Abstract
The cadherin superfamily encodes more than 100 receptors with diverse functions in tissue development and homeostasis. Classical cadherins mediate adhesion by binding interactions that depend on their N-terminal extracellular cadherin (EC) domains, which swap N-terminal β-strands. Sequence alignments suggest that the strand-swap binding mode is not commonly used by functionally divergent cadherins. Here, we have determined the structure of the EC1–EC2 domains of cadherin 23 (CDH23), which binds to protocadherin 15 (PCDH15) to form tip links of mechanosensory hair cells. Unlike classical cadherins, the CDH23 N terminus contains polar amino acids that bind Ca2+. The N terminus of PCDH15 also contains polar amino acids. Mutations in polar amino acids within EC1 of CDH23 and PCDH15 abolish interaction between the two cadherins. PCDH21 and PCDH24 contain similarly charged N termini, suggesting that a subset of cadherins share a common interaction mechanism that differs from the strand-swap binding mode of classical cadherins.
Keywords: hair cell, tip link, hair cell, PCDH21, PCDH24
The organization of cells into tissues and organs depends on cadherin molecules that regulate such diverse processes as cell adhesion, synapse formation, and the development and function of sensory cells in the inner ear and retina (1–4). The defining feature of the cadherin superfamily is the extracellular cadherin (EC) domain, which occurs in varying repetitions in all cadherins. Based on sequence homology and domain structure, cadherins are divided into subfamilies including the classical cadherins, desmosomal cadherins, seven transmembrane cadherins, and protocadherins. Classical cadherins and desmosomal cadherins are the best-studied superfamily members and have well-documented roles in mediating cell adhesion and the formation of desmosomes, respectively (1–4). The function and adhesive properties of other cadherin superfamily members are less well studied.
Crystallographic and biochemical studies have provided insights into the structure of cadherin extracellular domains and the mechanism that mediates adhesion between classical cadherins (5–13). The EC domain forms a protein module of the Ig-like fold consisting of seven β-strands that are arranged as two opposed β-sheets. The N- and C termini reside on opposite ends of the EC domain, facilitating the formation of tandem repeats. The connections between successive EC domains are rigidified by coordination of three Ca2+ ions, which is mediated by amino acids that are conserved in all cadherins (5, 9, 14). Adhesion specificity resides in the EC1 domains. The crystallographic structures show that the adhesive binding interface is formed by “swapping” of the amino-terminal β-strands of opposite EC1 domains, whereby the strand of one monomer replaces the strand of the other. Critical for this interaction are the side chains of conserved Trp residues, which fit into hydrophobic pockets on the EC1 domain of the binding partner (5, 8, 10).
Recent findings suggest that the formation of cadherin adhesion complexes is a multistep process, where cadherins establish a so called “X-dimer” intermediate that facilitates the subsequent formation of the strand-swapped dimer (7). The intermediate is called X-dimer because the dimeric assembly of the elongated molecules resembles the shape of an X. Residues near the EC1–EC2 Ca2+ binding sites mediate X-dimer formation. Earlier crystallographic studies of E-cadherin fragments containing small N-terminal extensions revealed a similar X-dimer structure (9, 11), which likely reflected these binding intermediates. T-cadherin contains the conserved residues near the EC1–EC2 Ca2+ binding sites but lacks strand-swapping signatures. Crystallographic studies of EC1–EC2 of T-cadherin reveal an X-dimer structure, suggesting that this cadherin interacts by this binding mode alone (6).
We have recently shown that two cadherin superfamily members, CDH23 and PCDH15, interact to form tip links, which gate mechanotransduction channels in hair cells of the inner ear (15). The two cadherins are also expressed in retinal photoreceptors, and mutations in their genes lead to deaf-blindness in humans (1, 16). Unlike adhesion complexes between cells, tip links do not contain thousands of cadherin molecules and are instead formed by one CDH23 homodimer interacting in trans with one PCDH15 homodimer (15). The EC1 domains of CDH23 and PCDH15 mediate binding interactions, but CDH23 and PCDH15 lack strand-swapping signatures, suggesting that they interact by a mechanism different from classical cadherins. We therefore set out to gain insights into the structure and binding mechanism of tip-link cadherins.
Results
Structure of the N-terminal EC1–EC2 Domains of CDH23 Reveals Unique Features Not Shared with Classical Cadherins.
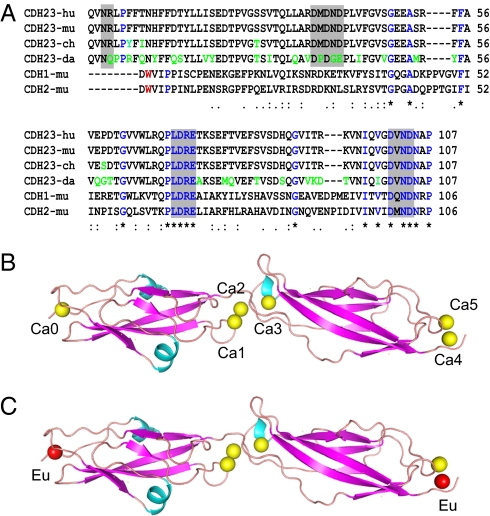
CDH23 from different species lacks Trp residues that are conserved in classical cadherins, but contains an N-terminal extension (Fig. 1A), suggesting that CDH23 mediates adhesive interactions by a mechanism different from classical cadherins. To test this model, we first determined the crystal structure of the EC1–EC2 fragment of mouse CDH23 at a resolution of 1.1 Å (Fig. 1 B and C and Table S1; PDB ID 3MVS). Numberings in sequences throughout the manuscript start with the first amino acid of the mature CDH23 EC1 domain (Glu1). The EC1–EC2 fragment was expressed in Escherichia coli with an affinity tag, which was removed before crystallization. Analysis by size exclusion chromatography showed that the protein fragment behaved as a monomer in solution. EC1 and EC2 domains of CDH23 have 22.8% sequence identity, and our structural analyses show that they have characteristic cadherin domain topology consisting of a sandwich of two β-sheets possessing three (A, G, and F) and four (B, E, D, and C) β-strands, respectively. Unlike in previously reported EC domain structures, there is also an α-helix between strands C and D of EC1 (Fig. 1 B and C).
Fig. 1.
Structure of the CDH23 EC1–EC2 domains. (A) Sequence alignment of the mature CDH23 EC1–EC2 domains from different species (hu, human; mu, murine; ch, chicken; da, zebrafish) with murine CDH1 and CDH2. Ca2+ binding motifs are highlighted with gray boxes. Sequence differences between CDH23 molecules are indicated in green. The conserved Trp2 residue in CDH1 and CDH2 is indicated in red. (B and C) Ribbon diagrams of CDH23 EC1–EC2: Ca2+-bound (B) and Eu2+-bound (C). Ca2+ is shown in yellow, Eu2+ in red.
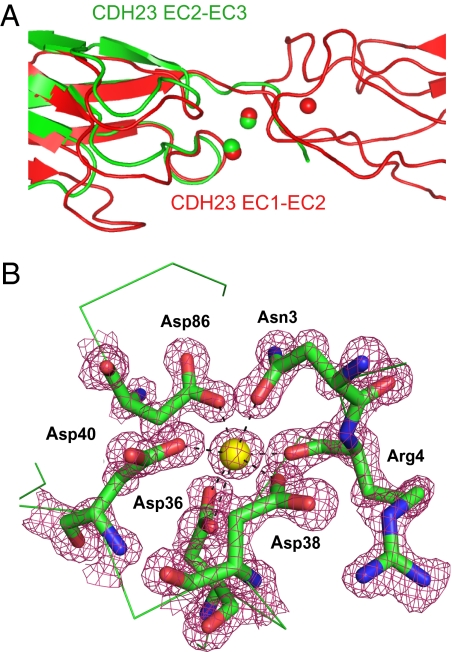
Typical of cadherins, the Ca2+ binding sites in the interdomain region between EC1 and EC2 are conserved and contain three Ca2+ ions (Ca1, Ca2, and Ca3) (Fig. 1B). Two additional Ca2+ ions are observed at the C-terminal end of EC2 (Ca4 and Ca5) (Fig. 2B). Superimposition of EC2 onto EC1 indicates that the positions of Ca4 and Ca5 correspond to those of Ca1 and Ca2, suggesting that these constitute interdomain Ca2+ binding sites between EC2 and EC3 (Fig. 2A). Importantly, an additional Ca2+ ion (Ca0) is observed on the very tip of EC1 (Figs. 1B and 2B). The structure of the CDH23 EC1–EC2 domains cocrystallized with 2 mM europium (III) nitrate had Ca0 replaced by Eu atoms (Fig. 1C), indicating that this Ca2+ ion is bound with lower affinity than the Ca2+ ions between EC domains. Ca4 was also replaced by Eu (Fig. 1C), but lower binding affinity might be a consequence of the expression of a CDH23 molecule truncated after EC2.
Fig. 2.
Calcium binding motifs in CDH23. (A) Superimposition of the EC1–EC2 junction of CDH23 (red) with the Ca2+ binding motifs at the C terminus of EC2 (green). (B) Detail of the Ca2+ binding motif Ca0 at the N terminus of EC1 shown with corresponding 2Fo-Fc electron density at 1.1 Å resolution and contoured at 1.5 σ (red).
The presence of Ca0 at the N terminus is a unique feature not reported for any other cadherin. The residues that ligand Ca2+ are Asn3, Arg4, Asp36, Asp38, Asp40, and Asp86 (Fig. 2B) and resemble Ca3 in the linker region between EC1 and EC2. The N-terminal Ca2+ ion functions to “lock down” the N terminus that in classical cadherins is free to interact with a neighboring EC1 in strand exchange, ruling out a common trans interaction mechanism for classical cadherins and CDH23.
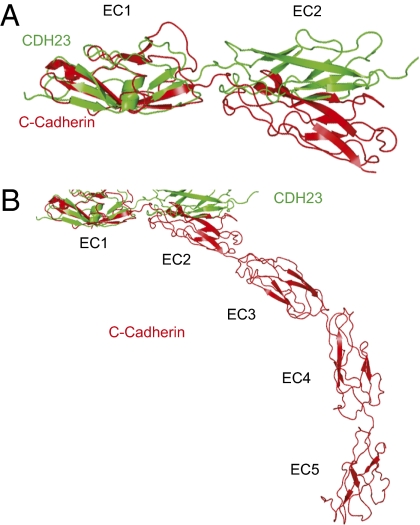
An additional unique feature of our structure compared with previously published structures of classical cadherins is that the CDH23 EC1–EC2 junction is essentially linear (Fig. 3 A and B). Classical cadherins such as C-cadherin have a significant bend angle between EC domains, which gives a pronounced twist to the molecule (Fig. 3 A and B) (5). As tip links appear in the electron microscope as linear molecules (17), the linear alignment of CDH23 EC1–EC2 might have important functional implications. However, it should be noted that in native tip links, the extracellular domains of two CDH23 domains align in cis, which could lead to structural changes that affect the bend angle between the EC1 and EC2 domains.
Fig. 3.
Axial alignment of the EC1–EC2 domain fragment. (A) Superimposition of the EC1–EC2 domain of CDH23 and C-cadherin. C-cadherin but not CDH23 has a significant bend angle between EC domains. (B) The bend angle between EC domains gives a pronounced twist to C-cadherin.
Amino Acids That Bind Calcium Are Required for trans Interactions Between CDH23 and PCDH15.
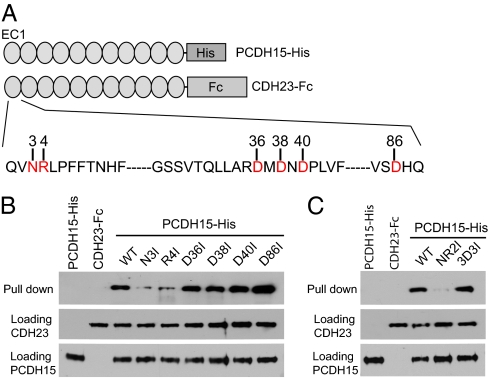
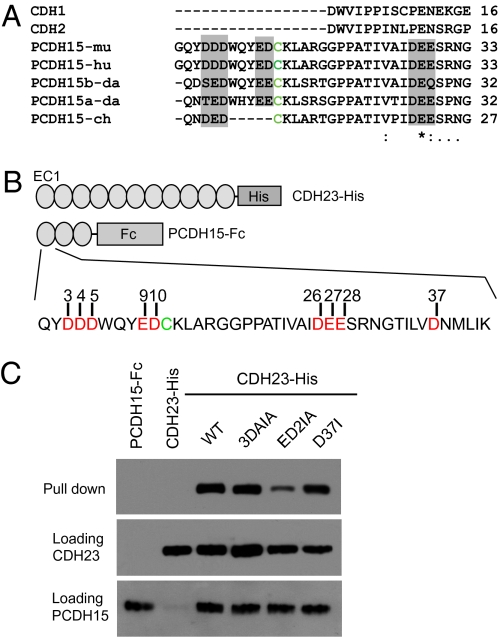
Tip links are formed by CDH23 homodimers, which interact in trans with PCDH15 homodimers to form extended filaments (15). As the EC1 domains of CDH23 and PCDH15 mediate interaction, we reasoned that amino acids binding Ca2+ at the N-terminal tip of EC1 might be important to stabilize interactions between the two cadherins. To test this model, we carried out protein interaction assays with wild-type and mutant proteins following our previously established procedures (15). We generated six recombinant CDH23 molecules carrying mutations that replaced each of the amino acids binding Ca2+ (Asn3, Arg4, Asp36, Asp38, Asp40, and Asp86) with Ile (referred to as N3I, R4I, D36I, D38I, D40I, and D86I) (Fig. 4A). We also engineered a double mutation replacing both Asn3 and Arg4 with Ile (referred to as NR2I), and a triple mutation replacing Asp36, Asp38, and Asp40 with Ile (referred to as 3D3I) (Fig. 4A). These mutations were engineered in CDH23 constructs consisting of the EC1–EC11 domain of CDH23 fused to the Fc domain of the Ig heavy chain (CDH23-Fc). For interaction studies, we used an additional construct consisting of the extracellular domain of PCDH15 fused to a His-tag (PCDH15-His) (Fig. 4A).
Fig. 4.
Mutational analysis of the EC1 domain of CDH23. (A) Diagram of the PCDH15-His and CDH23-Fc constructs used for pull-down experiments. CDH23 amino acids that were mutated are shown in red. (B) PDCH15-His was incubated with the indicated CDH23 constructs. Lanes 1 and 2 show incubation of PCDH15-His without CDH23-Fc and vice versa. The Top row shows pull-downs after detection with Fc antibodies. The Middle and Bottom lanes are loading controls for CDH23-Fc as detected with antibodies to Fc and PCDH15-His as detected with antibodies to PCDH15. (C) Pull-downs were carried out with double and triple mutants. For nomenclature see main text.
The fusion constructs were independently expressed in HEK293 cells and harvested from their supernatant. The proteins were then mixed and protein complexes were isolated using Ni-NTA beads, which bind to the His-tag but not the Fc-tag. In agreement with earlier findings (15), Western blot analysis revealed that wild-type CDH23-Fc efficiently interacted with PCDH15-His (Fig. 4B). In contrast, single mutations in Asn3 and Arg4 of CDH23 reduced interactions; a double mutation of Asn3 and Arg4 nearly abolished interactions (Fig. 4 B and C). However, single mutations in Asp36, Asp38, Asp40, and Asp86 had no effect (Fig. 3B). Finally, the triple mutation in Asp36, Asp38, and Asp40 did not affect interactions either (Fig. 3B). We therefore conclude that Asn3 and Arg4 are essential for interactions between CDH23 and PCDH15.
The N terminus of the EC1 domain of PCDH15 also contains an N-terminal extension containing several polar amino acids that are conserved across species (Fig. 5A; note that the WxYEx motif is encoded by a mini exon; the respective splice variant might have so far escaped detection in chickens). We reasoned that these amino acids might participate in Ca2+ binding and might be required for trans interactions between CDH23 and PCDH15. We therefore mutated polar amino acids in the PCDH15 EC1 domain to Ile or Ala (Fig. 5B). The mutations were engineered in a PCDH15-Fc construct consisting of the first three EC domains of PCDH15 (Fig. 5B) and expressed in HEK293 cells. Only some of the constructs were expressed successfully and we used them for pull-down experiments. In one construct, we replaced both Glu9 and Asp10 with Ile and Ala (ED2IA). One additional construct contained triple mutations replacing Asp3, Asp4, and Asp5 with Ala and Ile (3DAIA). A final construct that was successfully expressed contained a point mutation converting Asp37 to Ile (D37I). Mutation of Glu10 and Asp11 reduced interactions with CDH23, whereas the other mutations had no detectable effect.
Fig. 5.
Mutational analysis of the EC1 domain of PCDH15. (A) Sequence alignment showing the N terminus of PCDH15 in different species and compared with murine CDH1 and CDH2. Note that chicken PCDH15 lacks the WHYEE motif. However, it is expressed in mice by a mini exon, and a splice variant containing this exon might have so far escaped detection. (B) Diagram of the CDH23-His and PCDH15-Fc constructs used in pull-down experiments. The sequence of the PCDH15 N terminus is shown. Putative Ca2+ binding amino acids that were mutated are indicated in red; a conserved Cys is shown in green. (C) CDH23-His was incubated with the indicated PCDH15 constructs. Lanes 1 and 2 show incubation of PCDH15-Fc without CDH23-His and vice versa. The Top row shows pull-downs after detection with Fc antibodies. The Middle and Bottom lanes are loading controls for CDH23-His as detected with antibodies to CDH23 and PCDH15-FC as detected with antibodies to Fc. For nomenclature see main text.
Binding Interactions of PCDH15 with Cr-2 Cadherins.
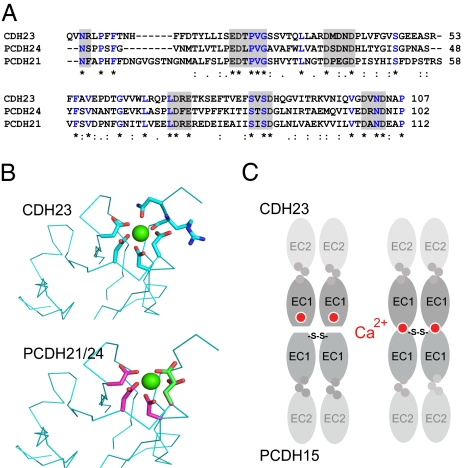
Cadherins have recently been grouped into subfamilies based on sequence homology between EC1 domains (18). In this alignment, CDH23 groups together with PCDH21 and PCDH24 in the Cr-2 branch of the superfamily. Analysis of the aligned sequences revealed that Asn3, which is required for efficient interactions between CDH23 and PCDH15, as well as other amino acids that potentially bind Ca2+, are conserved at the N terminus of the Cr-2 cadherins (Fig. 6A). Homology modeling of the sequence of the EC1–EC2 domains of PCDH21 and PCDH24 on the structure of the CDH23 EC1–EC2 domains supports high conservation in the folding of EC1–EC2. The Ca2+ binding motif at the N terminus and structurally important side chains of amino acids that point to the interior of the EC domains are conserved between CDH23, PCDH21, and PCDH24 (Fig. 6B). These findings suggest that Cr-2 cadherins might use a common binding mechanism during the establishment of adhesion complexes.
Fig. 6.
Conserved Ca2+ binding motive at the N terminus of PCDH21 and PCDH24. (A) Sequence alignment of the N terminus of the mature CDH23 protein with PCDH21 and PCDH24. Putative Ca2+ binding amino acids are highlighted with gray boxes. (B) Models generated (using Swiss PDB Viewer) by threading PCDH21 and PCDH24 sequences onto the CDH23 structure as per the alignments in A, indicate structural conservation that allows for Ca2+ binding. (C) Model for the interaction domain between CDH23 and PCDH15. The Ca2+ binding motif at the tip of EC1 in CDH23 either exposes a binding surface (Left) or Ca2+ contributes directly in the binding surface (Right).
Discussion
We have previously shown that CDH23 and PCDH15 interact to form tip links in mechanosensory hair cells of the inner ear (15). These filaments differ in several important ways from adhesion complexes formed by classical cadherins, suggesting that they might interact by a specialized mechanism. First, whereas adhesion complexes between cells consist of hundreds if not thousands of molecules, a tip link only contains one CDH23 homodimer interacting with one PCDH15 homodimer (15). Second, unlike homodimers of classical cadherins, which align in cis by contacts formed only between the EC1 and EC2 domains, two CDH23 molecules align along large parts of their extracellular domain to form closely intertwined dimers; a similar cis alignment is observed for the extracellular domains of PCDH15 molecules (15). Third, tip links likely sustain substantial mechanical force during stimulation of hair cells, suggesting that their structure and adhesive surface is designed to sustain such forces.
The findings that we present here now support the view that CDH23 and PCDH15 interact by a mechanism that differs from the mechanism by which classical cadherins form trans interactions. Key signature features of the strand-swap binding interface such as Trp2 and their binding pockets that are characteristic for classical cadherins are not conserved in the EC1 domains of CDH23 and PCDH15. In contrast, both CDH23 and PCDH15 contain N-terminal extensions that contain polar amino acids. PCDH15 also contains a cysteine close to the N terminus. Our structural studies on CDH23 show that the polar residues near the N terminus of the EC domain bind a Ca2+ ion. When the most N-terminal polar amino acids of CDH23 and PCDH15 are substituted with a hydrophobic amino acid (Ile), interactions between CDH23 and PCDH15 are affected. The effect of the mutations can be explained in different ways (Fig. 6C). As one possibility, binding of Ca2+ might be required to clamp down the N terminus of the EC1 domains and expose a specific binding surface. In support of this model, mutations in Asn3 or Arg4, which are positioned on the same surface of the N-terminal EC1 domain, affect binding to PCDH15. Asn3 and Arg4 might therefore form an intimate part of the interaction surface between CDH23 and PCDH15 (Fig. 6A).
However, an exciting alternative mechanism would be that during the formation of a stable complex between CDH23 and PCDH15, a rearrangement occurs in the N terminus of both CDH23 and PCDH15, leading to exposure of the Ca2+ ion and its binding by polar amino acids in CDH23 and PCDH15 across the adhesion surface. The Cys near the N terminus of PCDH15 (Fig. 5A) might help to align the two EC1 domains of PCDH15 by disulfide bonding, leading to the proper positioning of polar amino acids (Fig. 6C). This model would in some regards resemble the strand-swapping model for classical cadherins, where Trp residues become exposed during the formation of adhesion complexes to stabilize trans interactions (19). It should also be noted that we have previously shown that PCDH15 and CDH23 bind to each other even in the presence of as little as 0.1 mM Ca2+, the lowest Ca2+ concentration tested (15). The binding experiments between mutant version of CDH23 and PCDH15 that we have described here were carried out in 1 mM Ca2+. It can at present not be excluded that some of the mutations that did still mediate interactions in 1 mM Ca2+, might lead to adhesive defects when tested at lower Ca2+ concentration. These experiments, as well as further crystallographic experiments and NMR studies will be essential to ultimately define the precise binding mechanism that mediates interactions between CDH23 and PCDH15.
The presence of a Ca2+ binding motif at the N terminus of CDH23 could play important roles in the dynamic control of the adhesive properties of tip-link cadherins. Unlike Ca2+ ions at the junction between EC1 and EC2, the N-terminal Ca2+ ion is readily replaced with Eu, suggesting that it is bound with lower affinity. The Ca2+ concentration could therefore significantly affect the formation and stability of adhesion complexes, especially in the endolymph that surrounds tip links, which has a comparatively low Ca2+ concentration (20, 21). The N-terminal Ca2+ binding motif might under certain conditions only be partially occupied. The presence of the Ca2+ motif at the N terminus might also explain why tip links are lost upon treatment with BAPTA (22). However, BAPTA treatment is expected to affect Ca2+ molecules at the interface between EC domains throughout the cadherin extracellular domains as well.
Previous sequence alignments that were based on the EC1 domains of cadherins have placed CDH23, PCDH21, and PCDH24 into the Cr-2 branch of the cadherin superfamily (18). Relative to the EC1 domain of classical cadherins, all Cr-2 cadherins contain an extension at the N terminus of EC1 that contains polar amino acids. Our sequence alignments and molecular modeling studies suggest that the EC1–EC2 domains of CDH23, PCDH21, and PDCH24 fold in similar ways with a strategically positioned Ca2+ binding site at the tip of EC1, reinforcing the concept that CDH23, PCDH21, and PCDH24 form a structural subclass within the cadherins superfamily. PCDH15 belongs to the Cr-3 subfamily, which also contains an N-terminal extension (18). It will be important to determine whether other Cr-3 cadherins also bind Cr-2 cadherins. Interestingly, mutations in CDH23, PCDH15, and PCDH21 have been linked to retinal disease in humans (23–28). An intriguing possibility is that there might be a functional link between these cadherins, possibly through the formation of heteromeric adhesion complexes between Cr-2 and Cr-3 family members. Mutations in CDH23 and PCDH15, but not in PCDH21 and PCDH24, have also been linked to auditory impairment (23, 26, 27, 29), suggesting that some of these heterophilic adhesion complexes might have tissue-specific functions.
Materials and Methods
Protein Preparation.
A DNA fragment containing the N-terminal amino acid residues 24–236 (EC1–EC2) of cadherin 23 was obtained by PCR using the forward primer 5′-GGAATTCCATATGCAGGTGAACCGACTACCTTTCTTC-3′ and the reverse primer 5′-GGTGGTTGCTCTTCCGCAGAAGATAGGATCCATGTCTTGCATGTCC-3′. The fragment was cloned into the pTXB1 vector (New England Biolabs) using the restriction sites NdeI and SapI. The pTXB1 attached the Mxe GyrA intein tag at the C terminus and of EC2. The tag contains an N-terminal cysteine residue that allows for thiol-induced cleavage. The construct was expressed in T7 Express Competent Cells (New England Biolabs). Pellets were lysed in 20 mM Tris-HCl, pH 8.5, 200 mM NaCl, 1 mM CaCl2, 0.1% Triton X-100, 100 μM PMSF, and 1/50 mL tablet of Complete EDTA-free protease inhibitor tablets (Roche). Lysates were sonicated and purified with chitin beads (New England Biolabs) and the intein tag was cleaved overnight in 20 mM Tris-HCl, pH 8.5, 200 mM NaCl, 1 mM CaCl2, and 50 mM DTT. CDH23 EC1–EC2 was purified and concentrated in a desalting buffer of 1 mM CaCl2 and 50 mM Tris-HCl, pH 8.5 in ddH2O to a concentration of 6.9 mg/mL.
Crystallization and Data Collection.
CDH23 EC1–EC2 was crystallized using the nanodrop vapor diffusion method (30). Drops (200 nL of 6.9 mg/mL protein in 1 mM CaCl2, 50 mM Tris-HCl, pH 8.5 plus 200 nL crystallant) were dispensed into 96-well Innovaplate SD-2 crystallization plates using an Innovadyne liquid-handling robot. Within 10 days at 20 °C, crystals (100 μm by 30 μm by 10 μm) were obtained in 22.5% ethylene glycol and 0.2 M NDSB-201. A native 1.1-Å data set (at a wavelength of 0.95369 Å) was collected on beamline 9–2 at the Stanford Synchrotron Radiation Laboratory. Native protein was cocrystallized with europium (III) nitrate hexahydrate (2 mM final concentration) in 10 mM betaine HCl and 10 mM β-nicotinamide adenine dinucleotide hydrate. Crystals were flash frozen in liquid nitrogen without additional cryoprotection. Single-wavelength anomalous diffraction (SAD) data were collected to 2.5 Å on a Eu-derivatized crystal at the same beam line at a wavelength of 1.7759 Å (peak wavelength of the Eu L-3 absorption edge). The BLU-ICE (31) software interface was use for data collection (remote access), and diffraction data were processed with HKL2000 (32). The space group of the crystals was P21. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 3MVS).
Structure Determination and Refinement.
Initial phases were obtained by SAD phasing with the 2.5-Å europium derivative data set at the Eu peak wavelength using the program SOLVE (33). Two Eu sites were found with 44% occupancy. The resulting phases had a figure of merit of 0.76 after density modification using RESOLVE. The SAD phases were merged, improved, and extended using the 1.1-Å native data set using the programs CAD and DM within the CCP4 package (34), assuming 1 monomer in the asymmetric unit (Matthews coefficient 2.35, solvent content 47.7%). Automated model building using Arp/wARP in CCP4 (34) traced most of the structure. The rest was manually built into the density using Coot (35) and refined against the high-resolution native data with iterative rounds of model building and refinement using Refmac5 within CCP4 (34). A summary of data collection and refinement statistics is shown in Table S1. The stereochemical quality of the final refined model was checked using the structure validation suite Quality Control Check v2.6 (http://smb.slac.stanford.edu/jcsg/QC/), and ribbon diagrams were made using Pymol (10). The coordinates and the structure factors have been deposited in the PDB.
Binding Assays.
DNA constructs were generated by PCR and expressed in HEK293 cells as described (15). For binding assays, His-tagged proteins were incubated with Fc-tagged proteins, complexes were purified with Ni-NTA beads, and analyzed by Western blotting as described (15).
Supplementary Material
Acknowledgments
We thank members of the U.M. and P. Kuhn laboratories for comments and criticisms. This work was supported by National Institutes of Health Grants DC005965 and DC007704 (to U.M.) and 5U54GM074961 (to P.K.), the Skaggs Institute for Chemical Biology (to U.M.), and the Dorris Neuroscience Center (to U.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 3MVS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006284107/-/DCSupplemental.
References
- 1.Muller U. Cadherins and mechanotransduction by hair cells. Curr Opin Cell Biol. 2008;20:557–566. doi: 10.1016/j.ceb.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson WJ. Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem Soc Trans. 2008;36:149–155. doi: 10.1042/BST0360149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro L, Love J, Colman DR. Adhesion molecules in the nervous system: Structural insights into function and diversity. Annu Rev Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki SC, Takeichi M. Cadherins in neuronal morphogenesis and function. Dev Growth Differ. 2008;50(Suppl 1):S119–S130. doi: 10.1111/j.1440-169X.2008.01002.x. [DOI] [PubMed] [Google Scholar]
- 5.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 6.Ciatto C, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol. 2010;17:339–347. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison OJ, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17:348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Häussinger D, et al. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 10.Patel SD, et al. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 11.Pertz O, et al. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro L, et al. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 14.Pokutta S, Herrenknecht K, Kemler R, Engel J. Conformational changes of the recombinant extracellular domain of E-cadherin upon calcium binding. Eur J Biochem. 1994;223(3):1019–1026. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 15.Kazmierczak P, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 16.Sakaguchi H, Tokita J, Müller U, Kachar B. Tip links in hair cells: Molecular composition and role in hearing loss. Curr Opin Otolaryngol Head Neck Surg. 2009;17:388–393. doi: 10.1097/MOO.0b013e3283303472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kachar B, Parakkal M, Kurc M, Zhao Y, Gillespie PG. High-resolution structure of hair-cell tip links. Proc Natl Acad Sci USA. 2000;97:13336–13341. doi: 10.1073/pnas.97.24.13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci USA. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosher SK, Warren RL. Very low calcium content of cochlear endolymph, an extracellular fluid. Nature. 1978;273:377–378. doi: 10.1038/273377a0. [DOI] [PubMed] [Google Scholar]
- 21.Salt AN, Inamura N, Thalmann R, Vora A. Calcium gradients in inner ear endolymph. Am J Otolaryngol. 1989;10:371–375. doi: 10.1016/0196-0709(89)90030-6. [DOI] [PubMed] [Google Scholar]
- 22.Assad JA, Shepherd GM, Corey DP. Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron. 1991;7:985–994. doi: 10.1016/0896-6273(91)90343-x. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed ZM, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alagramam KN, et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 25.Bolz H, Ebermann I, Gal A. Protocadherin-21 (PCDH21), a candidate gene for human retinal dystrophies. Mol Vis. 2005;11:929–933. [PubMed] [Google Scholar]
- 26.Bolz H, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 27.Bork JM, et al. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson RH, et al. Biallelic mutation of protocadherin-21 (PCDH21) causes retinal degeneration in humans. Mol Vis. 2010;16:46–52. [PMC free article] [PubMed] [Google Scholar]
- 29.Alagramam KN, et al. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 30.Santarsiero BD, et al. An approach to rapid protein crystallization using nanodroplets. J Appl Crystallogr. 2002;35:278–281. [Google Scholar]
- 31.McPhillips TM, et al. Blu-Ice and the Distributed Control System: Software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger T. SOLVE and RESOLVE: Automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.