Abstract
Pseudomonas aeruginosa quorum control of gene expression involves three LuxR-type signal receptors LasR, RhlR, and QscR that respond to the LasI- and RhlI-generated acyl-homoserine lactone (acyl-HSL) signals 3OC12-HSL and C4-HSL. We found that a LasR-RhlR-QscR triple mutant responds to acyl-HSLs by regulating at least 37 genes. LuxR homolog-independent activation of the representative genes antA and catB also occurs in the wild type. Expression of antA was influenced the most by C10-HSL and to a lesser extent by other acyl-HSLs, including the P. aeruginosa 3OC12-HSL and C4-HSL signals. The ant and cat operons encode enzymes for the degradation of anthranilate to tricarboxylic acid cycle intermediates. Our results indicate that LuxR homolog-independent acyl-HSL control of the ant and cat operons occurs via regulation of antR, which codes for the transcriptional activator of the ant operon. Although P. aeruginosa has multiple pathways for anthranilate synthesis, one pathway—the kynurenine pathway for tryptophan degradation—is required for acyl-HSL activation of the ant operon. The kynurenine pathway is also the critical source of anthranilate for energy metabolism via the antABC gene products, as well as the source of anthranilate for synthesis of the P. aeruginosa quinolone signal. Our discovery of LuxR homolog-independent responses to acyl-HSLs provides insight into acyl-HSL signaling.
Keywords: anthranilate, gene regulation, quorum sensing
The metabolically versatile bacterium Pseudomonas aeruginosa is ubiquitous and can adapt to diverse habitats. For example, it can be found in soil and water or as an opportunistic pathogen in a wide range of hosts, including plants and humans. The expression of a large number of P. aeruginosa genes, including virulence genes, is controlled by a signaling mechanism called quorum sensing. Quorum sensing involves production and sensing of chemical signals that allow bacteria to regulate gene expression in response to alterations in cell density (1–4). The archetypical LuxI-R quorum-sensing system that controls luminescence in Vibrio fischeri uses the enzyme LuxI to generate a diffusible acyl-homoserine lactone (acyl-HSL) signal, 3-oxo-hexanoyl-HSL, which binds to the acyl-HSL receptor and transcription factor LuxR (5, 6). Similar quorum-sensing systems have been found in about 100 species of Proteobacteria, where they function as regulators of a range of functions. Acyl-HSL quorum sensing commonly controls extracellular products, such as exoproteases, exopolysaccharides, antibiotics, and aggregation factors (7–10).
P. aeruginosa has two acyl-HSL synthases and three receptors (11–14). The LasI synthase produces 3-oxo-C12-HSL, for which there are two receptors, LasR and QscR. The RhlI synthase produces C4-HSL, for which the receptor is RhlR. Together these quorum-sensing systems regulate hundreds of P. aeruginosa genes. Different elements of the P. aeruginosa quorum sensing circuit also influence each other at multiple levels; for example, LasR-3OC12-HSL activates rhlR and rhlI transcription, and QscR influences expression of a subset of las- and rhl- controlled genes (15, 16). In fact, the regulons of LasR, RhlR, and QscR are partially overlapping. From previous work, it is evident that many factors, both known and unknown, influence the control of gene expression by acyl-HSLs in P. aeruginosa (17, 18).
Integrated into the acyl-HSL quorum-sensing circuits is a third signal, 2-heptyl-3-hydroxy-4-quinolone, known as the Pseudomonas quinolone signal (PQS) (19). Transcriptome analyses have shown that quinolone signaling directly or indirectly controls the expression of at least 90 genes (20, 21). The acyl-HSL and PQS signaling systems influence each other; the las system activates synthesis of PQS, which in turn activates rhlI expression (22, 23). In addition, LasR, RhlR, and QscR influence expression of genes that can potentially alter intracellular levels of the PQS biosynthesis precursor anthranilate (18).
Here we report acyl-HSL regulation of gene expression in P. aeruginosa that does not follow the classical quorum-sensing tenet, in that it is not mediated by a LuxR-type transcription factor. The LuxR homolog-independent acyl-HSL regulon includes the antABC and catBCA genes, which encode enzymes for the degradation of anthranilate. We show that acyl-HSL regulation of the ant and cat operons correlates with altered expression of the transcriptional regulator AntR. In P. aeruginosa, anthranilate is a pivotal branch-point metabolite; it can be directed either via the TCA cycle for energy metabolism or into the PQS synthesis pathway (18). The flux of anthranilate into the diverging pathways is tightly controlled by transcriptional and posttranscriptional regulatory mechanisms that are dictated variously by iron and anthranilate availability, as well as quorum sensing (18, 24). Our study reveals another layer in the extensive control that P. aeruginosa places on the metabolism of anthranilate.
Results
Signal Receptor-Independent Response to Acyl-HSLs.
As a control for experiments designed to determine the transcriptome response to QscR and C10-HSL, we performed microarray analyses on a P. aeruginosa PAO LasR-RhlR-QscR triple mutant (DA15) with or without added C10-HSL. To our surprise, we identified 37 genes that showed a significant C10-HSL response (Table S1). Of these, 13 genes were induced and 24 genes were repressed. Many, but not all, of the genes that we identified are not predicted to be parts of operons. Examination of cases in which only part of an operon showed a C10-HSL response invariably revealed that the remaining genes also showed an appropriate response, but not of a magnitude to provide statistical significance. We were particularly interested in cases where all genes in an operon showed similar statistically significant C10-HSL responses. Most likely the responses of such genes are biologically important; thus, for our subsequent experiments, we concentrated on the ant and cat operons. All of the genes in these operons were activated by C10-HSL, and the responses were somewhat greater than the responses of other genes (Table S1).
The P. aeruginosa PAO1 genome contains an ORF (PA1136) coding for a protein with sequence similarity to LasR, RhlR, QscR, and other LuxR homologs (25). There is a notable similarity in the acyl-HSL–binding regions (Fig. S1), and PA1136 shows the same extent of identity to each of the other LuxR homologs shown in Fig. S1 as they do to one another other; however, PA1136 has only five of the seven residues conserved among all characterized LuxR homologs (3). Nevertheless, PA1136 might code for a C10-HSL receptor. Therefore, we constructed a signal receptor null strain lacking lasR, rhlR, qscR, and PA1136 (SC20) and measured transcript levels of the first genes in the cat and ant operons, catB and antA, in the null mutant. The null mutant and the LasR-RhlR-QscR triple mutant showed the same response to C10-HSL (Table 1). Thus, PA1136 does not encode a LasR homolog responsible for mediating the C10-HSL–dependent expression of antA and catB.
Table 1.
Transcript abundance of selected C10-HSL–regulated genes
| Transcript level* |
|||
| Strain | catB | antA | PA1897 |
| DA15 (ΔlasR, ΔrhlR, ΔqscR) | 0.02 | 0.74 | 0.06 |
| DA15 (ΔlasR, ΔrhlR, ΔqscR) plus 2 μM C10-HSL | 0.92 (55) | 3.2 (4.3) | 0.06 (0.9) |
| SC20 (ΔlasR, ΔrhlR, ΔqscR, ΔPA1136) | 0.01 | 0.43 | 0.06 |
| SC20 (ΔlasR, ΔrhlR, ΔqscR, ΔPA1136) plus 2 μM C10-HSL | 1.6 (93) | 4.8 (6.5) | 0.06 (1) |
| PAO1 (WT) | 0.08 | 1.5 | 1.2 |
| PAO1 (WT) plus 2 μM C10-HSL | 1.7 (20) | 7.2 (5) | 2.7 (2.4) |
*Numbers in parentheses indicate fold changes in transcript abundance on growth in the presence of C10-HSL. Transcript abundance was measured by real-time RT-PCR.
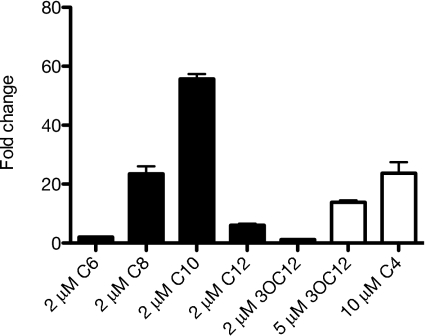
Do other acyl-HSLs substitute for C10-HSL as LuxR homolog-independent activators of gene expression? To answer this question, we measured transcript levels of catB in the LasR-RhlR-QscR triple mutant grown in the presence of a number of different acyl-HSLs (Fig. 1). Various acyl-HSLs activated catB, and the response was most sensitive to C10-HSL. Although not as sensitive to the P. aeruginosa LasR and RhlR signals 3OC12-HSL and C4-HSL, a response to physiological levels of C4-HSL (∼10 μM) and a small response to physiological levels of 3OC12-HSL (∼5 μM) were noted (26).
Fig. 1.
The influence of various acyl-HSLs on catB expression in the LasR-RhlR-QscR triple-mutant DA15. Black bars indicate signals tested at 2 μM; white bars, signals tested at higher concentrations. Transcript abundance was measured by real-time RT-PCR.
Gene Regulation by C10-HSL Occurs in WT P. aeruginosa.
In P. aeruginosa, the acyl-HSL quorum-sensing circuits are interconnected with LasR-, RhlR-, and QscR-controlling overlapping regulons. C10-HSL as well as 3OC12-HSL, can serve as a signal for gene activation by QscR. To test whether LuxR homolog-independent C10-HSL gene regulation is relevant in the context of an intact quorum-sensing circuit, we measured the abundance of the catB and antA transcripts in WT P. aeruginosa PAO1 (Table 1). Real-time RT-PCR analysis showed that transcript levels of both catB and antA were stimulated when the WT was grown with added C10-HSL. Controls demonstrated that the QscR-dependent PA1897 did not respond to C10-HSL in the LasR-RhlR-QscR triple mutant. As expected, given its functional QscR and production of 3OC12-HSL, the WT had a much higher PA1897 transcript level than the triple mutant.
C10-HSL–Dependent Induction of catB Relies on Anthranilate Provided via the Kynurenine Pathway for Tryptophan Degradation.
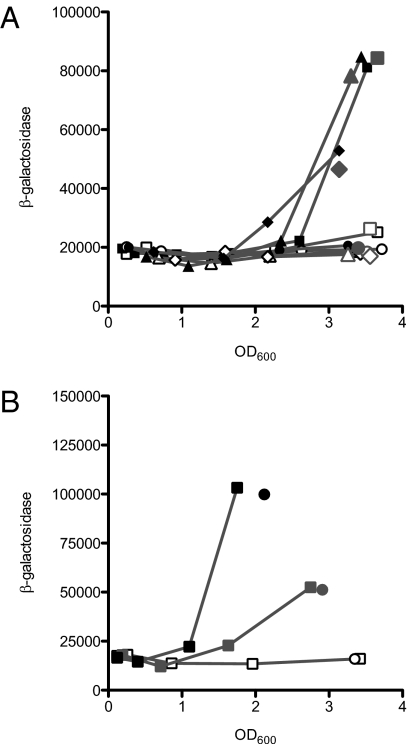
Although a number of factors affect antABC expression, the regulation of this operon cannot be fully explained based solely on the influence of these factors, suggesting the presence of additional factors that contribute to this regulation (18, 24). Previous work has shown that the transcriptional activator AntR activates the expression of antABC in response to the inducer anthranilate (18). P. aeruginosa can acquire anthranilate from the environment or synthesize it via one of three pathways: the kynurenine pathway, the tryptophan synthesis pathway, or the PQS pathway (Fig. S2) (27–30). We hypothesized that acyl-HSL activation of the ant and cat operons proceeds through AntR, which requires anthranilate as a coactivator. To begin to test this hypothesis, we generated strains with mutations in each of the three anthranilate synthesis pathways in P. aeruginosa DA15, into which we engineered a chromosomally integrated catB-lacZ transcriptional fusion. Expression of catB–lacZ fusion required a functional kynA, but a pqsR mutation and a trpE mutation did not eliminate C10-HSL induction of catB–lacZ (Fig. 2A). C10-HSL induction in the kynA mutant was recovered by exogenous addition of anthranilate (Fig. 2B). These data support the view that anthranilate and AntR mediate C10-HSL induction of cat and ant genes. Under our experimental conditions, the source of the anthranilate is the kynurenine pathway. A previous study showed that the kynurenine pathway is the primary source of anthranilate for PQS biosynthesis in rich media (27) similar to the medium that we used in the present study.
Fig. 2.
Transcriptional activation of a catB-lacZ chromosomal fusion. β-galactosidase activity is given as relative light units normalized to OD600. For one of the replicate experiments, only the final values are given for clarity. (A) β-galactosidase activity with (closed symbols) or without (open symbols) 2 μM C10-HSL in the following strains: LasR-RhlR-QscR triple-mutant SC24 (squares), SC24ΔpqsR (triangles), SC24ΔtrpE (diamonds), and SC24ΔkynA (circles). The gray symbols in large type represent final values from an independent replicate. (B) β-galactosidase activity of the SC24ΔkynA strain (open symbols), with 0.2 mM anthranilate (gray symbols), and with 0.2 mM anthranilate and 2 μM C10-HSL (black symbols). Circles indicate the final values of an independent replicate.
C10-HSL Advances antR Expression.
Our findings with anthranilate synthase mutants demonstrate that anthranilate is required for the C10-HSL induction of catB and presumably other genes affected by C10-HSL in the LasR-RhlR-QscR triple-mutant background. This effect could result from the concerted action of anthranilate and AntR, the anthranilate-dependent activator of antABC. In support of this idea, we found that transcription of catB in an AntR mutant is low and not influenced by C10-HSL.
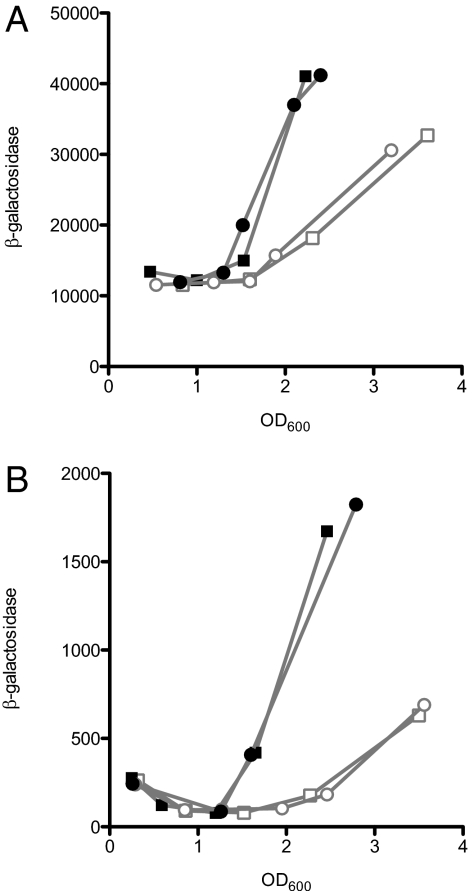
If C10-HSL affects ant and cat gene expression through AntR, then it also might be expected to influence antR expression. To explore this possibility, we measured β-galactosidase activity in the lasR-rhlR-qscR mutant strain DA15 with a WT antR plus either an engineered chromosomal antR-lacZ transcriptional or translational fusion. When grown in the presence of added C10-HSL, both fusion strains showed earlier induction of the reporter (Fig. 3). This pattern of expression also was observed when the fusions were in the WT. Control strains with a chromosomally integrated promoterless lacZ gene exhibited low basal β-galactosidase activity that was not influenced by C10-HSL.
Fig. 3.
Expression of chromosomal antR-lacZ transcriptional (A) and translational fusion (B) in the LasR-RhlR-QscR triple-mutant strain DA15. The results of two independent experiments are shown. Open symbols indicate activity in the absence of added acyl-HSL, and closed symbols indicate activity in the presence of 2 μM C10-HSL. β-galactosidase activity is given as relative light units normalized to OD600.
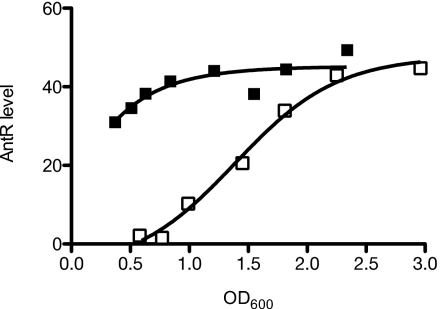
To track expression of AntR, we constructed strain SC30 by generating a chromosomal fusion of a sequence encoding the VSV-G epitope to antR in strain DA15. We found that adding the tag did not interfere with the function of AntR by monitoring expression of the chromosomal catB-lacZ reporter in strain SC31, a strain containing the epitope-tagged AntR and a catB-lacZ reporter in the chromosomal attB site. Consistent with results of the antR fusion studies, immunoblot analysis of strain SC30 showed that AntR accumulated at higher levels earlier during growth with C10-HSL (Fig. 4). Thus, our data are consistent with the idea that AntR mediates C10-HSL activation of the ant and cat operons.
Fig. 4.
A representative experiment showing AntR protein levels in the LasR-RhlR-QscR triple-mutant strain DA15 expressing AntR with an amino terminal VSV-G tag. Cells were grown with (closed squares) or without (open squares) 2 μM C10-HSL. Levels were monitored by Western blotting with VSV-G antiserum, and the intensity of the bands was quantitated as described in Materials and Methods. AntR levels are the band intensities in the Western blot.
Discussion
Acyl-HSLs have received considerable attention as signaling molecules that control community behavior in many Proteobacteria (2, 31, 32). Typically, acyl-HSLs regulate gene expression by binding to LuxR-type receptors and promoting their transcriptional regulatory activity. Here we have described acyl-HSL control of gene expression in P. aeruginosa that does not use a LuxR-type signal receptor and thus operates beyond the classical LuxI-R quorum regulation. To the best of our knowledge, this aspect of acyl-HSL control of gene expression has not been described for other bacteria. We have identified 37 genes that respond to C10-HSL in a P. aeruginosa signal receptor mutant strain, and provided evidence that this C10-HSL response is relevant in the WT strain as well. Although C10-HSL was the most effective of the signals that we tested, all signals were active to varying extents.
Our findings point to the need to reconsider the P. aeruginosa quorum-sensing paradigm on multiple levels. We have presented evidence suggesting the presence of at least one mechanism other than that involving a LuxR homolog through which both P. aeruginosa quorum-sensing signals 3OC12-HSL and C4-HSL can regulate gene expression. Our evidence indicates that this type of gene regulation also can be mediated by acyl-HSLs produced by other bacteria, thus allowing P. aeruginosa to potentially respond to certain cohabiting microbes. We do not yet know whether this type of activity might extend beyond P. aeruginosa.
We focused most of our attention on two particular operons induced by C10-HSL in the LasR-RhlR-QscR mutant, the antABC and catBCA operons. It is probably not a coincidence that the ant operon has been previously reported to show a curious pattern of quorum control (18, 33). There is considerable evidence linking its regulation to each of the key elements of the quorum-sensing circuit; however, a coherent picture of how the different connections are integrated is not clear from the available data. Before the present study, its acyl-HSL regulation had been examined only in the context of LasR, RhlR, and QscR dependence. Although we describe an additional layer of control, our understanding of this layer is limited at this time.
Classical quorum-sensing regulation of gene expression is a dynamic process. Different genes are activated at different times in the mid-logarithmic to late stationary phase of growth (33). This sort of continuum of responses could apply to LuxR-independent acyl-HSL regulation as well, with different genes responding at different times during growth. Our transcriptome analyses were performed at OD600 2; thus, it is possible that our findings provide only a snapshot of the C10-HSL response, and the genes identified in this study are a subset of the genes regulated by C10-HSL in the LasR, RhlR, QscR mutant. A case in point is antR, which codes for the transcriptional activator of the ant operon. Unlike the ant operon, antR was not identified in the initial set of C10-HSL–regulated genes; however, we provide evidence that antR expression is activated by C10-HSL. Our experiments indicate that the antR response is proximal to the C10-HSL induction of the ant and cat operons. How C10-HSL affects activation of antR remains unclear, however.
A key question is whether LuxR homolog-independent acyl-HSL control of P. aeruginosa metabolism is relevant in human infections. P. aeruginosa metabolism in infected patients is certain to be complex, and the effects of nutritional substrates on bacterial physiology in vivo are poorly understood. Thus, any assessment of the relevance of in vitro findings on bacterial physiology during an infection should be made with caution. Nonetheless, there is considerable evidence in the literature supporting our view that acyl-HSL control of P. aeruginosa metabolism is relevant in conditions extant in the pathogenic environment of the cystic fibrosis (CF) lung. Previous studies documenting the presence of PQS in CF sputum imply the availability of anthranilate, a PQS precursor (34, 35). Together with previously reported findings, our results demonstrate that the PQS biosynthesis pathway and the anthranilate degradation pathway for energy metabolism draw from the same pool of intracellular anthranilate (27). This pool derives from the degradation of the aromatic amino acid tryptophan via the kynurenine pathway. The high levels of aromatic amino acids in CF sputum and the availability of anthranilate as a growth substrate, coupled with the fact that P. aeruginosa accumulates at high densities in CF airways, makes acyl-HSL control of P. aeruginosa metabolism during chronic CF lung infections a distinct possibility (36–40). It is interesting to note that a number of the repressed genes in the C10-HSL regulon have known or putative tricarboxylic acid (TCA) transport functions (25). The regulation of these genes could be a consequence of activation of the anthranilate degradation pathway, which eventually feeds into the TCA cycle. The pattern of C10-HSL–regulated gene expression can be interpreted as reflecting a concerted shift in carbon source preference. If this is so, then our finding that acyl-HSLs can induce or facilitate this shift is of some physiological significance. The impact of nutritional cues on diverse phenotypes, such as biofilm development, surface motility, and cell-cell signaling, has been well documented, and metabolic pathways have been discussed as possible targets for drug development (40–42).
We do not yet know the precise mechanism of signal receptor-independent control of gene regulation by acyl-HSLs. A better understanding of LuxR homolog-independent responses to acyl-HSLs could provide some insight. We do know that AntR is required for the response. The function of AntR is either parallel to or downstream of the acyl-HSL control of antABC and catBCA; regardless, it mediates the response. The fact that C10-HSL triggers increased AntR expression early in growth suggests that AntR mediates the C10-HSL response. But how does C10-HSL influence AntR expression? Several possibilities can be envisioned. There may be a non-LuxR type of transcription factor that interacts with acyl-HSLs. Perhaps there is an acyl-HSL–responsive small regulatory RNA that affects AntR expression either directly or indirectly. We hope that a more detailed understanding of the scope and mechanism of this type of acyl-HSL control might provide important insight into the evolution of acyl-HSL quorum sensing. It is clear that LuxI family-dependent signal generation and LuxR family signal receptor systems have coevolved. But which appeared first? Did signal synthesis evolve first, with signals functioning in some as yet unknown way? Was this followed by coevolution of receptors, or was it the other way around?
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
The bacterial strains and plasmids used are listed in Table 2. For plasmid and strain constructions, bacteria were grown in LB broth, supplemented with antibiotics when appropriate to maintain plasmids and select for recombinants. For transcript profiling, real-time RT-PCR, β-galactosidase assays, and Western blot analyses, bacteria were grown in LB broth buffered with 50 mM 3-(N-morpholino) propanesulfonic acid (pH 7.0). The optical densities (OD600) at inoculation were 0.01 for strain DA15 and its derivatives and 0.005 for all other strains.
Table 2.
Bacterial strains used
| Strain or plasmid | Relevant properties | Source or reference |
| E.coli | ||
| DH5α | F−, φ80dlacZΔM15Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−, mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| S17-1 | recA pro hsdR RP4-2-Tc::Mu-km::Tn7 | (46) |
| P. aeruginosa | ||
| PAO1 | WT | (42) |
| DA15 | PAO1 derivative, ΔlasR, ΔrhlR, ΔqscR | (47) |
| SC20 | DA15 ΔPA1136 | This study |
| SC21 | SC24 ΔpqsR | This study |
| SC22 | SC24 ΔtrpE | This study |
| SC23 | SC24 ΔkynA | This study |
| SC24 | DA15 with a catB-lacZ transcriptional fusion at the attB site | This study |
| SC26 | DA15 with a antR-lacZ transcriptional fusion at the attB site | This study |
| SC27 | DA15 with a antR-lacZ translational fusion at the attB site | This study |
| SC28 | PAO1 with a antR-lacZ transcriptional fusion at the attB site | This study |
| SC29 | PAO1 with a antR-lacZ translational fusion at the attB site | This study |
| SC30 | DA15 with a VSV-G epitope sequence tagged to the N terminus of AntR | This study |
| SC31 | SC24 with a VSV-G epitope sequence tagged to the N terminus of AntR | This study |
| Plasmids | ||
| pEXG2 | Gene-replacement vector; sacB Gmr | (43) |
| mini-CTX-lacZ | Integrative reporter vector; Tcr | (44) |
| mini-CTX-lacZ-EB | Integrative reporter vector; Tcr | Brutinel and Yahr, University of Iowa |
| pFLP2 | Source of Flp recombinase; Apr/Cbr | (45) |
km, kanamycin resistance; Apr, ampicillin resistance; Cbr, carbenicillin resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance.
To construct chromosomal deletions, alleles were generated by PCR amplification of DNA flanking the gene of interest and cloned into pEXG2 (43). The resulting plasmids were used to transform Escherichia coli S17-1, and then mobilized into P. aeruginosa. Transconjugants were selected on Pseudomonas isolation agar (PIA) containing gentamicin (100 μg/mL), followed by recovery of deletion mutants on PIA plates containing 5% sucrose. Candidate deletion mutants were screened by PCR; real-time RT-PCR was used to demonstrate a lack of detectable mRNA for the deleted gene.
The catB-lacZ transcriptional fusion was constructed by directional cloning of a 281-bp PCR product beginning 190 bp upstream of the catB translational start site into the mini-CTX-lacZ plasmid (44). A 278-bp PCR product beginning 252 bp upstream of the antR translational start site was directionally cloned into mini-CTX-lacZ and mini-CTX-lacZ-EB to generate antR-lacZ transcriptional and translational fusions, respectively. The resulting plasmids were verified by DNA sequencing and mobilized into P. aeruginosa to allow chromosomal integration. Transconjugants were selected on PIA-containing tetracycline (100 μg/mL). The plasmid backbone was excised using the pFLP2-encoded Flp recombinase, and the pFLP2 plasmid was cured via sacB counterselection (45). PCR analysis was performed to verify that the insertions occurred at the chromosomal attB site. Growth of the two fusion strains was slightly affected by C10-HSL (Fig. S3). To construct a N-terminal vesicular stomatitis virus glycoprotein (VSV-G) epitope–AntR fusion, a PCR-generated fragment with the sequence 5′-TATACAGATATTGAAATGAATAGATTAGGAAAA-3′ inserted in-frame after the initiation codon of antR was cloned into pEXG2. The resulting plasmid was verified by sequencing and mobilized into strains DA15 and SC24 to generate strains SC30 and SC31, respectively. Allelic replacement was confirmed by PCR and Western blot analysis.
Microarray Experiments.
Cells were harvested at OD600 2. RNA isolation and cDNA synthesis, labeling, and P. aeruginosa GeneChip array (Affymetrix) processing were performed as described previously (33). Where appropriate, synthetic C10-HSL was added to the medium at a final concentration of 2 μM before inoculation. Each experiment was done in duplicate. Affymetrix GeneChip Operating Software was used for initial data analysis. Transcripts assigned an “absent” call in both conditions (with or without C10-HSL) were excluded from further analyses. Cyber-T (http://cybert.microarray.ics.uci.edu) was used for statistical analyses. The P value threshold was set at 0.007, and the fold change cutoff was set at >2.5.
Real-Time RT-PCR.
The procedures for RNA isolation and cDNA synthesis were similar to those used for the microarray experiments. Comparative RT-PCR was performed on an ABI Prism 7900 system using SYBR Green PCR Master Mix (Applied Biosystems). Each reaction used 5 ng of cDNA template in a 25-μL volume. Relative transcript levels were determined by the comparative standard curve method. Standard curves were generated using gene-specific primers with 10 ng to 0.1 pg of RNA-free genomic DNA purified from P. aeruginosa PAO1. A dissociation curve step was included at the end of each PCR to verify that a single specific product was amplified and confirm the absence of primer dimers. PA1897, a QscR-activated gene, was used as an internal control to verify the absence of significant nonspecific variation between samples.
β-Galactosidase Assays.
Galacto-Light Plus (Tropix) was used to measure β-galactosidase activity in lacZ reporter fusion strains. Results are given in units of β-galactosidase activity per OD600.
Western Blot Analysis.
At each time point, ∼3E+09 bacteria was harvested by centrifugation, and the cell pellet was suspended in 150 μL of 150 mM Tris-HCl (pH 7.5) and 150 μL of 2× Laemlli buffer. Samples were boiled for 5 min and sonicated with a Branson Microtip Sonifier (10 s at an output of 0.4). Protein concentration of the lysates was determined using a 660-nm protein assay (Pierce); 15 μg of protein for each sample was separated by SDS/PAGE, followed by polypeptide transfer to a nitrocellulose membrane. The membrane was probed with a 1:4,000 dilution of anti–VSV-G antibody (V4888; Sigma-Aldrich) and a 1:50,000 dilution of peroxidase conjugated goat anti-rabbit IgG, followed by detection of the conjugates with the Supersignal West Pico chemiluminescent substrate (Pierce). The intensity of the bands was determined using ImageQuant 5.1 (GE Healthcare).
Supplementary Material
Acknowledgments
We thank J. Mougous, C. Harwood, and C. Manoil for helpful discussions. This work was supported by the National Institutes of Health, National Institute of General Medical Sciences (Grant GM59026).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005909107/-/DCSupplemental.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: The LuxR-LuxI family of cell density–responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 4.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler BL. How bacteria talk to each other: Regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 6.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 7.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 8.Beck von Bodman S, Farrand SK. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puskas A, Greenberg EP, Kaplan S, Schaefer AL. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerkop BA, et al. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol. 2009;191:3909–3918. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chugani SA, et al. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesci EC, Iglewski BH. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997;5:132–134. doi: 10.1016/S0966-842X(97)01008-1. discussion, 134–135. [DOI] [PubMed] [Google Scholar]
- 16.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster M, Greenberg EP. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 18.Oglesby AG, et al. The influence of iron on Pseudomonas aeruginosa physiology: A regulatory link between iron and quorum sensing. J Biol Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bredenbruch F, Geffers R, Nimtz M, Buer J, Häussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol. 2006;8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 21.Déziel E, et al. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: Multiple quorum-sensing–regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 22.McGrath S, Wade DS, Pesci EC. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS) FEMS Microbiol Lett. 2004;230:27–34. doi: 10.1016/S0378-1097(03)00849-8. [DOI] [PubMed] [Google Scholar]
- 23.McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilderman PJ, et al. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 26.Whiteley M, Parsek MR, Greenberg EP. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrow JM, 3rd, Pesci EC. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J Bacteriol. 2007;189:3425–3433. doi: 10.1128/JB.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao H, et al. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci USA. 2001;98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Essar DW, Eberly L, Han CY, Crawford IP. DNA sequences and characterization of four early genes of the tryptophan pathway in Pseudomonas aeruginosa. J Bacteriol. 1990;172:853–866. doi: 10.1128/jb.172.2.853-866.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsek MR, Greenberg EP. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Williams P, et al. Quorum sensing and the population-dependent control of virulence. Philos Trans R Soc Lond B Biol Sci. 2000;355:667–680. doi: 10.1098/rstb.2000.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collier DN, et al. A bacterial cell-to-cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett. 2002;215:41–46. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- 35.Guina T, et al. Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc Natl Acad Sci USA. 2003;100:2771–2776. doi: 10.1073/pnas.0435846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barth AL, Pitt TL. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J Med Microbiol. 1996;45:110–119. doi: 10.1099/00222615-45-2-110. [DOI] [PubMed] [Google Scholar]
- 37.Hoiby N. Understanding bacterial biofilms in patients with cystic fibrosis: Current and innovative approaches to potential therapies. J Cyst Fibros. 2002;1:249–254. doi: 10.1016/s1569-1993(02)00104-2. [DOI] [PubMed] [Google Scholar]
- 38.Thomas SR, Ray A, Hodson ME, Pitt TL. Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax. 2000;55:795–797. doi: 10.1136/thorax.55.9.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohman DE, Chakrabarty AM. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun. 1982;37:662–669. doi: 10.1128/iai.37.2.662-669.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klausen M, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 42.Shrout JD, et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 43.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becher A, Schweizer HP. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques. 2000;29:948–950. doi: 10.2144/00295bm04. 952. [DOI] [PubMed] [Google Scholar]
- 45.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 46.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- 47.Siehnel R, et al. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2010;107:7916–7921. doi: 10.1073/pnas.0908511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.