Abstract
We recently showed that the Wiskott–Aldrich syndrome protein (WASP) family member, WASH, localizes to endosomal subdomains and regulates endocytic vesicle scission in an Arp2/3-dependent manner. Mechanisms regulating WASH activity are unknown. Here we show that WASH functions in cells within a 500 kDa core complex containing Strumpellin, FAM21, KIAA1033 (SWIP), and CCDC53. Although recombinant WASH is constitutively active toward the Arp2/3 complex, the reconstituted core assembly is inhibited, suggesting that it functions in cells to regulate actin dynamics through WASH. FAM21 interacts directly with CAPZ and inhibits its actin-capping activity. Four of the five core components show distant (approximately 15% amino acid sequence identify) but significant structural homology to components of a complex that negatively regulates the WASP family member, WAVE. Moreover, biochemical and electron microscopic analyses show that the WASH and WAVE complexes are structurally similar. Thus, these two distantly related WASP family members are controlled by analogous structurally related mechanisms. Strumpellin is mutated in the human disease hereditary spastic paraplegia, and its link to WASH suggests that misregulation of actin dynamics on endosomes may play a role in this disorder.
Keywords: actin dynamics and capping, endosome, WAVE regulatory complex, WASH regulatory complex, hereditary spastic paraplegia
Members of the Wiskott-Aldrich syndrome protein (WASP) family, which includes WASP, N-WASP, WAVE (1–3), WHAMM, JMY, and WASH, control actin cytoskeletal dynamics throughout biology. They act in large part by regulating the actin nucleating activity of the ubiquitous Arp2/3 complex. WASP proteins stimulate Arp2/3 complex using a conserved C-terminal VCA (Verprolin homologous, central hydrophobic, and acidic) region. They contain distinct N-terminal elements, which facilitate integration into unique macromolecular complexes. These complexes stabilize WASP proteins in cells, control their subcellular localization, and regulate the biochemical activity of the VCA.
In WASP and N-WASP, an N-terminal GTPase-binding domain (GBD) inhibits the VCA by binding to it intramolecularly. The Rho family GTPase, Cdc42, relieves this autoinhibition by binding the GBD and competitively displacing the VCA, enabling it to bind and activate Arp2/3 complex (1, 2). In contrast, WAVE proteins are intrinsically active, but are inhibited in cells by constitutive incorporation into a heteropentameric assembly called the WAVE regulatory complex (WRC, containing WAVE, SRA1, NAP1, ABI, and HSPC300). The WRC can be activated by the GTPase Rac1, which acts with low affinity in vitro and likely functions with other inputs to stimulate WAVE activity strongly in cells (3–6). It is not known whether the other WASP family members exist in regulatory complexes in cells, or how their activity toward Arp2/3 complex is controlled.
The recently discovered WASH protein (Wiskott–Aldrich syndrome protein and SCAR homologue) is conserved from simple eukaryotes such as Entamoeba and Dictyostelium to humans (7). We recently showed that WASH localizes to early endosomal subdomains, where it is associated with vesicular F-actin, and regulates retromer-mediated vesicle trafficking through an interaction with the protein FAM21 (8). Preliminary biochemical analyses showed that endogenous cytoplasmic WASH runs on a size exclusion chromatography column at an estimated mass of approximately 700 kDa (versus its actual mass, 55 kDa), suggesting that WASH exists in a macromolecular complex (8). Sorting of cargo from endosomes is a tightly regulated process, which plays a key role in receptor recycling, degradation, signaling and deregulation of this process contributes to human disease, including cancer and neurodegeneration. Thus, a further understanding of the mechanisms regulating WASH function is paramount to discerning its role in endosomal sorting/trafficking and its potential role in disease.
Results and Discussion
WASH Exists in a Large Macromolecular Complex.
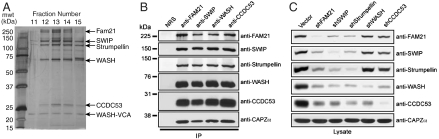
We used both conventional- and antibody-affinity chromatography to purify the WASH complex and identify its components from three different sources, bovine brain, HeLa cells, and Drosophila S2 cells. Bovine WASH copurified over five steps, including affinity chromatography on an anti-VCA antibody matrix, with four additional proteins (Fig. 1A). These were identified by mass spectrometry as bovine homologues of the human proteins FAM21, KIAA1033, Strumpellin, and coiled-coil domain containing protein 53 (CCDC53). Using a six-step conventional chromatography protocol, we also copurified these same four components with WASH from HeLa cell extracts (Fig. S1A). In some trials the CAPZ heterodimer (CAPZα/CAPZβ) also copurified with WASH, suggesting that CAPZ is also associated, but perhaps more weakly (Fig. S1A). Immunoprecipitation experiments confirmed the elements of the complex, with antibodies to FAM21, KIAA1033, WASH, and CCDC53 each precipitating all other members including CAPZα (Fig. 1B). Because of its association with Strumpellin (see below) and WASH, we designate the previously unnamed KIAA1033 as SWIP (Strumpellin- and WASH-interacting protein). Finally, we generated a Drosophila S2 cell line stably expressing Drosophila WASH with a C-terminal tandem affinity purification tag (9). We found that the fly homologues of FAM21, SWIP, Strumpellin, and CCDC53 all copurified with dWASH, indicating that the complex exists in organisms other than mammals (Fig. S1B). Therefore, using several approaches we have identified the same WASH-containing complex from three different sources, suggesting that this assembly is generally used in eukaryotic systems to control actin dynamics.
Fig. 1.
WASH functions within a multiprotein complex. (A) Silver stained 10% SDS-PAGE of the bovine WASH complex eluting from the final gel filtration column during its purification. (B) Jurkat T cell lysates were immunoprecipitated with control rabbit IgG (NRS) or the indicated WASH complex antibodies and immunoblotted as indicated. (C) HeLa cells were suppressed, lysed and immunoblotted as indicated.
Except for the CAPZ heterodimer, which is a well-understood complex that caps the fast-growing ends of actin filaments (10), little is known about the components of the WASH complex. The N-terminus of FAM21 contains a predicted coiled-coil (residues 50–150). The C-terminus of the protein is predicted to be largely disordered except for a region homologous to CAPZIP (residues 935–1047), a CAPZ binding protein (11) (see below). CCDC53 was previously shown to interact with WASH in Caenorhabditis elegans (where the proteins are named DDL-1 and DDL-2, respectively) in a large-scale yeast-two-hybrid screen (12). Suppression of both proteins increases the lifespan of worms in a manner dependent on the daf-16/FOXO transcription factor, although the mechanisms responsible for this effect are unknown (13). Strumpellin has no identifiable subdomains, but the protein is predicted to be highly helical. The gene encoding Strumpellin was recently found mutated in the neurodegenerative disease hereditary spastic paraplegia (HSP), but there is no understanding of Strumpellin function or how its mutations give rise to disease (14, 15). Finally, no functions have been reported for SWIP, which like Strumpellin has no recognizable subdomains and is predicted to be highly helical. The different components of the WASH complex have likely coevolved, as all WASH-containing genomes that we have surveyed also contain all other complex members, whereas genomes lacking WASH (e.g., yeast and Arabidopsis thaliana) do not.
We next examined pairwise colocalization of the elements of the WASH complex by using a suppression/reexpression vector system (8). We previously showed that endogenous WASH localizes to subdomains of early endosomes and can be observed as discrete WASH spots juxtaposed to EEA1-positive staining early endosomes. As shown in Fig. S2, every pair of the five core subunits that we examined showed strong colocalization with each other and with a subset of EEA1-positive endosomes. Because both WASP and WAVE family proteins are stabilized through their interactions with other binding partners, we examined the integrity of the WASH complex using shRNA. The expression of WASH and CCDC53 were decreased by knock down of all partners in the complex (Fig. 1C). In contrast, whereas the levels of FAM21, Strumpellin, and SWIP were all reduced by each other’s knock down, they were resistant to shRNAs against WASH and CCDC53 (Fig. 1C). In contrast, CAPZα levels did not change upon suppression of any WASH core complex members, suggesting that CAPZ is likely a peripheral component. Taken together, these data support the notion that CCDC53, FAM21, Strumpellin, and SWIP form a high-affinity WASH core complex. We propose to call this assembly the WASH regulatory complex (SHRC).
WASH and WAVE Complexes Show Structural Similarity.
Upon considering the SHRC components, we were struck by features that were grossly similar to the components of the WAVE complex. For example, both assemblies contain five components, and in each case two of the large components (SRA1/NAP1 and SWIP/Strumpellin) are predicted to be entirely helical without subdomains. This led us to examine the sequences of the various proteins in more detail. Standard sequence-against-sequence [e.g., BLAST (16)] and sequence-against-profile [psi-BLAST (17)] analyses yielded no obvious relatives for any of the components (except for the region of FAM21 homologous to CAPZIP). However, the profile-against-profile search in the HHPred package (18), which is designed to identify distant homology between sequences, revealed remarkable similarity between the WASH and WAVE complexes (Fig. S3). SWIP shows highly significant similarity throughout its entire length to SRA1 (identities = 14%, probability = 99.5, E-value = 8.3e-08). No other proteins in the human genome are similar to SWIP (with a cutoff E-value of 1). Further, Strumpellin and NAP1 also show significant similarity across much of their lengths (identities = 13%, probability = 97.8, E-value = 0.19). Thus, the SRA1/NAP1 dimer of the WAVE complex is related to the SWIP/Strumpellin dimer of the WASH complex, and is likely to adopt similar structure (and perhaps function) in the two systems. Interestingly, SRA1 and NAP1 are similar to each other as well (identities = 15%, probability = 98.3%, E-value = 0.019), suggesting that the individual dimers could have formed through a process of gene duplication and divergence. The N-termini of WASH, WAVE, CCDC53, and HSPC300 are all predicted to form coiled-coils (19). HHPred found similarity between the helical segments of WASH (aa 29–88) and WAVE (aa 33–92), and between those of CCDC53 (aa 26–78) and HSPC300 (aa 27–79) (Fig. S3 and Fig. S4 A and B). No similarity was detected between the N-terminus of FAM21 and any portion of ABI, so the analogy between the WASH and WAVE complexes may not be exact. Nevertheless, the sequence similarity of four out of five components suggests that the two assemblies have similar structural, and perhaps functional, organization.
WASH Has Intrinsic Activity that Is Inhibited Within the SHRC.
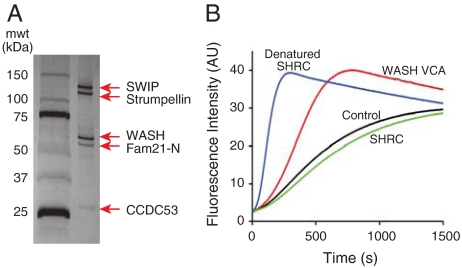
Assembly of WAVE into the WRC inhibits its intrinsic activity toward Arp2/3 complex. Because the WRC and SHRC share structurally conserved elements, we wondered whether the intrinsic activity of WASH might be similarly inhibited by complex assembly. To test this hypothesis, we reconstituted the human assembly in Sf9 insect cells using baculovirus-mediated expression. Initial attempts to produce the intact complex gave very low yields due to poor expression of full-length FAM21. However, an N-terminal fragment of FAM21 (residues 1–356) containing the WASH-interacting region (8) expressed much better, enabling generation of the five protein core complex that could be purified to homogeneity in high yield over four chromatographic steps (Fig. 2A). The production of a stable, high-affinity, biochemically well-behaved assembly strongly suggests that the five proteins are necessary and sufficient to form the SHRC [as they are for the WRC (6)].
Fig. 2.
Recombinant WASH complex is inhibited. (A) Silver stained 4–20% SDS-PAGE gel of the purified recombinant human WASH complex. All components are full length except FAM21 (residues 1–356 only). (B) Pyrene-actin assembly assays containing 10 nM Arp2/3 complex and 4 μM actin (5% pyrene labeled) in KMEI-15G buffer (control, black) plus 100 nM WASH-VCA (red), recombinant SHRC (green), or denatured recombinant SHRC (by prior heating at 60° for 10 min, blue).
We next tested if the reconstituted complex could activate Arp2/3 in pyrene-actin assembly assays. As reported previously (7), the WASH VCA strongly activated Arp2/3-mediated actin assembly, as indicated by a decrease in the lag time to polymerization and an increase in maximum polymerization rate (Fig. 2B). Full-length WASH was highly active, suggesting that WASH is not autoinhibited like WASP and N-WASP (20). In contrast to the WASH VCA, highly purified recombinant SHRC had no measurable activity toward Arp2/3 complex. Lack of activity was not the consequence of VCA degradation, as the complexed WASH was intact as determined by mass spectrometry. Furthermore, as previously observed for the WAVE complex (5, 6), the SHRC could be activated by heating to 60 °C, suggesting that the temperature-sensitive organization of the assembly functions to suppress activity of the VCA (Fig. 2B). While this manuscript was under review, Derivery et al. (21) reported that a WASH complex isolated from 3T3 cells is constitutively active toward Arp2/3 complex in pyrene-actin assembly assays. However, the SHRC used in these assays was obtained by overexpression of Protein C-tagged WASH followed by a single-step anti-Protein C affinity purification. This procedure would not separate SHRC from free WASH or SHRC subcomplexes. These impurities lack the inhibitory components of the intact assembly, explaining the high activity of this preparation. We note that high level purification and careful handling of the SHRC reconstituted here is critical to observing its true activity, as various subcomplexes that are present in partially purified samples are highly active. Thus, its inhibited activity is a more faithful representation of SHRC function and, like the WRC (6) the biochemical function of the SHRC is to inhibit WASH activity toward the Arp2/3 complex.
WASH, CCDC53, and SWIP Assemble into the SHRC Through Structurally Conserved Regions.
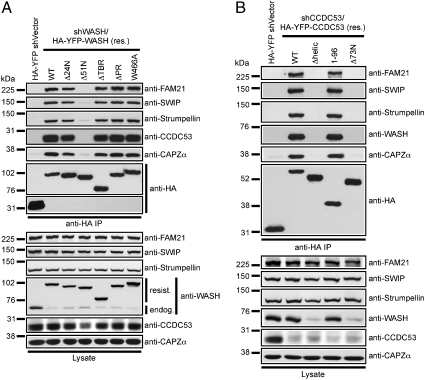
To determine if the SHRC is assembled in a similar manner to the WRC, we mapped associations in the SHRC using suppression/reexpression to replace endogenous proteins with various truncated fragments, and then examined pairwise immunoprecipitation of other components (Fig. 3). Like WAVE (22, 23), we found that the evolutionarily conserved N-terminal coiled-coil region of WASH contained within the WASH homology domain 1 (WAHD1) was necessary for association with all components of the complex (Fig. 3A and Fig. S4 A and C). In addition, the structurally conserved helical element identified in CCDC53, was necessary and sufficient for association with all other components (Fig. 3B and Fig. S4 B and D). Moreover, expression of the helical domain was sufficient for stabilizing WASH in cell lysates (Fig. 3B). Additionally, whereas full-length SWIP is required for complete SHRC assembly, we found that the C-terminus of SWIP mediated interaction with Strumpellin (Fig. S5 A and B). This interaction parallels the observation that SRA1 directly interacts with NAP1 through its C-terminal KETTE-binding domain (24). Taken together these data suggest that conserved structural elements in WASH, CCDC53, and SWIP are involved in SHRC complex assembly.
Fig. 3.
SHRC organization is analogous to that of the WAVE complex. HeLa cells were transfected with various shWASH/HA-YFP-WASH (A) or shCCDC53/HA-YFP-CCDC53 (B) reconstitution vectors, and mutant HA-YFP-fusion proteins were analyzed for integration into the WASH complex via immunoprecipitation. Lysates were immunoblotted to verify suppression.
HSP-Associated Strumpellin Mutants Support SHRC Assembly.
Several of the genes mutated in HSP, an autosomal dominant disease characterized by progressive motor neuron loss, are known to participate in endosomal trafficking or microtubule dynamics (14). Because WASH functions to regulate endosomal sorting, it is interesting that three distinct HSP-associated point mutations were recently described within the SHRC component, Strumpellin (15). Thus, we were interested in determining the assembly of Strumpellin into the SHRC as well as the consequence of HSP-associated Strumpellin mutations on SHRC assembly. As shown in Fig. S5 C and D, expression of WT Strumpellin rescued complex assembly whereas truncation of Strumpellin from either end disrupted complex assembly, suggesting that these deletions affected overall Stumpellin folding. The HSP-associated mutations also stabilized complex assembly (Fig. S5 C and E), indicating that the mechanism by which these described mutations manifest disease is not a result of their inability to assemble into the SHRC.
FAM21 Assembles with the SHRC Through its N-terminus and Interacts with CAPZ and Lipids Through its C-terminus.
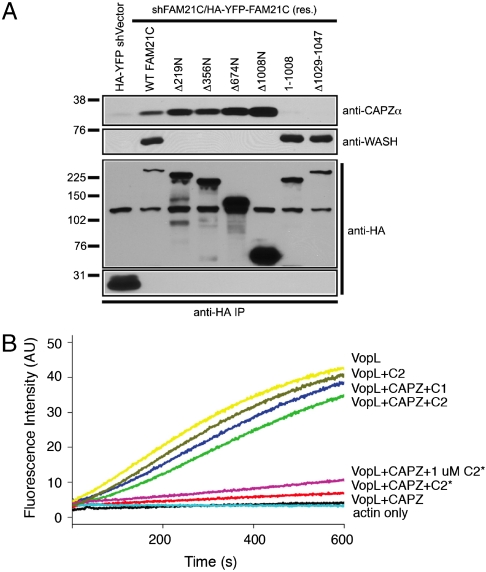
Of the WASH and WAVE complex members, FAM21 and ABI did not share any predicted structural conservation in HHpred analyses. However, similar to ABI (22, 23), which interacts with WAVE through its N-terminal coiled-coil region, our previous work demonstrated that FAM21 interacted with WASH through its N-terminus (residues 1–356). Here we show that the first 220 amino acids of FAM21 are necessary and sufficient for association with all complex components except CAPZα (Fig. S6A). In contrast, CAPZα interacted with the FAM21 mutant lacking the N-terminal region necessary for SHRC assembly suggesting that it is likely peripherally associated and not important for SHRC stability. Through mapping studies, we found that a fragment containing the C-terminal portion of FAM21 (Δ1008N) was necessary and sufficient for interacting with CAPZα (Fig. 4A and Fig. S6B). This region of FAM21 contains the region homologous to CAPZIP (residues 935–1047). Interestingly, a small deletion encompassing the CAPZ binding consensus motif (LXHX4RXKX6P) within full-length FAM21 abrogated binding to CAPZα (Fig. 4A and Fig. S6C). Furthermore, this FAM21-CAPZ interaction was direct (Fig. S6D).
Fig. 4.
FAM21 binds to CAPZ and inhibits its capping activity. (A) HeLa cells were transfected with various shFAM21/HA-YFP-FAM21 reconstitution vectors, and mutant HA-YFP-fusion proteins were analyzed for association with WASH and CAPZα via immunoprecipitation. (B) Inhibition of CAPZ capping activity by FAM21, monitored by pyrene-actin polymerization assay using VopL. FAM21 C1 fragment (residues 938–1067), C2 (residues 1010–1067), and C2* (residues 938–1067 with R1031D).
Several proteins containing the CAPZ binding element, such as Carmil and CKIP-1 inhibit capping activity of CAPZ. We tested whether FAM21 functions similarly using the pyrene-actin polymerization assay and the Vibrio type III effector VopL (25), which potently enhances actin assembly (Fig. 4B). This activity was substantially blocked by the addition of CAPZ (Fig. 4B). Addition of the FAM21 C1 or C2 fragments nearly restore the actin assembly activity of VopL presumably by preventing association of CAPZ with barbed ends. C2 does not affect the activity of VopL when CAPZ is missing (Fig. 4B). C2*, which contains an R1031D mutation in the highly conserved CAPZ binding motif, shows diminished CAPZ binding (Fig. S6D) and reduced anticapping activity even when used at a 10-fold excess (Fig. 4B). Our data demonstrate that CAPZ interacts directly with SHRC through a conserved element within the C-terminus of FAM21 and that FAM21 possesses strong anticapping activity.
It is intriguing that both WASP and WAVE family members have phospholipid-binding regions that help recruit them to areas enriched in specific phospholipids and WHAMM directly interacts with membranes through its N-terminal region. In contrast, WASH does not contain any recognizable phospholipid-binding region and we previously demonstrated that the C-terminus of FAM21 is sufficient for endosomal localization of WASH (8). Although the CAPZα/β dimer is known to bind phospholipids, we wondered whether FAM21 might interact with phospholipids directly. To test this, we generated a GST-fusion protein containing the C-terminus of FAM21 (aa 937–1341, Fig. S7A) and examined its ability to interact with phospholipids in a PIP-strip overlay assay. As shown in Fig. S7B, this region of FAM21 could directly interact with several phospholipid species as well as phosphatidylserine. PtdIns3P enrichment on early endosomes is thought to result in later accumulation of PtdIns(3,5)P2 during the early to late endosome transition (26). Significantly, we see intermediate binding to PtdIns3P and strong binding to PtdIns(3,5)P2, which is interesting in light of the fact that the WASH complex functions during recycling processes that may occur during this early to late endosome transition (e.g., retromer-mediated retrograde trafficking). We also see strong binding of PtdIns4P, which is enriched at the Golgi; and PtdIns5P, which is less well characterized. Thus, FAM21 has the capacity to directly link the WASH complex to endosomal domains enriched in these phospholipids.
The SHRC and the WRC Adopt Similar Structures.
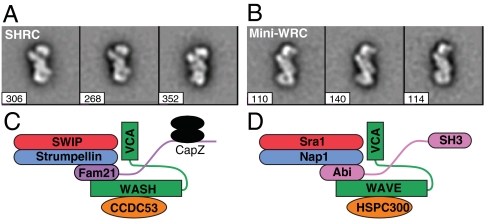
We next used electron microscopy to analyze the structures of the WASH and WAVE complexes. We previously described a reconstituted “miniWRC,” which recapitulates the biochemical properties of the complete WRC, but lacks the C-terminus of ABI and the proline-rich region of WAVE (6). We compared this material to the recombinant SHRC generated here, lacking the C-terminus of FAM21. In both cases, the two assemblies showed highly nonrandom orientation on the sample grid, making complete three-dimensional reconstruction difficult. Nevertheless, even from two-dimensional class averages, it was clear that the two assemblies were highly similar (Fig. 5 A and B and Fig. S8 A and B). Both appeared as elongated S- or Z-shapes (likely reflecting opposite faces of the complex) with dimensions of approximately 10 × 20 nm. In some orientations, additional densities could also be observed spanning diagonally across their centers. High-resolution structures of the complexes will be needed to understand how this organization leads to inhibition of VCA activity and an ability to respond to upstream signals.
Fig. 5.
The WRC and SHRC are structurally similar. Two-dimensional class averages of the recombinant SHRC (A) and miniWRC (B). The three highest ranked (most similar) classes out of 10 of each complex are shown. Number of particles in each class is listed. Cartoon models of SHRC (C) and WRC (D) assemblies.
Conclusion
A common regulatory theme of WASP family proteins is their ability to couple distinct intracellular signaling pathways to VCA activation (1). WASP and N-WASP do this in cis through an autoinhibitory interaction, and WAVE and WASH do it through an analogous mechanism in trans. Additionally, all four proteins are dependent on associated factors for stability—WASP and N-WASP on WIP (27), and WAVE and WASH on components of the WRC (4) and SHRC. Whereas numerous signaling proteins and lipids are known to control localization of WASPs and WAVEs to specific sites of action (1), the localization of WASH is not well-understood, aside from recent evidence suggesting that FAM21 may promote SHRC association with endosomes (8). Our data here suggest that FAM21 can directly interact with phospholipids through its C-terminus, while assembling into the SHRC through its N-terminus. Thus, unlike WASP, WAVE, and WHAMM, which contain intrinsic phospholipid-binding elements, WASH membrane targeting may be contained within a complex component. Lastly, we have found that FAM21 also couples the WASH complex to CAPZα/β through a conserved binding motif in its C-terminus. The CAPZ dimer binds barbed ends of actin filaments to prevent further elongation. Through this effect CAPZ controls filament length and branch density, and consequently force generation at areas of Arp2/3-mediated actin assembly (10). CAPZ was also recently shown to indirectly promote filament nucleation by Arp2/3 complex by shifting the balance of VCA activity from filament barbed end binding (28) to promotion of nucleation (29). Our data suggest that FAM21, like other CAPZ binding proteins inhibits actin-capping activity, suggesting that SHRC sequestration of CAPZ via FAM21 may promote the localized F-actin polymerization needed to promote vesicle scission at endosomes.
The most surprising discovery in our work here is that the complex controlling WASH is distantly related to that controlling WAVE. In the 7 years since the discovery of the WAVE complex (3), it had not been recognized that there were relatives in the database. In fact, the homology we have discovered here is sufficiently distant that had we not seen relationships in multiple components of the WASH complex, we might have disregarded this information. However, the facts that the SHRC proteins are united into a functional unit, and that relationships are observed for multiple components of that unit, strongly suggest that the two seemingly distinct systems are in fact related. Our observations that the conserved regions in both WASH and CCDC53 are required for complex assembly, as well as our preliminary EM analyses showing gross structural similarity between the two complexes, further support this idea. Knowledge of this relationship should help frame future studies in both systems.
Whereas the activities of WASPs and WAVEs toward Arp2/3 complex are controlled by the Rho GTPases Cdc42 and Rac, respectively (1), it is unclear whether GTPases analogously control WASH. Yet, the shared assembly and regulatory function of the WRC and SHRC suggests the possibility that WASH may be similarly activated. Interestingly, a recent report showed that WASH functions downstream of RhoA during Drosophila oocyte development (20). However, in contrast to this report, we have been unable to identify an interaction of RhoA with the assembled SHRC or activation (Fig. S9). Additionally, we did not observe an interaction with Cdc42, but did routinely see a weak interaction with Rac1 (Fig. S9). However, whereas Rac1 was able to activate the WRC, it was not capable of activating the SHRC (Fig. S9), suggesting that SHRC activation is likely regulated through other molecular interactions in vivo.
In conclusion, our data suggest that key components of the WASH and WAVE complexes likely developed through gene duplication. Whereas there has been significant divergence in the primary sequence of the complex members, they have evolutionarily maintained structural elements obligate for similar complex assembly and inhibition of activity, suggesting the possibility that they may be regulated analogously (Fig. 5 C and D and Fig. S8C). Despite the similarities, these two WASP family members display unique cellular functions (see Fig. S8C). Finally, given the association of Strumpellin with HSP (15), and the fact that the known mutations do not affect SHRC assembly, it will be important to examine the effect of these mutations on the cellular and biochemical activities of the SHRC.
Materials and Methods
Antibodies and Plasmids.
Reagents were from Sigma unless specified. Anti-HA Affinity Matrix and anti-HA-HRP were from Roche. We used monoclonal antibodies to EEA1 and CAPZα (BD Transduction Laboratories) and fluorescently conjugated secondary reagents from Molecular Probes. PIP strips were from Echelon Biosciences. Anti-FAM21 and anti-WASH were described (8). Antisera against SWIP, Strumpellin, and CCDC53 were generated by immunizing rabbits with GST-fusion proteins: SWIP (aa 1–93 and 928–1173), Strumpellin (aa 696–820), and CCDC53 (aa 1–194) (Cocalico Biologicals). Antisera against bovine WASH were raised against VCA (aa 316–471) and residues 120–133. The shRNA vectors, shWASH, and shFAM21 were described (8). We also used shSWIP (GGGCTATGTACGAATGATAA), shStrumpellin (GAGAGAGATAGTGGATAAA), and shCCDC53 (CATGCATAGGGGTACATTTA; 3′ UTR sequence). The SWIP (GGGtTAcGTgCGcATGATtA) and Strumpellin (GAGgGAaATcGTcGAcAAg) cDNAs were made shRNA-resistant by mutagenesis and used to generate suppression/reexpression vectors.
Chromatography.
All chromatography steps were performed at 4 °C. All columns and affinity resins were purchased from GE Healthcare except for hydroxyapatite (BioRad) and Calmodulin resin (Stratagene).
Purification of WASH Complex from Bovine Brain.
The bovine WASH complex was purified from calf brains after immunoactivity against the WASH-VCA (aa 1–167), or an N-terminal peptide from bovine WASH (aa 120–133). Two calf brains (purchased from Pel-freez) were homogenized in buffer (40 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 5 mM βME, protease inhibitors mix). The homogenate was centrifuged at 4,000 rpm for 30 min, followed by 1 hr ultracentrifugation (40,000 rpm). The supernatant was then subjected to an ammonium sulfate cut from 25% to 40% saturation. The resulting pellet was resuspended in Q buffer (20 mM Tris pH 8.5, 50 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 5 mM βME), dialyzed against the same buffer overnight, and then loaded onto an 8 mL Source 15 Q column. The WASH complex was eluted by a linear gradient (200–500 mM NaCl, 20 column volume) and came off around 320 mM NaCl. Affinity-purified anti-WASH-VCA antibody was crosslinked to CNBr-activated Sepharose 4B beads. The beads (approximately 1 mL) were incubated with the WASH-containing Source 15Q fractions overnight in 4 °C, and washed extensively by buffer (20 mM Tris pH 8.0, 500 mM NaCl, 0.1% (v/v) Tween-20). Complex was eluted with WASH-VCA and further purified by gel filtration chromatography.
Generation of Recombinant WASH Complex.
Untagged WASH, GST-tagged FAM21 (residues 1–356), and N-terminally His6-tagged Strumpellin, SWIP, and CCDC53 were coexpressed in Sf9 insect cells from individual baculoviruses. Cells were harvested 60 hr post infection, resuspended in lysis buffer (20 mM Tris pH 8.0, 20 mM imidazole pH 8, 200 mM NaCl, 1 mM MgCl2, 20% glycerol (w/v), 5 mM βME, protease inhibitors mix) and subjected to one freeze/thaw cycle. The complex was adsorbed from cleared lysate onto a Ni-NTA column and eluted with imidazole. Eluate was treated with tobacco etch virus protease (TEV) overnight and separated on successive Source 15Q and MonoS columns. Fractions were concentrated and separated on a Superose 6 gel filtration column in KMEI-20G buffer [10 mM imidazole pH 7, 20% glycerol (w/v), 100 mM KCl, 1 mM EGTA pH 8, 1 mM MgCl2, 5 mM βME]. All purification buffers contained 20% glycerol.
Actin Assembly Assay.
Actin was purified from rabbit muscle and the Arp2/3 complex was purified from bovine thymus. Actin assembly assays contained 4 μM actin (5% pyrene labeled), 10 nM Arp2/3 complex in KMEI-15G buffer (15% (w/v) glycerol), 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole pH 7.0), and were performed as described (6).
Capping Inhibition Assay.
The assays contained 0.5 μM actin (20% pyrene labeled), 5 nM VopL, 20 nM CAPZ (mouse α2β2) in the presence or absence of untagged FAM21 proteins, in KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole pH 7.0). VopL protein (residues 90–484) was purified as described (25).
Cell Culture, Transfection, Immunoprecipitation, and Immunoblot Analysis.
HeLa and Jurkat cells were passaged in RPMI 1640 medium with 5% fetal bovine serum, 5% fetal calf serum and 4 mM L-glutamine, and were transfected using electroporation as described (8). Immunoprecipitations (500–1000 μg of protein) and lysates (100 μg of protein) were prepared and analyzed as described (8).
Immunofluorescence.
HeLa cells were grown directly on coverslips, fixed in 4% paraformaldehyde, and prepared for immunofluorescence as described (8). Images were obtained with an LSM-710 laser scanning confocal microscope and analyzed using ZEN software package (Carl Zeiss).
Electron Microscopy.
Four μL of 10 μg/mL protein solution was applied to glow discharged carbon coated 300-mesh Cu/Rh grids (emsdiasum.com) and incubated for 30–60 sec. Excess solution was blotted off with filter paper (Whatman). The grid was washed and then stained with 2% uranyl acetate. Grids were imaged under low dose conditions (10–25 electrons/Å2) on a FEI Tecnai G2 Spirit BioTwin microscope equipped with a LaB6 filament operated at 120 kV at a nominal magnification of 30,000×. Images were recorded with a Gatan 2048x2048-pixel CCD camera (Gatan, Inc.) using 0.8–2.5 μm underfocus, with a final resolution of 3.63 Å/pixel on the object. Particles were picked using the boxer application in EMAN (30), normalized, and filtered to 22 Å. Ten class averages were generated using 9 iterations of reference-free classification (refine2d.py), using a common reference to orient the classes to show the “upright” view. Classes with fewer than 8 particles were discarded automatically after each iteration.
Supplementary Material
Acknowledgments.
We thank members of the Rosen and Billadeau laboratories for helpful discussions; in particular, Baoyu Chen, Chengzhu Chen, and Bingke Yu [University of Texas Southwestern Medical Center (UTSW)] for sharing reagents. We thank Dr. James Chen and Dr. Qinghua Liu for advice on biochemical purification, Dr. Masahide Kikkawa for advice on EM analyses and staff at the Protein Chemistry Core Research Facility (UTSW) and Protein Chemistry and Proteomics Shared Resource (Mayo) for protein identification. Work was supported by the Mayo Foundation, the Howard Hughes Medical Institute, National Institutes of Health Grants R01-AI065474 (D.D.B.) and R01-GM056322 (M.K.R.), Welch Foundation Grant I–1544 (M.K.R.), Allergic Diseases Training Grant NIH-T32-AI07047 (T.S.G.), and a Cancer Research Institute Fellowship to (D.J.). D.D.B is a Leukemia and Lymphoma Scholar.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913293107/-/DCSupplemental.
References
- 1.Takenawa T, Suetsugu S. The WASP–WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8(1):37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 2.Leung DW, Rosen MK. The nucleotide switch in Cdc42 modulates coupling between the GTPase-binding and allosteric equilibria of Wiskott–Aldrich syndrome protein. Proc Natl Acad Sci USA. 2005;102(16):5685–5690. doi: 10.1073/pnas.0406472102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418(6899):790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 4.Stradal TE, Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol. 2006;18(1):4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Derivery E, Lombard B, Loew D, Gautreau A. The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton. 2009;66(10):777–790. doi: 10.1002/cm.20342. [DOI] [PubMed] [Google Scholar]
- 6.Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16(5):561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linardopoulou EV, et al. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3(12):e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17(5):699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puig O, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24(3):218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 10.Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int Rev Cel Mol Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyers CE, et al. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem J. 2005;389(Pt 1):127–135. doi: 10.1042/BJ20050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1(1):119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7(12):1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 15.Valdmanis PN, et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am J Hum Genet. 2007;80(1):152–161. doi: 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 20.Liu R, et al. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136(16):2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17(5):712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Innocenti M, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6(4):319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 23.Echarri A, Lai MJ, Robinson MR, Pendergast AM. Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol Cell Biol. 2004;24(11):4979–4993. doi: 10.1128/MCB.24.11.4979-4993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogdan S, Grewe O, Strunk M, Mertens A, Klambt C. Sra-1 interacts with Kette and Wasp and is required for neuronal and bristle development in Drosophila. Development. 2004;131(16):3981–3989. doi: 10.1242/dev.01274. [DOI] [PubMed] [Google Scholar]
- 25.Liverman AD, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci USA. 2007;104(43):17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramesh N, Geha R. Recent advances in the biology of WASP and WIP. Immunol Res. 2009;44(1–3):99–111. doi: 10.1007/s12026-008-8086-1. [DOI] [PubMed] [Google Scholar]
- 28.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: Barbed end capture by N-WASP WH2 domains. Cell. 2007;128(5):901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133(5):841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.