Abstract
Iron-sulfur (Fe/S) cluster enzymes are crucial to life. Their assembly requires a suite of proteins, some of which are specific for particular subsets of Fe/S enzymes. One such protein is yeast Iba57p, which aconitase and certain radical S-adenosylmethionine enzymes require for activity. Iba57p homologs occur in all domains of life; they belong to the COG0354 protein family and are structurally similar to various folate-dependent enzymes. We therefore investigated the possible relationship between folates and Fe/S cluster enzymes using the Escherichia coli Iba57p homolog, YgfZ. NMR analysis confirmed that purified YgfZ showed stereoselective folate binding. Inactivating ygfZ reduced the activities of the Fe/S tRNA modification enzyme MiaB and certain other Fe/S enzymes, although not aconitase. When successive steps in folate biosynthesis were ablated, ∆folE (lacking pterins and folates) and ∆folP (lacking folates) mutants mimicked the ∆ygfZ mutant in having low MiaB activities, whereas ∆folE ∆thyA mutants supplemented with 5-formyltetrahydrofolate (lacking pterins and depleted in dihydrofolate) and ∆gcvP ∆glyA mutants (lacking one-carbon tetrahydrofolates) had intermediate MiaB activities. These data indicate that YgfZ requires a folate, most probably tetrahydrofolate. Importantly, the ∆ygfZ mutant was hypersensitive to oxidative stress and grew poorly on minimal media. COG0354 genes of bacterial, archaeal, fungal, protistan, animal, or plant origin complemented one or both of these growth phenotypes as well as the MiaB activity phenotype. Comparative genomic analysis indicated widespread functional associations between COG0354 proteins and Fe/S cluster metabolism. Thus COG0354 proteins have an ancient, conserved, folate-dependent function in the activity of certain Fe/S cluster enzymes.
Keywords: comparative genomics, oxidative stress, YgfZ protein, Iba57, COG0354
Iron-sulfur (Fe/S) clusters are versatile but labile prosthetic groups found in > 100 proteins from all domains of life (1 and 2). Fe/S cluster synthesis, assembly, and repair have consequently drawn much attention, and genetic and biochemical approaches have begun to dissect the multicomponent systems involved. It is now known that the required sulfur is generated in persulfide form by cysteine desulfurylases (e.g., IscS, SufSE) and then transferred to scaffold proteins (e.g., IscU, NfuA) on which clusters are assembled and from which they are mobilized to target apoproteins (1–4). However, many aspects of synthesis and assembly remain opaque, including the roles of auxiliary proteins that are needed for the activity of various subsets of Fe/S enzymes (1–4). Repair of Fe/S clusters is less understood but is clearly vital during oxidative stress (5 and 6).
A surprising new player in the Fe/S cluster arena is the protein family classified as COG0354 in the Clusters of Orthologous Groups database. COG0354 proteins occur in all domains of life and include yeast Iba57p and Escherichia coli YgfZ. Inactivating iba57, whose product is mitochondrial, resulted in activity loss in four Fe/S enzymes and a petite phenotype (7 and 8). Similarly, ablating ygfZ decreased the MiaB-mediated methylthiolation of N6-isopentenyladenosine in tRNA (9), MiaB being an Fe/S protein of the radical S-adenosylmethionine (SAM) family whose activity drops in mutants defective in Fe/S cluster synthesis (10). The selectivity of these activity losses places Iba57p, and possibly YgfZ, in the category of proteins affecting particular subsets of Fe/S enzymes. Both Iba57p and YgfZ form complexes with IscA-type proteins (7 and 11). Like other proteins involved in Fe/S cluster synthesis or repair, YgfZ is induced by oxidative stress (12). Some bacterial COG0354 genes are apparently essential, although E. coli ygfZ is not (9 and 13).
COG0354 proteins are structurally similar to folate-dependent enzymes including glycine decarboxylase T protein (GcvT), dimethylglycine and sarcosine oxidases, and TrmE (14 and 15), all of which mediate one-carbon (C1) transfer reactions involving tetrahydrofolate (THF) (Fig. 1). Like the three-dimensional structures of these proteins, that of YgfZ predicts a folate-binding site (14).
Fig. 1.
Structure of tetrahydrofolate (THF) and its C1 derivatives. The monoglutamyl form is shown; a short γ-linked polyglutamyl tail is usually attached to the γ-carboxyl of the glutamate. C1 groups at various levels of oxidation are attached to N-5 and/or N-10 of THF as shown.
In this study, we used structural, biochemical, and mutational approaches to show that YgfZ binds folate in vitro, that folate synthesis mutations mimic the effect of ablating YgfZ, and that YgfZ can be functionally replaced by COG354 proteins from all domains of life.
Results
YgfZ Shows Folate-Binding in Vitro.
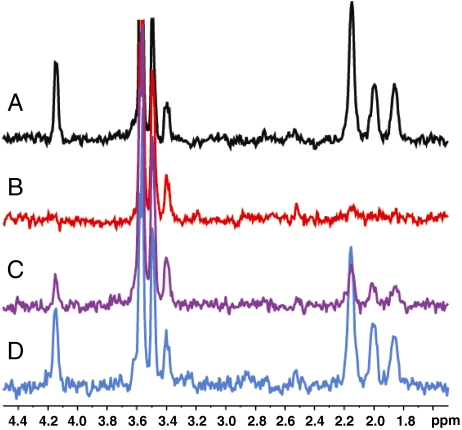
NMR was used to monitor the interaction of purified YgfZ with the natural (6S) form of 5-formyltetrahydrofolate (5-formyl-THF) or folic acid, both labeled in the glutamate moiety with 13C. Clear interaction was observed with (6S)-5-formyl-THF; this interaction was abolished by adding a 9-fold excess of unlabeled (6S)-5-formyl-THF but not (6R)-5-formyl-THF (Fig. 2, Fig. S1, Fig. S2). The small effect of the 6R form may be due partly to contaminating 6S. These data indicate stereoselective binding of 5-formyl-THF; no interaction was observed with folic acid. The complete loss of resonances in Fig. 2B (5-formyl-THF plusYgfZ) results from intermediate exchange. Assuming the difference between free and YgfZ-bound chemical shifts of 5-formyl-THF to be 1 ppm, the data would fit with an exchange rate of ∼3,700 Hz. Reasonable estimates of the equilibrium dissociation constant (KD) for (6S)-5-formyl-THF binding to YgfZ would accordingly be as low as 0.1 mM for an association rate constant (kon) limited by diffusion (107 M-1 s-1) or as high as 3 mM if kon were an order of magnitude slower (106 M-1 s-1). Tryptophan fluorescence quenching (14) also indicated that YgfZ binds folates, but the data were not corrected for the inner filter effect and so required corroboration.
Fig. 2.
NMR evidence for folate binding by E. coli YgfZ. Samples for each spectrum contained 1 mM (6S)-5-formyl-THF with 13C labels in the glutamate moiety. One-dimensional 13C-HSQC experiments obtained with 512 scans provided a filter to observe only 1H resonances directly bonded to 13C. (A) (6S)-5-Formyl-THF alone; (B) (6S)-5-formyl-THF plus 1 mM YgfZ; (C) (6S)-5-formyl-THF plus 1 mM YgfZ and 9 mM unlabeled (6R)-5-formyl-THF; (D) (6S)-5-formyl-THF plus 1 mM YgfZ and 9 mM unlabeled (6S)-5-formyl-THF. Corresponding full one-dimensional 1H spectra are shown in Fig. S1, and the 2D HSQC-TOCSY spectrum of 5-formyl-THF is in Fig. S2. The large cluster of resonances from ∼3.4 to 3.6 ppm is attributable to glycerol and triethylene glycol in the buffer. No interaction with YgfZ was seen when 13C-labeled folic acid (1 mM) replaced labeled (6S)-5-formyl-THF.
Ablating YgfZ in E. coli Impacts Multiple Fe/S Enzymes.
To test whether deleting ygfZ has effects beyond that on MiaB (9) we compared the activities of six diverse Fe/S enzymes in wild type and ΔygfZ strains (Table 1) grown without or with oxidative stress imposed with the redox-cycling agent plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) (12). Succinate dehydrogenase, fumarase, and dimethylsulfoxide reductase activities were all significantly lower in the ΔygfZ strain, especially under oxidative stress, and the same is presumably true of 6-phosphogluconate dehydratase as this strain failed to use gluconate as carbon source when plumbagin was present (Table 1). Aconitase B was unaffected by the deletion, and sulfite reductase activity was mildly increased, showing that the negative effect of deleting ygfZ is specific to certain enzymes.
Table 1.
Effect of deleting ygfZ on Fe/S enzyme activities in E. coli
| Enzyme |
Activity, nmol min-1 mg-1 protein |
|||||
| Control |
+Plumbagin 25 μM |
|||||
| Wild type | ΔygfZ | Wild type | ΔygfZ | |||
| Succinate dehydrogenase | 63.0 ± 3.4 | 33.1 ± 11.1* | (−47%)* | 57.4 ± 6.9 | 16.8 ± 2.6** | (−71%) |
| Fumarase A + B | 1,092 ± 110 | 742 ± 86** | (−32%) | 1,431 ± 176 | 862 ± 179*** | (−40%) |
| DMSO reductase† | 48.0 ± 7.9 | 0.8 ± 0.1 | (−98%) | 16.5 ± 2.4 | < 0.005* | (−100%) |
| 6-P-Gluconate dehydratase | 319 ± 74 | 363 ± 98 | 195 ± 60 | ND‡ | ||
| Aconitase B | 46.1 ± 4.2 | 49.4 ± 4.3 | 53.6 ± 1.1 | 45.4 ± 9.5 | ||
| Sulfite reductase | 680 ± 30 | 805 ± 36* | (+18%) | 1,281 ± 91 | 1,673 ± 122* | (+31%) |
Data are means of 2–6 independent replicates ± SE. *, **, and *** denote differences between wild type and deletant that are significant at P < 0.05, < 0.01, and < 0.001, respectively.
*Values in parentheses are percent change in activity in the deletant relative to the wild type.
†Measured in vivo in aerobically grown cultures; values are consequently not directly comparable to others in the table.
‡Not determined; the ΔygfZ strain was unable to grow on the gluconate-containing induction medium in the presence of 25 μM plumbagin.
Mutational Ablation of Folate Pools.
The folate requirement of E. coli YgfZ could in principle be probed by blocking various steps in folate biosynthesis (Fig. 3A) and measuring the activity of a YgfZ-dependent “reporter” Fe/S enzyme. To validate this approach we analyzed folates in a set of strains respectively designed to eliminate: (i) folates and their pterin precursors (ΔfolE), (ii) folates but not pterins (ΔfolP), (iii) essentially all C1-substituted folates (ΔgcvP ΔglyA), or (iv) dihydrofolate (DHF) and pterins (a ΔfolE ΔthyA strain expressing Synechocystis folate carrier Slr0642 and given 5-formyl-THF; 16). Results were as predicted although the latter strain had 5-fold less total folate than the wild type (Table 2). The ΔgcvP ΔglyA strain lacked detectable C1 folates, and was functionally C1-folate deficient as it did not support synthesis of the tRNA base 5-methylaminomethyl-2-thiouridine (mnm5 s2 U), which requires a C1 folate (17) (Fig. 3B). Nor was mnm5 s2 U detected in other folate-deficient strains (Fig. 3B). Pterin analysis confirmed the presence of pterins in the ΔfolP strain, the most prominent being monapterin (18).
Fig. 3.
Mutational analysis of the folate requirement of E. coli YgfZ. (A) Outline of THF biosynthesis and C1-folate metabolism showing the mutational strategy and its predicted effects on folate pools. DHF, dihydrofolate; H2 pterins, dihydropterins; H2 pteroate, dihydropteroate; pABA, p-aminobenzoate. (B) LC-MS/MS quantification of the tRNA nucleoside 5-methylaminomethyl-2-thiouridine (mnm5s2U) in wild type and deletant strains grown in Antibiotic Medium 3 supplemented as described in Materials and Methods. Data are means and standard errors for three independent samples. (C) LC-MS/MS quantification of the tRNA nucleosides i6A and ms2i6A in wild type and deletant strains, and the ms2i6A/i6A ratio. Other details as in B.
Table 2.
Folate profiles of E. coli strains
| Strain |
Folates, pmol mg-1 protein* |
||||
| THF | CH3-THF | CH = THF + 10-CHO-DHF† | 5-CHO-THF | Total | |
| Wild type | 48.1 ± 10.7 | 10.6 ± 1.9 | 738 ± 93 | 68.9 ± 10.9 | 866 ± 114 |
| ΔfolE | < 0.05 | < 0.05 | < 0.05 | < 0.05 | < 0.2 |
| ΔfolP | < 0.05 | < 0.05 | < 0.05 | < 0.05 | < 0.2 |
| ΔgcvP ΔglyA | 845 ± 171 | < 0.05 | < 0.05 | < 0.05 | 845 ± 171 |
 ‡ ‡
|
152 ± 100 | 7.1 ± 0.7 | 14.4 ± 3.5 | 5.8 + 1.5 | 180 ± 98 |
*Means and standard errors of 3–7 replicates. THF, tetrahydrofolate; CH3-THF, 5-methyl-THF; CH = THF, 5,10-methenyl-THF; 10-CHO-DHF, 10-formyl-DHF; 5-CHO-THF, 5-formyl-THF. The detection limit for each folate was 0.05 pmol mg-1 protein.
†10-CHO-THF is converted to CH = THF and 10-CHO-DHF during analysis.
‡Dihydrofolate not detected (detection limit 0.25 pmol mg-1 protein, allowing for 20% recovery).
YgfZ Activity is Folate-Dependent.
Having confirmed the predicted effects on folate pools, we used MiaB as a reporter to assess whether YgfZ activity requires a folate and, if so, which one(s). MiaB mediates posttranscriptional methylthiolation of N6-isopentenyladenosine (i6A) in tRNAs, giving 2-methylthio-N6-isopentenyladenosine (ms2i6A), so that reducing MiaB activity decreases the ms2i6A/i6A ratio (9 and 10). This ratio was measured in the four strains described above and in ygfZ and miaB deletants as benchmarks; the latter forms no ms2i6A (Fig. 3C). Deleting folE lowered MiaB activity almost as much as deleting ygfZ itself, which establishes pterin or folate dependency. A role for pterins was excluded because: (i) the folP deletant—which has pterins but no folates—showed greatly lowered MiaB activity, and (ii) the ΔfolE ΔthyA strain given 5-formyl-THF—which has folates but no pterins—did not. MiaB activity in the ΔfolE ΔthyA strain may have been below wild type due to low folate levels (Table 2).
The ΔfolE ΔthyA strain given 5-formyl-THF cannot synthesize DHF enzymatically (Fig. 3A) and none was detected (Table 2). The high MiaB activity in this strain (Fig. 3C) therefore argues against a DHF requirement, and so narrows the field of possibilities to THF or one of its C1 forms. That the requirement can be met by THF itself is suggested by the intermediate MiaB activity of the ΔgcvP ΔglyA strain, which contains THF but no C1 folates (Table 2 and Fig. 3B). MiaB activity—and hence YgfZ activity—in this strain may not have reached that of the wild type because its 18-fold elevated THF level (Table 2) caused substrate inhibition (19).
ygfZ Deletion Impairs Oxidative Stress Resistance and Growth on Minimal Media.
As YgfZ is induced by oxidative stress (12) we tested the sensitivity of the ΔygfZ strain to plumbagin or paraquat added to LB medium. The deletant showed only a slight growth defect on LB alone but was more sensitive to both stress agents (Fig. S3A). It also grew poorly on minimal A salts or M9 medium with glucose, glycerol, or acetate as carbon source (Fig. S3B); this fits with the lowered activities of the Krebs cycle enzymes succinate dehydrogenase and fumarase (Table 1).
Eukaryotic and Prokaryotic COG0354 Genes Complement the ygfZ Mutation.
The above growth phenotypes allowed complementation tests of COG0354 genes from bacterial, archaeal, protist, fungal, animal, and plant sources. Two plant genes were tested, Arabidopsis At4g12130 and At1g60990, whose products are respectively predicted to be mitochondrial and plastidial. All seven genes tested complemented the oxidative stress hypersensitivity phenotype (Fig. 4A). The bacterial, Leishmania, mouse, and Arabidopsis genes also complemented the growth phenotype on minimal medium, but the archaeal gene did not and the yeast gene did so only weakly. As the complementation results were obtained in darkness, COG0354 action cannot depend upon light.
Fig. 4.
Complementation of the ygfZ mutation by diverse COG0354 genes. (A) Tests with the ΔygfZ strain harboring pBAD24 alone (Vector) or expressing E. coli YgfZ or COG0354 proteins from a bacterium phylogenetically distant from E. coli (Bartonella henselae), from the eukaryotes Arabidopsis (m, predicted mitochondrial; c, predicted plastidial), mouse, Leishmania major, and Saccharomyces cerevisiae, and from the archaeon Haloferax volcanii. Three independent clones were streaked of each construct. Plates contained LB medium minus or plus 30 μM plumbagin, or M9 medium with 0.2% glycerol, 0.02% arabinose, and appropriate antibiotics. Incubation was at 37 °C for 2 d (LB) or at 22 °C for 5 d (M9). (B) LC-MS/MS quantification of i6A and  , and the ms2i6A/i6A ratio, in tRNA of the ΔygfZ or ΔfolP ΔygfZ strains harboring vector alone or expressing mouse or Leishmania major COG0354, grown in Antibiotic Medium 3 plus 300 μM thymidine, 0.02% arabinose and appropriate antibiotics. Data are means and standard errors for three independent samples.
, and the ms2i6A/i6A ratio, in tRNA of the ΔygfZ or ΔfolP ΔygfZ strains harboring vector alone or expressing mouse or Leishmania major COG0354, grown in Antibiotic Medium 3 plus 300 μM thymidine, 0.02% arabinose and appropriate antibiotics. Data are means and standard errors for three independent samples.
To check that eukaryotic COG0354 genes also complemented the MiaB activity phenotype, the ms2i6A/i6A ratio was determined for cells harboring the genes from mouse or Leishmania (Fig. 4B, left). Both genes greatly increased the ratio compared to the vector-only control. To probe folate dependency, the mouse and Leishmania genes were tested in a ΔfolP ΔygfZ background. The ms2i6A/i6A ratio for both remained very low, thereby implicating folates in their activity (Fig. 4B, right). Parenthetically, this assay does not test the activity of the mouse or Leishmania proteins with these species’ native pterin cofactor tetrahydrobiopterin since E. coli has tetrahydromonapterin as its predominant pterin (18 and 20).
Comparative Genomics Links COG0354 to Fe/S Clusters, Folates, and Oxygen.
Analysis of the phyletic distribution and chromosomal clustering of prokaryotic COG0354 genes showed that they cooccur (i.e., are present or absent together) with genes encoding Fe/S assembly proteins of the IscA-SufA family in 90% of the genomes examined, and never occur without them (Fig. S4A). IscA-SufA genes themselves occur in only 69% of genomes while other genes of the Isc and Suf systems are nearly universal (Fig. S3A). COG0354 genes also often cluster with genes for Fe/S enzymes and related proteins (Table S1 and Fig. S4B). Connecting COG0354 to folates, COG0354 genes in Archaea are confined to Halobacteria (Fig. S4A), which are almost the only Archaea with folates (21 and 22). Finally, making a link with oxidative stress, COG0354 genes occur in 86% of aerobes having a full complement of Fe/S proteins but in only 15% of anaerobes. Furthermore, the E. coli ygfZ gene contains a marbox structure and is SoxS regulated (12).
Discussion
Our data establish a unique role for folate as a cofactor for E. coli YgfZ in the assembly or repair of Fe/S clusters, and confirm a similar role in mouse and Leishmania COG0354 proteins. The data also demonstrate the criticality of YgfZ during oxidative stress, in agreement with its induction in such conditions (12) and with the rarity of COG0354 genes in anaerobes. The Fe/S enzyme activity data extend the known protein repertoire of COG0354 proteins (7 and 9), and the chromosomal clustering data suggest this repertoire may be wider still. The complementation results extend the species repertoire by showing that eukaryotic, archaeal, and bacterial COG0354 genes can replace E. coli ygfZ in some conditions. A human gene was similarly shown to replace iba57 in yeast (7).
The NMR evidence for folate binding came from (6S)-5-formyl-THF, THF itself being too unstable for such studies. While not a C1 donor, (6S)-5-formyl-THF binds to (and inhibits) most folate-dependent enzymes and is often used as a stable surrogate for THF (15). The estimated KD (≥0.1 mM) for (6S)-5-formyl-THF is higher than typically seen for binding of folate substrates. This higher KD could arise because 5-formyl-THF is not the natural YgfZ substrate, or due to the absence of polyglutamylation, which typically lowers KD values. Moreover, since YgfZ normally occurs in complexes with IscA-type proteins in vivo (7 and 11), the free protein could adopt a nonnative conformation, as seen with the paralogous folate-binding protein GcvT (23). A nonnative conformation would be likely to affect ligand binding kinetics.
Thus we conclude that COG0354 has a folate-dependent function that impacts diverse Fe/S proteins and is at least partly conserved in all domains of life. Our results shed light on the nature of this folate-dependent function: First, the genomic codistribution data imply that COG0354 proteins cooperate with IscA-SufA family proteins, in agreement with the finding that yeast and E. coli COG0354 proteins complex with IscA homologs (7 and 11). Second, the experimental and genomic data connecting COG0354 proteins with oxidative stress, and the nonuniform effects on various Fe/S enzyme activities, point to a role in repair of specific enzymes as opposed to de novo synthesis of all Fe/S clusters. Third, while both YgfZ and Iba57p clearly affect only subsets of the Fe/S enzymes in their respective host organism, these subsets appear not to be identical; most notably, aconitase was impacted by COG0354 ablation in yeast (7) but not E. coli. Another hint of variation in target enzyme specificity is that all the COG0354 genes tested complemented the ΔygfZ mutant under oxidative stress but not all did so on minimal medium. Different Fe/S proteins are presumably crucial to growth in these contrasting conditions. Supposing a repair role, the enzymes targeted in any given organism could reflect idiosyncrasies in the susceptibility of different Fe/S clusters to damage, as seen in other studies (5, 6, 10).
What role might THF play in the function of COG0354? One possibility that we consider remote is a purely structural role, as with folate hexaglutamate in the assembly of bacteriophage T4 (24). A second and more likely possibility is that THF acts as an electron donor, there being reductions in Fe/S cluster assembly and repair whose electron donor is unknown (3 and 5). THF can be oxidized readily (standard redox potential = -230 mV for the DHF/THF pair) and the redox properties of the THF ring are exploited in nature in thymidylate synthesis, where 5,10-methylenetetrahydrofolate acts as both electron donor and C1 donor (25). There is also precedent for THF acting solely as an electron donor in the case of bacterial phenylalanine hydroxylase (26). A third possibility would involve C1 metabolism, perhaps with THF acting as an acceptor for a C1 moiety. A C1 adduct on an Fe/S cluster-liganding cysteine residue would block cluster formation, and might be removed via COG0354-mediated transfer to THF in a mechanism analogous to that of the glycine cleavage T protein or oxidative demethylases. Future studies will address these alternative propositions.
Materials and Methods
Bioinformatics.
Prokaryote genomes were analyzed using the SEED database and its tools (27). Full results are available at http://theseed.uchicago.edu/FIG/ in the YgfZ and Iron-sulfur cluster assembly subsystems. Eukaryote genomes and ESTs were searched using BLAST algorithms at NCBI (http://www.ncbi.nlm.nih.gov/). COG0354 members were identified using the NCBI Conserved Domain algorithm and the KGCYxGQE motif (14).
E. coli Strains, Plasmids, and Media.
Strains, plasmids, and primers are listed in Table S2 and Table S3. Deletions from the Keio collection (28) and a ΔfolP∷Kan allele (29) were transferred to E. coli K12 MG1655 by P1 transduction; for double deletants, Kan cassettes were removed by flippase-mediated recombination. All deletions were PCR-verified. Strains for complementation studies harbored pACYC-RP. Cells were cultured at 37 °C in Antibiotic Medium 3 (Difco) plus 300 μM thymidine, LB medium, minimal A salts (30) or M9 medium (16) as indicated. The ΔfolE ΔthyA strain harboring plasmid-borne slr0642 was given 50 μM 5-formyl-THF, 1 mM DTT, and 0.1% (w/v) ascorbate. Antibiotic concentrations (μg/mL) were: kanamycin, 25; chloramphenicol, 10; ampicillin, 25; tetracycline, 10; erythromycin, 200. Gene expression was induced with 0.02% (w/v) arabinose or 1 mM isopropyl-β-D-thiogalactoside. COG0354 sequences are described in Table S4.
Fe/S Enzyme Assays.
Wild type and ΔygfZ strains were grown aerobically, minus or plus 25 µM plumbagin, to an OD600 of ∼1 in LB medium except for 6-phosphogluconate dehydratase, which was induced in minimal A salts containing 0.2% (w/v) K-gluconate. Enzymes were assayed at 22 °C. Succinate dehydrogenase was assayed in inverted membrane vesicles (31) after activating in 0.6 M K-phosphate, pH 7.0, at 30 °C for 20 min. Assays contained 0.1 M K-phosphate, pH 7.8, 10 mM succinate, 0.75 mM phenazine methosulfate, 30 µM 2,6-dichlorophenolindophenol, and 1 mM KCN; dye reduction was monitored at 600 nm (ε = 21 mM-1 cm-1). Soluble extracts were prepared as described (32). Aconitase was assayed spectrophotometrically in mixtures containing 50 mM Tris-HCl, pH 7.6, 25 mM Na citrate, 0.25 mM NADP+, 0.6 mM MnCl2, 1-2 units of isocitrate dehydrogenase in a total volume of 0.1 mL (33). The activity sensitive to 1 mM EDTA was taken as that of aconitase B (34). Fumarase A plus B activity was assayed as described (35) in 50 mM Tris-HCl, pH 7.6. 6-Phosphogluconate dehydratase was assayed as described (36). Sulfite reductase was assayed spectrophotometrically in 50 mM Tris-HCl, pH 7.5, containing 0.25 mM NADPH and 0.3 mM ferricyanide (37). Dimethylsulfoxide reductase was assayed in vivo by growing cultures supplied with 14 mM DMSO to stationary phase, and determining the rate of dimethylsulfide production by headspace analysis using gas-chromatography/mass spectrometry. Protein was measured by the bicinchoninic acid method with bovine serum albumin as standard.
Folate and Pterin Analysis.
Folates were extracted from cultures grown to an OD600 of 1.0, deglutamylated, isolated by affinity chromatography, and analyzed by HPLC with electrochemical detection as described (20) except that, for DHF analysis, initial extraction was at pH 2.0. Total pterins from cultures grown to an OD600 of 1.0 were analyzed as described (20).
Nucleoside Analysis.
Bulk nucleic acids were isolated from stationary phase cells and enriched for tRNA (38) before Nucleobond AXR 400 column chromatography purification (Machery-Nagel). Pure tRNA was then hydrolyzed and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described (39). These procedures differ from those of Ote et al. (9), which may account for differences in our ms2i6A/i6A ratios.
NMR Analyses.
(6S)-5-Formyl-THF [ -glutamate] was from Merck Eprova; other folates were from Schircks. 1-mm NMR tubes were used in a 5-mm cryogenic probe on a Bruker AvanceII 600 MHz instrument. Sample volumes were 10 μL for each experiment; temperature was 20 °C. One-dimensional 13C-heteronuclear single quantum coherence (HSQC) experiments were used to select for protons directly bonded to 13C. Due to the low volume, 512 scans were collected for each experiment. One-dimensional 1H spectra were collected with 16 scans per sample. For more details, see SI Text.
-glutamate] was from Merck Eprova; other folates were from Schircks. 1-mm NMR tubes were used in a 5-mm cryogenic probe on a Bruker AvanceII 600 MHz instrument. Sample volumes were 10 μL for each experiment; temperature was 20 °C. One-dimensional 13C-heteronuclear single quantum coherence (HSQC) experiments were used to select for protons directly bonded to 13C. Due to the low volume, 512 scans were collected for each experiment. One-dimensional 1H spectra were collected with 16 scans per sample. For more details, see SI Text.
Supplementary Material
Acknowledgments.
We thank G. Swedberg for the ΔfolP∷Kan strain; D. Tieman and M. Bailly for assistance; K. Austin and D. Fremont for efforts to test folate binding by surface plasmon resonance; J. Peng for advice on binding constant estimates; and S. Jang for criticism of the manuscript. This work was supported in part by the National Science Foundation award MCB-0839926 (to A.D.H.), by the Department of Energy award FG02-07ER64498 (to V.d.C.-L.), by National Institutes of Health Grant AI21903 (to S.M.B.), by EMBO fellowship ALTF 106-2005 (to T.J.V.), and by an endowment from the C.V. Griffin, Sr. Foundation. NMR studies were supported by the National Science Foundation’s National High Magnetic Field Laboratory User Program in the Advanced Magnetic Resonance Imaging and Spectroscopy (AMRIS) Facility in the McKnight Brain Institute of the University of Florida.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911586107/-/DCSupplemental.
References
- 1.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.Lill R. Function and biogenesis of iron-sulfur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 3.Fontecave M, Ollagnier de Choudens S. Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Arch Biochem Biophys. 2008;474:226–237. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Barras F, Loiseau L, Py B. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv Microb Physiol. 2005;50:41–101. doi: 10.1016/S0065-2911(05)50002-X. [DOI] [PubMed] [Google Scholar]
- 5.Djaman O, Outten FW, Imlay JA. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem. 2004;279:44590–44599. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 6.Justino MC, Almeida CC, Teixeira M, Saraiva LM. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J Biol Chem. 2007;282:10352–10359. doi: 10.1074/jbc.M610656200. [DOI] [PubMed] [Google Scholar]
- 7.Gelling C, Dawes IW, Richhardt N, Lill R, Mühlenhoff U. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol Cell Biol. 2008;28:1851–1861. doi: 10.1128/MCB.01963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinmetz LM, et al. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 9.Ote T, et al. Involvement of the Escherichia coli folate-binding protein YgfZ in RNA modification and regulation of chromosomal replication initiation. Mol Microbiol. 2006;59:265–275. doi: 10.1111/j.1365-2958.2005.04932.x. [DOI] [PubMed] [Google Scholar]
- 10.Skovran E, Lauhon CT, Downs DM. Lack of YggX results in chronic oxidative stress and uncovers subtle defects in Fe-S cluster metabolism in Salmonella enterica. J Bacteriol. 2004;186:7626–7634. doi: 10.1128/JB.186.22.7626-7634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:929–947. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JW, Sun CM, Sheng WL, Wang YC, Syu WJ. Expression analysis of up-regulated genes responding to plumbagin in Escherichia coli. J Bacteriol. 2006;188:456–463. doi: 10.1128/JB.188.2.456-463.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes S, et al. Essential genes on metabolic maps. Curr Opin Biotechnol. 2006;17:448–456. doi: 10.1016/j.copbio.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Teplyakov A, et al. Crystal structure of the YgfZ protein from Escherichia coli suggests a folate-dependent regulatory role in one-carbon metabolism. J Bacteriol. 2004;186:7134–7140. doi: 10.1128/JB.186.21.7134-7140.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scrutton NS, Leys D. Crystal structure of DMGO provides a prototype for a new tetrahydrofolate-binding fold. Biochem Soc Trans. 2005;33:776–779. doi: 10.1042/BST0330776. [DOI] [PubMed] [Google Scholar]
- 16.Klaus SM, et al. Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J Biol Chem. 2005;280:38457–38463. doi: 10.1074/jbc.M507432200. [DOI] [PubMed] [Google Scholar]
- 17.Scrima A, Vetter IR, Armengod ME, Wittinghofer A. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J. 2005;24:23–33. doi: 10.1038/sj.emboj.7600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikemoto K, et al. (6R)-5,6,7,8-Tetrahydro-L-monapterin from Escherichia coli, a novel natural unconjugated tetrahydropterin. Biol Chem. 2002;383:325–330. doi: 10.1515/BC.2002.035. [DOI] [PubMed] [Google Scholar]
- 19.Nijhout HF, Reed MC, Budu P, Ulrich CM. A mathematical model of the folate cycle: New insights into folate homeostasis. J Biol Chem. 2004;279:55008–55016. doi: 10.1074/jbc.M410818200. [DOI] [PubMed] [Google Scholar]
- 20.Pribat A, et al. FolX and FolM are essential for tetrahydromonapterin synthesis in Escherichia coli and Pseudomonas aeruginosa. J Bacteriol. 2009;192:475–482. doi: 10.1128/JB.01198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White RH. Distribution of folates and modified folates in extremely thermophilic bacteria. J Bacteriol. 1991;173:1987–1991. doi: 10.1128/jb.173.6.1987-1991.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchenau B, Thauer RK. Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch Microbiol. 2004;182:313–325. doi: 10.1007/s00203-004-0714-0. [DOI] [PubMed] [Google Scholar]
- 23.Rébeillé F, Neuburger M, Douce R. Interaction between glycine decarboxylase, serine hydroxymethyltransferase and tetrahydrofolate polyglutamates in pea leaf mitochondria. Biochem J. 1994;302:223–228. doi: 10.1042/bj3020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozloff LM. A structural role for dihydropteroyl hexaglutamate in the tail baseplate of various bacteriophages. Adv Exp Med Biol. 1983;163:359–374. doi: 10.1007/978-1-4757-5241-0_26. [DOI] [PubMed] [Google Scholar]
- 25.Matthews RG. Are the redox properties of tetrahydrofolate cofactors utilized in folate-dependent reactions? Fed Proc. 1982;41:2600–2604. [PubMed] [Google Scholar]
- 26.Fujisawa H, Nakata H. Phenylalanine 4-monooxygenase from Chromobacterium violaceum. Methods Enzymol. 1987;142:44–49. doi: 10.1016/s0076-6879(87)42007-7. [DOI] [PubMed] [Google Scholar]
- 27.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fermér C, Swedberg G. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: Kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli. J Bacteriol. 1997;179:831–837. doi: 10.1128/jb.179.3.831-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JH. Experiments in Molecular Genetics. NY: Cold Spring Harbor Laboratory Press; 1972. p. 432. [Google Scholar]
- 31.Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol. 1998;180:5989–5996. doi: 10.1128/jb.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orsomando G, et al. Plant γ-glutamyl hydrolases and folate polyglutamates: characterization, compartmentation, and cooccurrence in vacuoles. J Biol Chem. 2005;280:28877–28884. doi: 10.1074/jbc.M504306200. [DOI] [PubMed] [Google Scholar]
- 33.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 34.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gort AS, Imlay JA. Balance between endogenous superoxide stress and antioxidant defenses. J Bacteriol. 1998;180:1402–1410. doi: 10.1128/jb.180.6.1402-1410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giró M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152:1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- 37.Eschenbrenner M, Covès J, Fontecave M. The flavin reductase activity of the flavorprotein component of sulfite reductase from Escherichia coli. A new model for the protein structure. J Biol Chem. 1995;270:20550–20555. doi: 10.1074/jbc.270.35.20550. [DOI] [PubMed] [Google Scholar]
- 38.Bailly M, et al. tRNA-dependent asparagine formation in prokaryotes: Characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNAAsn. Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Phillips G, et al. The biosynthesis of the 7-deazaguanosine modified tRNA nucleosides: A new role for GTP cyclohydrolase I. J Bacteriol. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.