Abstract
Differential distribution of the plant hormone auxin within tissues mediates a variety of developmental processes. Cellular auxin levels are determined by metabolic processes including synthesis, degradation, and (de)conjugation, as well as by auxin transport across the plasma membrane. Whereas transport of free auxins such as naturally occurring indole-3-acetic acid (IAA) is well characterized, little is known about the transport of auxin precursors and metabolites. Here, we identify a mutation in the ABCG37 gene of Arabidopsis that causes the polar auxin transport inhibitor sensitive1 (pis1) phenotype manifested by hypersensitivity to auxinic compounds. ABCG37 encodes the pleiotropic drug resistance transporter that transports a range of synthetic auxinic compounds as well as the endogenous auxin precursor indole-3-butyric acid (IBA), but not free IAA. ABCG37 and its homolog ABCG36 act redundantly at outermost root plasma membranes and, unlike established IAA transporters from the PIN and ABCB families, transport IBA out of the cells. Our findings explore possible novel modes of regulating auxin homeostasis and plant development by means of directional transport of the auxin precursor IBA and presumably also other auxin metabolites.
Keywords: PDR9, PDR8, IBA transport, auxin synthesis
Plants have evolved outstanding capacities to adapt their metabolism and development to respond to their environment. Changes in the availability and distribution of endogenous signaling molecules—plant hormones—play important roles in these responses (1). The phytohormone auxin, perceived by TIR1/AFB receptor proteins and interpreted by downstream nuclear signaling pathway, is an important signal that mediates transcriptional developmental reprogramming (reviewed in refs. 2 and 3). The differential distribution of auxin within tissues is essential for many adaptive responses including embryo and leaf patterning, root and stem elongation, lateral root initiation, and leaf expansion (4). Differential distribution of the major active auxin, IAA, depends on its intercellular transport and metabolic processes that involve biosynthesis by several pathways and release from storage forms including amide- or ester-linked conjugates with amino acids, peptides, and sugars (reviewed in ref. 5). The role of another endogenously occurring auxinic compound IBA is still unclear. It has been proposed that IBA acts independently of IAA (6), but a number of recent genetic findings suggest that IBA functions as an important precursor to IAA during conversion resembling peroxisomal fatty acid β-oxidation (5, 7). Besides metabolism, a crucial process controlling cellular auxin levels is the directional, intercellular auxin transport that depends on specialized influx and efflux carriers (reviewed in ref. 8). IAA transporters include amino acid permeases-like AUXIN RESISTANT1 (AUX1) mediating auxin influx (9–11), the PIN-FORMED (PIN) efflux carriers (12–14), and the MULTIDRUG RESISTANCE/P-GLYCOPROTEIN (PGP) class of ATP-Binding Cassette (ABC) auxin transporters (15–18). Despite the demonstrated importance and wealth of knowledge on the transport of IAA, the mechanism and physiological relevance of transport of its precursors and metabolites remain elusive.
Results
pis1 Mutant Is Hypersensitive to Exogenous IBA.
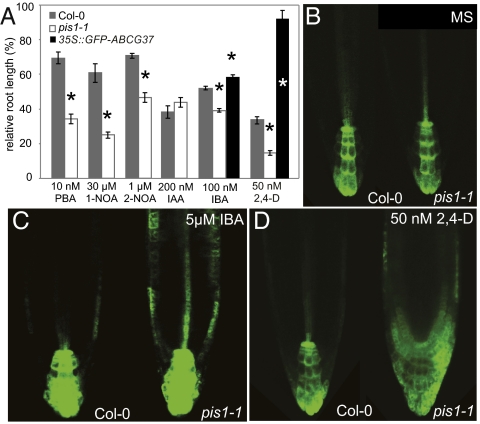
To understand the mechanisms of auxin homeostasis regulation, we analyzed one of the early characterized mutants, polar auxin transport inhibitor sensitive1 (pis1), of Arabidopsis thaliana that is hypersensitive to different auxinic (and/or auxin transport interfering) compounds, but not to the active, natural auxin IAA (19). pis1 mutant roots show strongly enhanced sensitivity to auxinic compounds including synthetic auxins (2,4-D and 2-NOA) and inhibitors of auxin transport (1-NOA, NPA, PBA, and TIBA) (19) (Fig. 1A). When naturally occurring auxins were tested, pis1 showed normal sensitivity to IAA and PAA, but increased sensitivity to IBA (Fig. 1A and Fig. S1A). To test whether the increased pis1 sensitivity to auxins is also reflected at the level of auxin signaling, we introduced DR5rev::GFP auxin response reporter (20, 21) into pis1-1 plants. Whereas no obvious changes in DR5 activity were observed on control medium (Fig. 1B), application of 2,4-D, NPA, or IBA, but not IAA, led to a broad activation of DR5 expression in pis1 roots at concentrations markedly lower than in wild-type seedlings (Fig. 1 C and D, and Fig. S1 B and C). Thus, DR5-monitored auxin signaling in pis1 shows increased sensitivity to auxinic compounds similarly to other phenotypic aspects.
Fig. 1.
Loss-of-function pis1 mutant is hypersensitive to auxinic compounds including natural auxin IBA. (A) abcg37 (pis1-1 allele) root growth is hypersensitive to different auxinic compounds (PBA, 1-NOA, 2-NOA, and 2,4-D) and the natural auxin precursor IBA, but not to the active auxin IAA; overexpression of GFP-ABCG37 in pis1-1 background confers resistance to IBA and 2,4-D. *, different from Col-0 control, P < 0.01 by ANOVA. (B) DR5rev::GFP in the pis1-1 mutant does not show any detectable difference compared with the wild-type on the control medium. (C and D) Hypersensitivity of pis1-1 to auxinic compounds is reflected by increased induction of DR5rev::GFP auxin response reporter at suboptimal concentrations of IBA (C) and 2,4-D (D).
PIS1 Codes for Polarly Localized ABCG37 ATP-Binding Cassette Transporter.
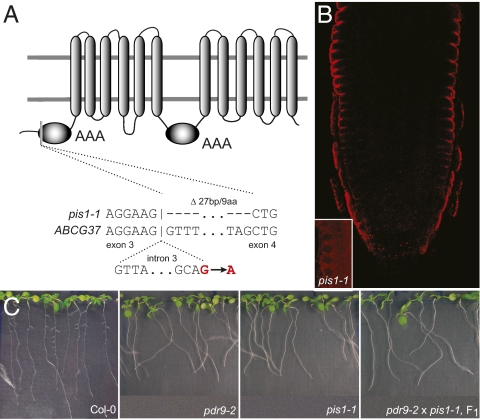
We mapped the pis1-1 mutation using 2,800 chromosomes to an 80-kb region on the lower arm of chromosome 3. Sequencing candidate genes revealed a mutation that leads to an altered splicing and deletion of 9 amino acids in the gene coding for the previously characterized protein ABCG37/PDR9 (22, 23), a member of the G-subgroup of ATP-binding cassette (ABC) transporters (24) (Fig. 2A). The altered splicing was confirmed by sequencing the ABCG37 cDNA from pis1-1 seedlings (Fig. 2A and Fig. S1D). Allelic complementation analyses of pis1-1 with the abcg37 T-DNA insertion mutant (pdr9-2) (22) confirmed that the auxin hypersensitivity of pis1 results from loss of ABCG37 function (Fig. 2C). Moreover, ABCG37 overexpression in 35S::GFP-ABCG37 lines complemented the pis1-1 mutation and conferred increased resistance of roots to IBA and 2,4-D (Fig. 1A). These and previous (22, 23) findings on changed auxin sensitivity in loss- and gain-of-function abcg37 alleles suggest a role of ABCG37 as exporter for auxinic compounds, but this function has not so far been demonstrated directly.
Fig. 2.
pis1 mutant carries mutation in the ABCG37 gene for ATP-binding cassette transporter. (A) The G to A substitution in pis1-1 affects ABCG37 splicing, resulting in a 9-aa deletion in the first ATP-binding AAA domain. (B) ABCG37 is expressed in the lateral root cap and epidermis, and shows outer polar plasma membrane localization (immunostaining of a primary root tip with anti-ABCG37). (Inset) Absence of the signal in the pis1-1 mutant. (C) pis1-1 fails to complement the pdr9-2 mutant allele of ABCG37 (seedlings germinated on 200 nM NPA; note the oversensitivity to NPA manifested by reduced root elongation, lateral root formation, and gravitropism).
We localized ABCG37 in planta using polyclonal anti-ABCG37 antibodies and detected the ABCG37 signal exclusively at the outermost sides of lateral root cap and epidermal cells of the wild-type (25), but not pis1-1 (Fig. 2B Inset), root tips. In the abcg37 gain-of-function allele pdr9-1 (22), the ABCG37 localization pattern was normal as in the wild type (Fig. S2C). This outer polar localization was confirmed by visualizing GFP-ABCG37 in 35S::GFP-ABCG37 roots (25) (Fig. S2 A and B).
ABCG36 and ABCG37 Act Redundantly.
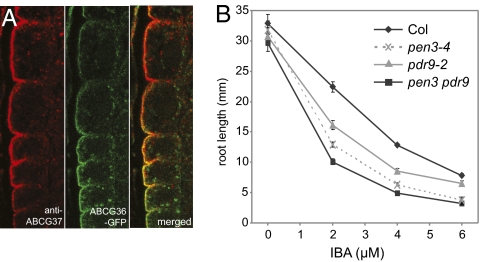
Notably, the ABCG37 transporter shows an almost identical localization pattern as the homologous ABCG36/PDR8/PEN3 transporter (Fig. 3A), which functions in pathogen responses (26), cadmium transport (27), and also in regulation of IBA sensitivity and IAA homeostasis (28). To uncover possible common roles of these transporters, we generated a double mutant lacking function of both ABCG36 and ABCG37. Root growth assays showed increased sensitivity to IBA of both single mutants and even stronger hypersensitivity of the double mutant (Fig. 3B). Nonetheless, the specificity of ABCG36 and ABCG37 action to different compounds does not overlap completely, in particular for synthetic compounds. For example, abcg37 (Fig. 1A), but not abcg36 (28), conveys increased sensitivity to the synthetic auxin 2,4-D, but both act redundantly on its analog with a longer side chain, 2,4-DB (Fig. S3D). Furthermore, these ABC transporters are important for normal development, including root hair elongation (Fig. S3 A and B), and cotyledon expansion (Fig. S3C). Not all aspects of development show similar genetic interactions between abcg36 and abcg37, however. Whereas the double mutant is more sensitive to IBA than either parent in root elongation assays (Fig. 3B), the double mutant does not show additive defects in root hair growth (Fig. S3B) and shows antagonistic action in cotyledon expansion (Fig. S3C). In addition, the pdr9-1 gain-of-function mutant (22) shows opposite phenotypes in root hair growth as compared with the loss-of-function mutant (Fig. S3 B and C). Altogether, these data suggest that ABCG36 and ABCG37, despite having not completely overlapping properties and showing complex contributions in different tissues, redundantly act on IBA sensitivity and multiple aspects of primary root development.
Fig. 3.
Localization and functional overlap of ABCG37 and ABCG36. (A) ABCG37 colocalizes with ABCG36-GFP in immunolocalization experiments. (B) The abcg36 abcg37 (pen3-4 pdr9-2) double mutant shows enhanced sensitivity to IBA compared with either single mutant as manifested by inhibition of root growth (sensitivity of each line was significantly different from others at 2 and 4 μM IBA; P < 0.05 by ANOVA).
ABGC36 and ABCG37 Regulate IBA Accumulation in Planta.
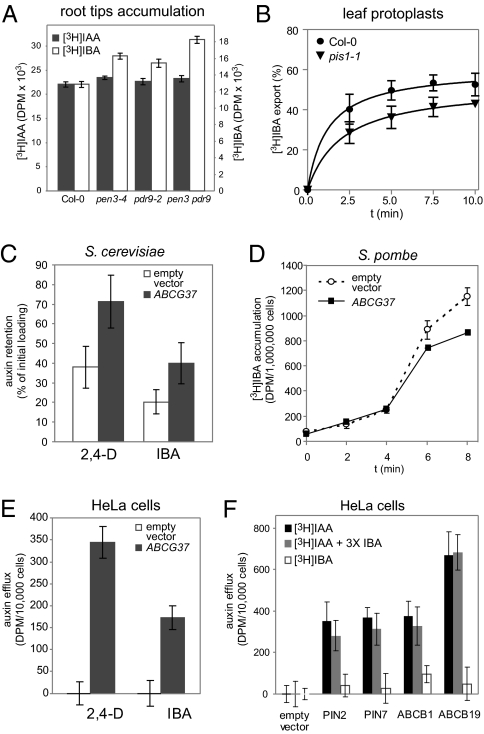
To address the function of ABCG36 and ABCG37 in regulating IBA homeostasis more directly in the place where their localization overlaps, we compared [3H]IAA and [3H]IBA accumulation in root tips excised from abcg36 and abcg37 single and double mutants. As reported previously (23, 28), abcg36 and abcg37 root tips displayed wild-type accumulation of [3H]IAA but hyperaccumulated [3H]IBA in this assay (Fig. 4A). Importantly, root tips of abcg36 abcg37 double mutants accumulated even more [3H]IBA than single mutants (Fig. 4A), consistent with the enhanced sensitivity of the double mutant to IBA in the root elongation assay (Fig. 3B). These results from root were corroborated by transport assays using protoplasts derived from pis1-1 mutant leaves. pis1 protoplasts exported significantly less [3H]IBA, [3H]2,4-D, and [3H]NPA than wild-type protoplasts, but showed unchanged [3H]IAA export (Fig. 4B and Fig. S4A). The activity of ABCG37 in leaves protoplasts is in line with altered cotyledon area in various abcg37 mutants (Fig. S3C); however, it remains unclear what would be the exact physiological role and relevant endogenous substrates for the ABCG37-mediated transport in the aerial tissues.
Fig. 4.
ABCG37 transports IBA and other auxinic compounds. (A) The absence of both ABCG36 and ABCG37 leads to increased [3H]IBA accumulation (P < 0.05 by ANOVA) in root tips, but does not affect [3H]IAA accumulation. (B) abcg37 (pis1-1) leaf mesophyll protoplasts show significantly lower export of [3H]IBA as compared with the wild-type protoplasts (P < 0.05 by ANOVA). (C) Expression of ABCG37 in S. cerevisiae leads to ABCG37 accumulation in the endoplasmic reticulum and increased retention of [3H]2,4-D and [3H]IBA (significantly different from the vector control, Student's t test, P < 0.05). (D) Expression of ABCG37 in S. pombe cells results in a decreased [3H]IBA accumulation, significant after 6 min (P < 0.05 by ANOVA). [3H]IBA concentration was 250 μM. (E) ABCG37 expression in HeLa cells confers active export of [3H]2,4-D and [3H]IBA compared with the empty vector (P < 0.05 by ANOVA). (F) When expressed in HeLa cells, PIN2, PIN7, ABCB1, and ABCB19 show a clear [3H]IAA transport (P < 0.005 by ANOVA), no significant [3H]IBA transport or IBA competition with [3H]IAA transport was observed. Auxin concentrations were 60 nM [3H]IAA, 60 nM [3H]IBA, and 180 nM unlabeled IBA (3× IBA). Values shown are means from three replicate experiments.
These results demonstrate that ABCG37 acts redundantly with ABCG36 in regulating IBA but not IAA accumulation, presumably acting as exporters of IBA (and other synthetic auxinic compounds) from cells.
ABCG37 Transports IBA in Heterologous Systems.
To directly test the ability of ABCG37 to export IBA and synthetic auxins, we examined transport activity of ABCG37 expressed in heterologous systems. Expression of ABCG37 in the budding yeast Saccharomyces cerevisiae, where it localizes to the endoplasmic reticulum (Fig. S4B), led to increased retention of [3H]2,4-D and [3H]IBA (Fig. 4C), suggesting transport activity of ABCG37 for IBA and other auxinic compounds.
Because the non-plasma membrane localization of ABCG37 in S. cerevisiae impedes direct interpretations, we expressed ABCG37 in a recently established Schizosaccharomyces pombe transport system (29), where it localized to the plasma membrane (Fig. S4C). No significant [3H]IAA transport was found in cells expressing ABCG37 (Fig. S4D), even at concentrations five times higher than previously shown for the PIN and ABCB auxin exporters (29). In assays with lower [3H]IBA concentrations, no difference in net accumulation was seen in a cell expressing ABCG37 as compared with controls (Fig. S4E). However, [3H]IBA saturation of the system resulted in a significant decrease in net accumulation in cells expressing ABCG37 (Fig. 4D), consistent with ABCG37 acting as exporter for IBA. The longer activity lag phase in the assay (Fig. 4D) can be explained by more rapid diffusive uptake of [3H]IBA in S. pombe cells as compared with [3H]IAA.
We also examined the ability of ABCG37 to transport auxinic compounds in mammalian HeLa cells, which do not contain endogenous ABCG-type proteins (24). ABCG37 conferred significant export of [3H]2,4-D and [3H]IBA (Fig. 4E). As reported previously for other ABC-type transporters and PIN proteins (13, 16), ABCG37 showed decreased substrate specificity when expressed in heterologous systems and was able to transport other weak organic acids, including IAA (Fig. S4F). Nonetheless, the unchanged sensitivity to IAA of abcg37 loss- and gain-of-function mutants and lack of transport activity for IAA in root and protoplast assays strongly suggest that IAA is not an endogenous substrate of ABCG37. We also tested the IBA transport activity of the well established IAA transporters PIN1, PIN7 (13), ABCB1, and ABCB19 (16). For those proteins, we did not detect any [3H]IBA transport activity (Fig. 4F), indicating that IBA and IAA use different efflux transporters.
In summary, the data from root, protoplast, and heterologous systems directly demonstrate that ABCG37 acts as a broad substrate specificity exporter for synthetic auxinic compounds, which also transports the endogenous auxin precursor IBA but not IAA.
ABCG37 Function Influences Auxin Transport and Homeostasis in the Root Tip.
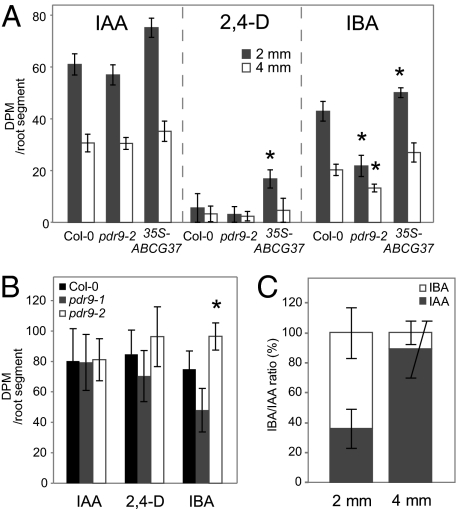
Next, we tested the relevance of ABCG37 transport function to intercellular auxin distribution in the root tip. We applied [3H]IAA, [3H]2,4-D, and [3H]IBA to the root columella cells of intact roots of wild-type and abcg37 gain- and loss-of-function seedlings (pdr9-1 and pdr9-2, respectively). Consistent with an export function for ABCG37, the whole-root uptake assays (subsequently excised 400-μm root tip segment) showed that uptake of IBA and probably 2,4-D, but not IAA, decreased in the pdr9-1 gain-of-function mutant and increased in the pdr9-2 loss-of-function mutant (Fig. 5B).
Fig. 5.
ABCG37 is involved in regulation of auxin homeostasis in the root tip. (A) Basipetal transport of columella-applied auxins: GFP-ABCG37 overexpression results in an increase in apparent diffusive movement of [3H]IBA and its nonpolarly transported analog [3H]2,4-D into the 2-mm segment adjoining the region of application, whereas loss of ABCG37 function results in decreased basipetal movement of the signal derived from [3H]IBA application, indicating more specific exclusion of IBA (*, P < 0.05 by ANOVA). (B) Uptake of columella-applied auxins: In a replicate assay, gain (pdr9-1) or loss (pdr9-2) of ABCG37 function leads to reduced or increased, respectively, uptake of [3H]2,4-D and [3H]IBA, but not [3H]IAA (*, significantly different from Col-0, P < 0.05 by ANOVA). (C) HPLC determination of radiolabeled IAA and IBA obtained from serial sections (0.4–2.4 mm and 2.4–4.4 mm from the root apex) 2 h after application of 100 fmol of [3H]IBA to root columella cells (ratio in 2-mm section significantly different from that in 4-mm section, P < 0.001 by ANOVA). The results indicate that [3H]IBA is converted to [3H]IAA.
We also tested basipetal auxin distribution using a discontinuous media microscale assay (30). Whereas there were no significant changes in transport of [3H]IAA, the abcg37 loss-of-function mutant roots showed less transport capacity for [3H]IBA, and the GFP-ABCG37 overexpressor showed more transport capacity for [3H]2,4-D and [3H]IBA (Fig. 5A).
Because IBA is proposed to be an IAA precursor (5), we tested whether IBA is converted to IAA as it moves from the root apex. HPLC analysis of auxins extracted from root segments 2 h after IBA application on the columella cells revealed that most of the [3H]IBA is converted into the [3H]IAA by the time it reaches the region 2.4–4 mm above the root apex (Fig. 5C). We conclude that ABCG37 regulates auxin distribution and homeostasis in roots by excluding IBA from the root apex, but does not act directly in basipetal transport. Given the rapid conversion of IBA to IAA in the root tip, we hypothesize that ABCG37 might be an additional regulator of auxin homeostasis there.
Discussion
Differential distribution of the plant hormone auxin within tissues mediates a large variety of developmental processes in plants (1, 4). Here, we show that in addition to local biosynthesis (31–33), subcellular compartmentalization (34), and cell-to-cell transport (4) of active IAA, auxin distribution can also be influenced by directional transport of the IAA precursor IBA across the plasma membrane. The established auxin exporters (13, 16) do not seem to use IBA as a substrate. Physiological data and transport assays from the heterologous systems establish the G-class ATP-binding cassette protein ABCG37 as exporter for IBA. ABCG37 shows broad substrate specificity for various auxinic compounds, including synthetic auxins and auxin transport inhibitors, but not the endogenous auxin IAA. It is possible that ABCG37 also transports other auxin metabolites, but this remains to be determined. Given the typical lower specificities of ABCG transporters (ref. 24 and references therein), it is also possible that ABCG37 plays a role in the transport of other, auxin-unrelated molecules. ABCG37 acts redundantly with ABCG36 in mediating root auxin homeostasis and development. Both proteins show a remarkable polar localization at the outermost side of root cells (25, 28) that implies IBA transport from the root into the surrounding environment. Notably, some microorganisms, including plant symbionts, produce IBA (35, 36), raising the intriguing possibility that the ABCG37-dependent transport of IBA, and/or structurally similar compounds, mediates interactions between the root and complex soil microflora.
Materials and Methods
Material and Growth Conditions.
Arabidopsis seedlings were grown under a 16-h photoperiod, 22/18 °C, on 0.5× MS medium with sucrose as described in ref. 21, unless indicated otherwise. The following mutants, transgenic plants, and constructs were described previously: pis1-1 (19), pdr9-2 (22), pen3-4 (26), and DR5rev::GFP (21). For 35S::GFP-ABCG37, the ABCG37 genomic fragment was cloned into a pBluescript-derived pEPA vector (37). The fusion construct was subcloned into binary pML-BART (38) and transformed into pis1-1 mutants.
The following chemicals were used: 2-(1-pyrenoyl)benzoic acid (PBA), 1- and 2-naphtoxyacetic acid (1- and 2-NOA), indole-3-acetic acid (IAA), 2,4-dichlorophenoxyacetic acid (2,4-D), 2,4-dichlorophenoxybutyric acid (2,4-DB), indole-3-butyric acid (IBA), benzoic acid, quercetin, p-aminobenzoic acid (PABA), phenylacetic acid (PAA), salicylic acid (all from Sigma), and brefeldin A (Molecular Probes).
Localization Analysis and Confocal Microscopy.
DR5rev::GFP signal in 5-d-old seedlings in Arabidopsis was observed as described in ref. 21. For 2,4-D and NPA experiments, plants were germinated on selected compound. For IBA and IAA DR5rev::GFP observations, to minimize potential metabolic conversions, seedlings were incubated in the auxin-supplemented liquid medium for 4 h.
Immunolocalizations in 5-d-old seedlings were done as described in ref. 39 with anti-ABCG37 (22) (1:500) and CY3-conjugated anti-rabbit (1:600; Dianova) antibodies. For confocal laser scanning microscopy, Leica TCS SP2 equipment was used. Images were processed in Adobe Photoshop.
Mutation Characterization and Double Mutant Isolation.
The predicted pis1-1 mutation in the donor splicing site was confirmed by RT-PCR and sequencing of the prevailing misspliced product.
All mutants were in the Col-0 accession. The pdr9-2 (SALK_050885) mutant (22) was crossed to the pen3-4 (SALK_000578) mutant (26). PCR analysis of F2 plants was used to identify double mutants. pdr9-2 was identified as described in ref. 23. Amplification of ABCG36/PDR8 with PDR8-1 (GTATCACCCAACTAAATCCTCACG) and PDR8-2 (ATCTGTTACACGGCCAAAGTTAG) yields a 1,450-bp product in wild type and no product in pen3-4. Amplification with PDR8-1 and LB1-SALK yields an ∼450-bp product in pen3-4 and no product in wild type.
Phenotype Analysis.
The root growth compound sensitivity of pis1-1 was tested at 3–8 concentrations on 6-d-old seedlings, and root length on control media reached ≈20 mm [corresponds to 100%, pis1-1 root length was not significantly different from wild type (19)]. At least 15 seedlings were processed for each concentration, and at least three independent experiments were done, giving the same statistically significant results; representative experiments are presented. Equal variances of values were verified by the Levene test, and the Kruskal–Wallis nonparametric test was performed simultaneously with ANOVA. Data were statistically evaluated with NCSS 2007.
For IAA-, IBA-, and 2,4-DB-responsive root elongation assays of double mutants, primary root lengths of seedlings grown for 8 d with the indicated auxin concentration were measured. Seedlings were grown at 22 °C under continuous illumination through yellow long-pass filters to slow indolic compound breakdown (40).
For cotyledon expansion assays, cotyledons of 7-d-old seedlings grown under continuous white light at 22 °C were removed and mounted. Cotyledons were imaged using a dissecting microscope, and cotyledon area was measured using NIH Image software (http://rsb.info.nih.gov/ij).
For root hair assays, roots of 5-d-old vertically grown seedlings grown under continuous white light at 22 °C were imaged using a dissecting microscope and root hair lengths from 4 mm of root adjacent to the root-shoot junction and measured using NIH Image software.
Transport Measurements.
Auxin accumulation in excised root tips was measured as described in ref. 28. Leaf protoplast transport assays were performed as described in ref. 16. Available microarray databases predict a moderate leaf ABCG37 expression (41), at similar levels as examined in PIN proteins (16).
S. cerevisiae assays were performed as described in ref. 16 with ABCG37 cDNA cloned as the HA-tagged version into the NotI site of the yeast shuttle vector, pNEV (42). Relative IAA export was calculated from retained radioactivity as follows: (radioactivity in the yeast at t = 10 min) − (radioactivity in the yeast at t = 0) · (100%)/(radioactivity in the yeast at t = 0 min).
S. pombe assays were performed as described in ref. 29, where a pTM isolated cDNA fragment of ABCG37 was subcloned into the pREP41 vector. The results show the accumulation of the radioactivity in cells. Determination of ABCG37 plasma membrane localization was done by two-phase partitioning followed by Western blot analysis as described in ref. 43.
Transport activities in HeLa cells were determined as described in refs. 13 and 16. Net efflux is expressed as DPM/10,000 cells divided by the amount of auxin retained by cells transformed with empty pTM1 vector minus the amount of auxin retained by cells transformed with ABCG37. The data presented are averaged datasets from three independent experiments. Student's t tests were run for individual pairwise comparisons and then compared by ANOVA using the Newman–Keuls post hoc test, followed for P values close to 0.05 by Dunnett's and Tukey's tests.
The IBA transport measurements of auxin exporters were conducted as described in ref. 44, using 60 nM [3H]IAA (21 Ci·mmol−1; American Radiolabeled Chemicals), 60 nM [3H]IBA (18.9 Ci·mmol−1, HPLC-purified; American Radiolabeled Chemicals), and 180 nM unlabeled IBA (Sigma).
Root tip-applied auxin transport was measured as described in refs. 16 and 30, with the following modifications: a 10-nL (root tip uptake) or 6-nL (basipetal root transport) droplet with 1 μM radioactively labeled auxin ([3H]IAA, 18 Ci·mmol−1; [3H]2,4-D, 21 Ci·mmol−1; [3H]IBA, 21 Ci·mmol−1; American Radiolabeled Chemicals) was applied on the third tier of columella cells. After 2 h, the root cap was removed and radioactivities of excised 2-mm root tip segments were measured. [3H]IBA was repurified by HPLC to remove contaminants before use.
The HPLC analysis of transported [3H]IBA signal was accomplished by extraction in methanol and separation in a 10–100% methanol/2% formic acid gradient with radiodetection compared with IAA and IBA standards. Significance was tested by ANOVA followed by the Newman–Keuls test.
Supplementary Material
Acknowledgments
We thank J. Mravec for discussions; S. Somerville (Department of Plant and Microbial Biology, UC-Berkeley, Berkeley, CA) for PEN3/ABCG36-GFP; V. Gaykova, Y. Cheng, A. Ahmed, and B. Titapiwatanakun for technical assistance; and Y. Helariutta for support during preparing the manuscript. This work was supported by the Odysseus Programme of the Research Foundation–Flanders (K.R., J.F., and Ł.Ł.), Grant Agency of the Academy of Sciences of the Czech Republic Grant IAA601630703 (to J.F.), Ministry of Education of the Czech Republic Grants LC06034 and MSM0021622415 (to E.N. and J.H.), the Forschungskredit of the University of Zurich (A.B.), the Novartis Foundation (M.G.), the Swiss National Fonds (M.G. and E.M.), the Japanese Society for the Promotion of Science (H.I.), Robert A. Welch Foundation Grant C-1309 (B.B.), U.S. Department of Energy Grant DE-FG02-06ER15804 (A.S.M.), and National Institutes of Health Grants 1K99GM089987 (L.C.S.) and GM067203 (W.M.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005878107/-/DCSupplemental.
References
- 1.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 2.Parry G, Estelle M. Auxin receptors: A new role for F-box proteins. Curr Opin Cell Biol. 2006;18:152–156. doi: 10.1016/j.ceb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 4.Vanneste S, Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig-Müller J. Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 2000;32:219–230. [Google Scholar]
- 7.Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics. 2008;180:237–251. doi: 10.1534/genetics.108.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Bennett MJ, et al. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Swarup K, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- 12.Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrásek J, et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- 14.Wiśniewska J, et al. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- 15.Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13:2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisler M, et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44:179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 17.Bandyopadhyay A, et al. Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans. 2007;35:137–141. doi: 10.1042/BST0350137. [DOI] [PubMed] [Google Scholar]
- 18.Mravec J, et al. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development. 2008;135:3345–3354. doi: 10.1242/dev.021071. [DOI] [PubMed] [Google Scholar]
- 19.Fujita H, Syōno K. PIS1, a negative regulator of the action of auxin transport inhibitors in Arabidopsis thaliana. Plant J. 1997;12:583–595. doi: 10.1046/j.1365-313x.1997.00583.x. [DOI] [PubMed] [Google Scholar]
- 20.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 22.Ito H, Gray WM. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 2006;142:63–74. doi: 10.1104/pp.106.084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strader LC, Monroe-Augustus M, Rogers KC, Lin GL, Bartel B. Arabidopsis iba response5 suppressors separate responses to various hormones. Genetics. 2008;180:2019–2031. doi: 10.1534/genetics.108.091512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verrier PJ, et al. Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Łangowski Ł, Růžička K, Naramoto S, Kleine-Vehn J, Friml J. Polar trafficking to the outer domain defines the root-soil interface. Curr Biol. 2010 doi: 10.1016/j.cub.2010.03.059. in press. [DOI] [PubMed] [Google Scholar]
- 26.Stein M, et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D-Y, Bovet L, Maeshima M, Martinoia E, Lee Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007;50:207–218. doi: 10.1111/j.1365-313X.2007.03044.x. [DOI] [PubMed] [Google Scholar]
- 28.Strader LC, Bartel B. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell. 2009;21:1992–2007. doi: 10.1105/tpc.109.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang H, Murphy AS. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 2009;59:179–191. doi: 10.1111/j.1365-313X.2009.03856.x. [DOI] [PubMed] [Google Scholar]
- 30.Peer WA, Murphy AS. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007;12:556–563. doi: 10.1016/j.tplants.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 32.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mravec J, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Morales LJ, Soto-Urzúa L, Baca BE, Sánchez-Ahédo JA. Indole-3-butyric acid (IBA) production in culture medium by wild strain Azospirillum brasilense. FEMS Microbiol Lett. 2003;228:167–173. doi: 10.1016/S0378-1097(03)00694-3. [DOI] [PubMed] [Google Scholar]
- 36.Badenoch-Jones J, et al. Mass-spectrometric identification of indole compounds produced by Rhizobium strains. Biomed Mass Spectrom. 1982;9:429–437. [Google Scholar]
- 37.Dhonukshe P, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17:520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 38.Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 39.Sauer M, Paciorek T, Benková E, Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc. 2006;1:98–103. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]
- 40.Stasinopoulos TC, Hangarter RP. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990;93:1365–1369. doi: 10.1104/pp.93.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauer N, Stolz J. SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- 43.Premsler T, Zahedi RP, Lewandrowski U, Sickmann A. Recent advances in yeast organelle and membrane proteomics. Proteomics. 2009;9:4731–4743. doi: 10.1002/pmic.200900201. [DOI] [PubMed] [Google Scholar]
- 44.Blakeslee JJ, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19:131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.