high preponderance of dominant-negative alleles. We first considered the possibility that this might be an artifact of the in vivo system, perhaps driven by the high levels of expression of mutant mRNA/protein. However, equimolar injections of WT/mutant mRNA attenuated but did not extinguish the dominant-negative effect, and expression even at suboptimal concentrations produced severe defects. There are three likely mechanisms to explain these observations. First, an excess of dominant-negative mutations was found in second-site modifier relationships, suggesting that the epistatic modification is autosomal dominant, whereas the disease per se remains recessive. Second, we predict that carriers of dominant-negative alleles will manifest subclinical BBS-associated phenotypes. Consistent with this notion, parents of patients who have BBS have been reported to be more prone to obesity and retinal dysfunction (43, 44). Intriguingly, we found the BBS6 A242S mutation to be a dominant-negative finding; this mutation has been associated with obesity in the heterozygous state in a population-based study (45). Finally, we suspect that dominant-negative alleles may exert their effects at the biochemical level, perhaps by disrupting the BBSome (31) or by perturbing the canonical/noncanonical Wnt signaling balance, as was the case for BBS4 L327P. It is possible that such interactions of mutant proteins exacerbate the disease phenotype but that interaction of mutant and WT protein, although detrimental to WT protein function, does not by itself result in disease. This formulation is consistent with the recent report of a dominant-negative mutation in β-amyloid precursor protein in recessively inherited Alzheimer disease (46).
Our data also indicate that common alleles can be detrimental to protein function and that, in several instances, the context-dependent interaction of rare and common alleles can determine the phenotype. We speculate that such interactions are probably abundant across genetic disorders and that the presence and relative contribution of alleles with high minor allele frequency should be evaluated carefully in the context of additional rare variants.
Finally, the study of BBS mutations has benefited from a combination of strong evolutionary conservation and a priori understanding of some of the pathways involved in disease pathogenesis. Nonetheless, it is likely that this approach is conceptually feasible for a broad range of other clinical phenotypes. It will be important to develop experimental tools that can evaluate the impact of variation on the output of distinct disease mechanisms or, more broadly, on the phenotypic consequences of whole systems. The availability of large-scale resequencing will necessitate the development and application of bioassays such as those described here to assess the functional relevance of variants identified in individuals with phenotypes of interest.
Materials and Methods
MOs and Embryo Manipulations.
MOs against zebrafish bbs1–12 (SI Appendix, Table S1) and control MO were obtained from Gene Tools. One nanoliter of each solution was injected into WT zebrafish embryos at the one- to two-cell stage. Human WT and mutant mRNA for BBS1–12 were transcribed in vitro with the SP6 Message Machine kit (Ambion). MO and mRNA concentrations (SI Appendix, Table S3) were determined based on the combination by which WT mRNA efficiently rescued the morphant phenotype. The same concentrations were used for rescue with mutant mRNA or injection of mRNA alone. Full dose–response curve data for each MO are available on request.
Classification and Scoring of Embryos.
Affected embryos were classified into two graded phenotypes on the basis of the relative severity compared with age-matched uninjected controls from the same clutch, as described in SI Appendix. Comparisons between injections of MO alone, mRNA alone, mutant rescue, and WT rescue were performed by means of the χ2 test.
Whole-Mount RNA in Situ Hybridization.
Whole-mount in situ hybridization was carried out, followed by flat mounting, clearing in methyl salicylate, photographing at a magnification of ×10, and measurement, as described in SI Appendix.
Monitoring of Gastrulation Movements.
Gastrulation movements were monitored by single blastomere injections of 10,000-Mr dextran-conjugated green fluorescent lineage tracer (Invitrogen) at the 16-cell stage and monitoring of positioning of fluorescent cells through epiboly stages.
Mammalian Cell Culture.
Cells were grown on glass coverslips in DMEM/F12 media (10% vol/vol FBS). Following transient transfection with Fugene 6 (Roche), cells were fixed in methanol 48 h later and immunostained with anti-Myc and anti-γ-tubulin rabbit polyclonal antibodies (Sigma).
Supplementary Material
Acknowledgments
We thank Andy McCallion, Jim Lupski, and the Katsanis laboratory for critical review of the manuscript. This work was supported by Grant R01HD04260 from the National Institute of Child Health and Human Development; Grants R01DK072301, R01DK075972 (to N.K.), and R01DK52431 (to R.L.L.) from the National Institute of Diabetes and Digestive and Kidney Diseases; the Macular Vision Research Foundation (N.K.); a Visual Neuroscience Training Program fellowship (N.A.Z.); and the Russell Berrie Foundation (R.L.L.). N.K. is the Jean and George W. Brumley Professor.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000219107/-/DCSupplemental.
References
- 1.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badano JL, et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 3.Slavotinek AM, et al. Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- 4.Katsanis N, et al. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- 5.Mykytyn K, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura DY, et al. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2) Hum Mol Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 7.Mykytyn K, et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 8.Chiang AP, et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Y, et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 10.Ansley SJ, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 11.Badano JL, et al. Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JB, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura DY, et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoetzel C, et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. and correction (2006) 38:727. [DOI] [PubMed] [Google Scholar]
- 15.Chiang AP, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc Natl Acad Sci USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoetzel C, et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. and correction (2008) 40:927. [DOI] [PubMed] [Google Scholar]
- 18.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 19.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: Results of a population survey. J Med Genet. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- 20.Ross AJ, et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes JM, et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
- 22.Cantagrel V, et al. International Joubert Syndrome Related Disorders Study Group Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna H, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbit KC, et al. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 26.Katsanis N. Ciliary proteins and exencephaly. Nat Genet. 2006;38:135–136. doi: 10.1038/ng0206-135. [DOI] [PubMed] [Google Scholar]
- 27.Fischer E, et al. Defective planar cell polarity in polycystic kidney disease. Nat Genet. 2006;38:21–23. doi: 10.1038/ng1701. [DOI] [PubMed] [Google Scholar]
- 28.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirayama S, et al. MKKS is a centrosome-shuttling protein degraded by disease-causing mutations via CHIP-mediated ubiquitination. Mol Biol Cell. 2008;19:899–911. doi: 10.1091/mbc.E07-07-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi T, et al. Biochemical characterization of missense mutations in the Arf/Arl-family small GTPase Arl6 causing Bardet-Biedl syndrome. Biochem Biophys Res Commun. 2009;381:439–442. doi: 10.1016/j.bbrc.2009.02.087. [DOI] [PubMed] [Google Scholar]
- 31.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 32.Kim JC, et al. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 33.Kim JC, et al. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J Cell Sci. 2005;118:1007–1020. doi: 10.1242/jcs.01676. [DOI] [PubMed] [Google Scholar]
- 34.Badano JL, et al. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Hum Mol Genet. 2003;12:1651–1659. doi: 10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- 35.Katsanis N, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 36.Katsanis N, et al. BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet. 2002;71:22–29. doi: 10.1086/341031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beales PL, et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72:1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beales PL, et al. Genetic and mutational analyses of a large multiethnic Bardet-Biedl cohort reveal a minor involvement of BBS6 and delineate the critical intervals of other loci. Am J Hum Genet. 2001;68:606–616. doi: 10.1086/318794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter LTPL, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chien SRL, Reiter LT, Bier E, Gribskov M. Homophila: Human disease gene cognates in Drosophila. Nucleic Acids Res. 2002;30:149–151. doi: 10.1093/nar/30.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf MJ, et al. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci USA. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis RE, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croft JBMD, Chase CL, Swift M. Obesity in heterozygous carriers of the gene for the Bardet-Biedl syndrome. Am J Med Genet. 1995;55:12–15. doi: 10.1002/ajmg.1320550105. [DOI] [PubMed] [Google Scholar]
- 44.Kim LSFG, Fishman GA, Seiple WH, Szlyk JP, Stone EM. Retinal dysfunction in carriers of bardet-biedl syndrome. Ophthalmic Genet. 2007;28:163–168. doi: 10.1080/13816810701537440. [DOI] [PubMed] [Google Scholar]
- 45.Andersen KL, et al. Variation of the McKusick-Kaufman gene and studies of relationships with common forms of obesity. J Clin Endocrinol Metab. 2005;90:225–230. doi: 10.1210/jc.2004-0465. [DOI] [PubMed] [Google Scholar]
- 46.Di Fede G, et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323:1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.